Abstract
TRP channels are polymodal signal detectors that respond to a wide range of physical and chemical stimuli. Elucidating how these channels integrate and convert physiological signals into channel opening is essential to understanding how they regulate cell excitability under normal and pathophysiological conditions. Here we exploit pharmacological probes (a peptide toxin and small vanilloid agonists) to determine structures of two activated states of the capsaicin receptor, TRPV1. A domain (S1-S4) that moves during activation of voltage-gated channels remains stationary in TRPV1, highlighting differences in gating mechanisms for these structurally related channel superfamilies. TRPV1 opening is associated with major structural rearrangements in the outer pore, including the pore helix and selectivity filter, as well as pronounced dilation of a hydrophobic constriction at the lower gate, suggesting a dual gating mechanism. Allosteric coupling between upper and lower gates may account for rich physiologic modulation exhibited by TRPV1 and other TRP channels.
The capsaicin (vanilloid) receptor, TRPV1, is a heat-activated cation channel that is modulated by inflammatory agents and contributes to acute and persistent pain 1. To understand the structural basis whereby TRPV1 responds to disparate physiological stimuli, it is necessary to view the channel in distinct functional states. This is an exceedingly challenging goal for eukaryotic membrane channels, having been achieved in only a handful of instances 2–5. Such attempts are hampered by lack of pharmacological tools with which to capture channels in specific states, and difficulties in achieving conformational uniformity required for X-ray crystallography. Among TRP channels, TRPV1 enjoys the richest pharmacology, including small molecule agonists and antagonists, as well as larger peptide toxins 6,7. Moreover, as described in the accompanying study, we have used single-particle electron cryo-microscopy (cryo-EM) to ascertain the structure of TRPV1 in its unliganded (apo), closed state without crystallization 8. Together, these advantages enhance possibilities for examining this channel in multiple functional states.
TRPV1 channels are homo-tetramers whose 3D structure resembles that of voltage-gated ion channels (VGICs), wherein an ion permeation pathway is formed by transmembrane helices S5 and S6 and the intervening pore loop region 8. This central pore is surrounded by four independently folded S1-S4 domains, which in the case of VGICs contain voltage sensors and undergo substantial movement during gating 9–11. Despite these architectural similarities, it remains unknown whether TRPs and VGICs are activated through common conformational rearrangements. One line of evidence to suggest differential gating mechanisms comes from analysis of toxins that function as gating modifiers for these channels. Spiders produce a multitude of peptide toxins 12, including some that antagonize voltage-gated potassium (Kv) channels by binding to and impeding movement of the S3-S4 voltage sensor 13,14. Others activate TRPV1 to elicit pain as part of the spider’s chemical defense mechanism 15,16. One such ‘vanillotoxin’ (called double-knot toxin, DkTx) is a 75 amino acid-long peptide consisting of two independently folded toxin moieties connected by a short linker, a bivalent arrangement that enables DkTx to trap TRPV1 in its open state with near-irreversible kinetics 15. Mutational analysis suggest that DkTx binds to residues within the S5-P-S6 pore region 15, consistent with the notion that the outer pore of TRPV1 is conformationally dynamic and contributes directly to gating 15,17–19. In contrast, the analogous region of Kv channels is believed to remain relatively stationary during normal gating 10,20, providing a compelling explanation for why some gating modifier toxins evolved to target the outer pore versus voltage sensor domains of TRP versus Kv channels, respectively. To resolve questions pertaining to TRP gating mechanisms and pharmacology, we determined the structure of TRPV1 trapped in different conformational states by DkTx and/or small vanilloid ligands. These studies reveal differential gating mechanisms for TRPs and VGICs, while highlighting advantages of cryo-EM for capturing protein structures in distinct conformations.
Structures of TRPV1-ligand complexes
To analyze TRPV1 in activated state(s), we incubated purified ‘minimal’ channel protein 8 with the vanilloid agonists, resiniferatoxin (RTX) or capsaicin. For samples containing RTX, we also included DkTx to facilitate trapping of channels in their fully open state. We determined 3D reconstructions of TRPV1 with RTX/DkTx or capsaicin to resolutions of 3.8Å or 4.2Å, respectively, based on gold standard Fourier shell correlation (FSC) = 0.143 criteria 21 (Extended Data Figs. 1 – 4). Many side chain densities, particularly those along the pore, were clearly resolved, allowing for accurate model building based on the structure of apo TRPV1 (Extended Data Figs. 5 and 6) 8.
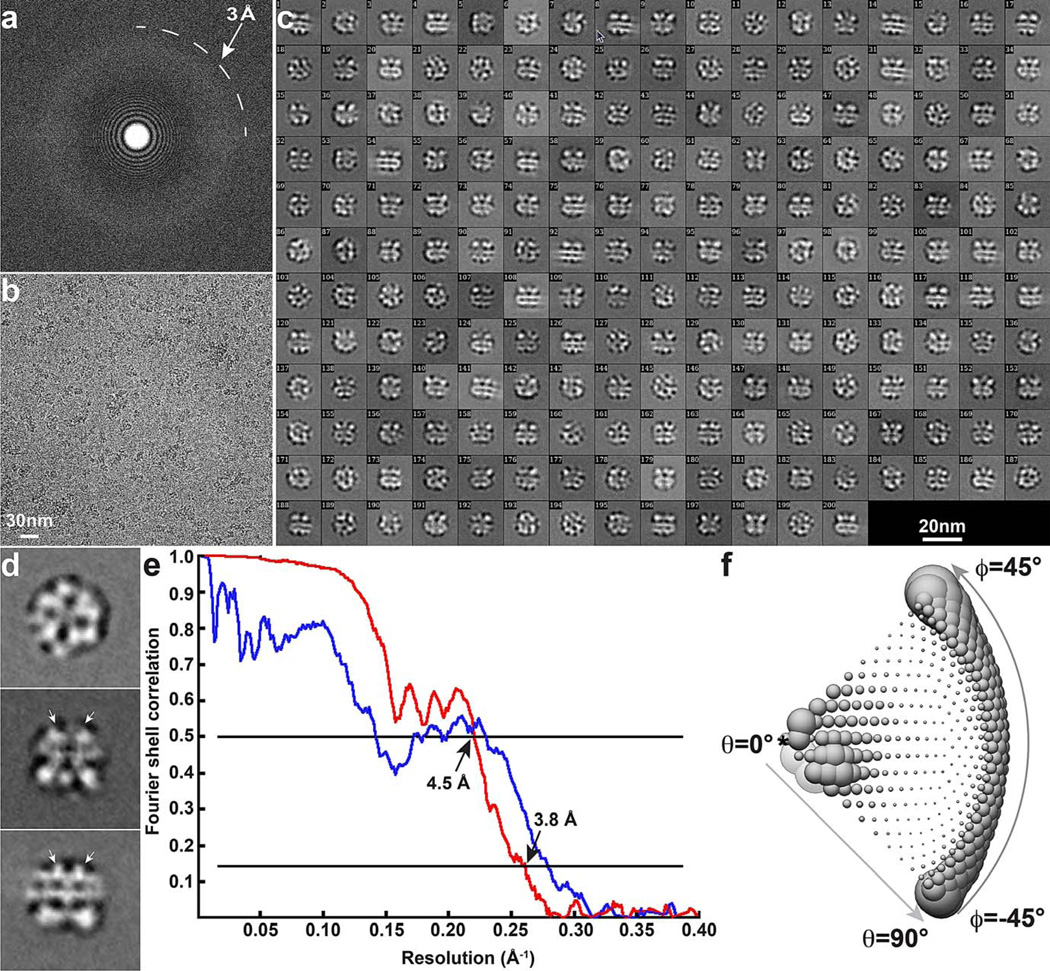
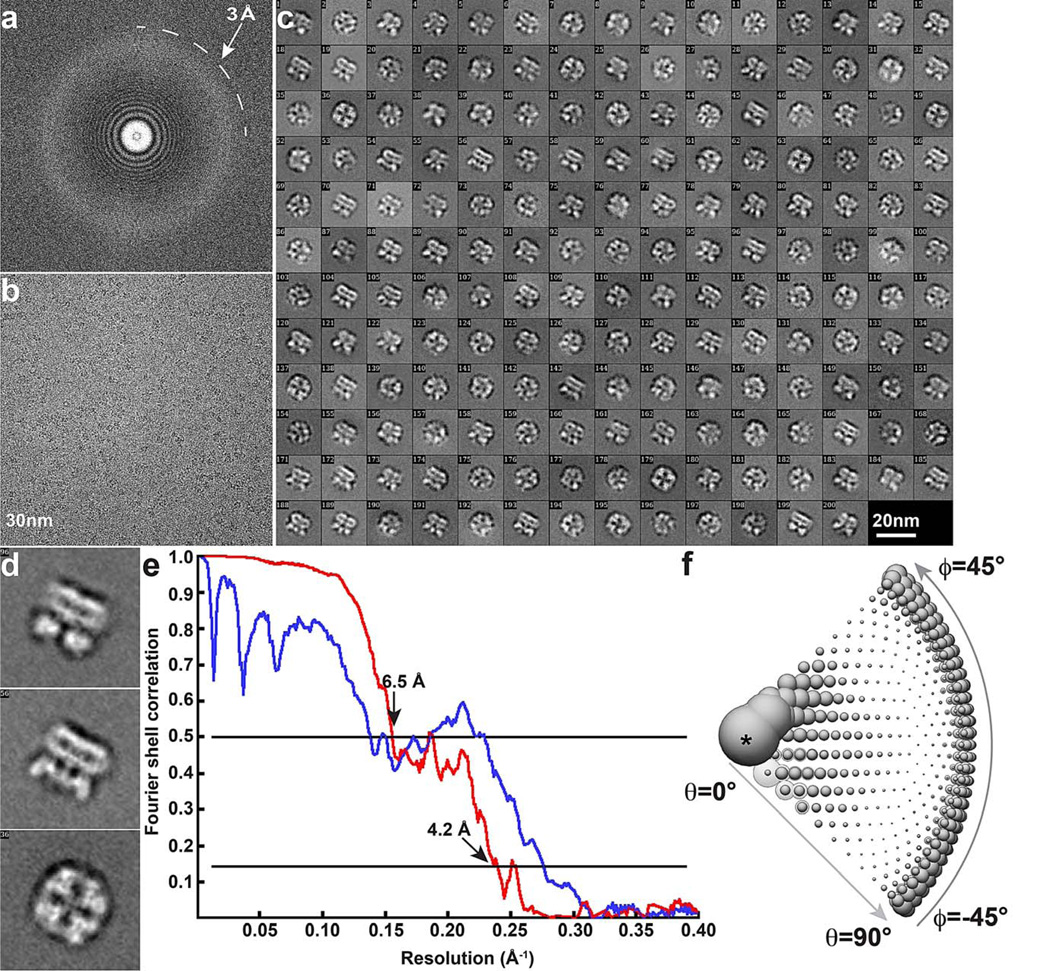
Extended Data Figure 1. Cryo-EM of TRPV1 in complex with RTX and DkTx.
a, b, Fourier transform (a) of a representative image (b). c, 2D class averages of cryo-EM particles. d, Enlarged view of three representative 2D class averages. Arrows indicate DkTx densities near the channel pore. e, Gold-standard FSC curve (red) of the final 3D reconstruction, marked with the resolutions that correspond to FSC = 0.5 and 0.143. The FSC curve between the final map and that calculated from the atomic model is shown in blue. f, Euler angle distribution of all particles used for calculating the final 3D reconstruction. The sizes of balls represent the number of particles. The accuracy of rotation is 5.213°, as reported by RELION.
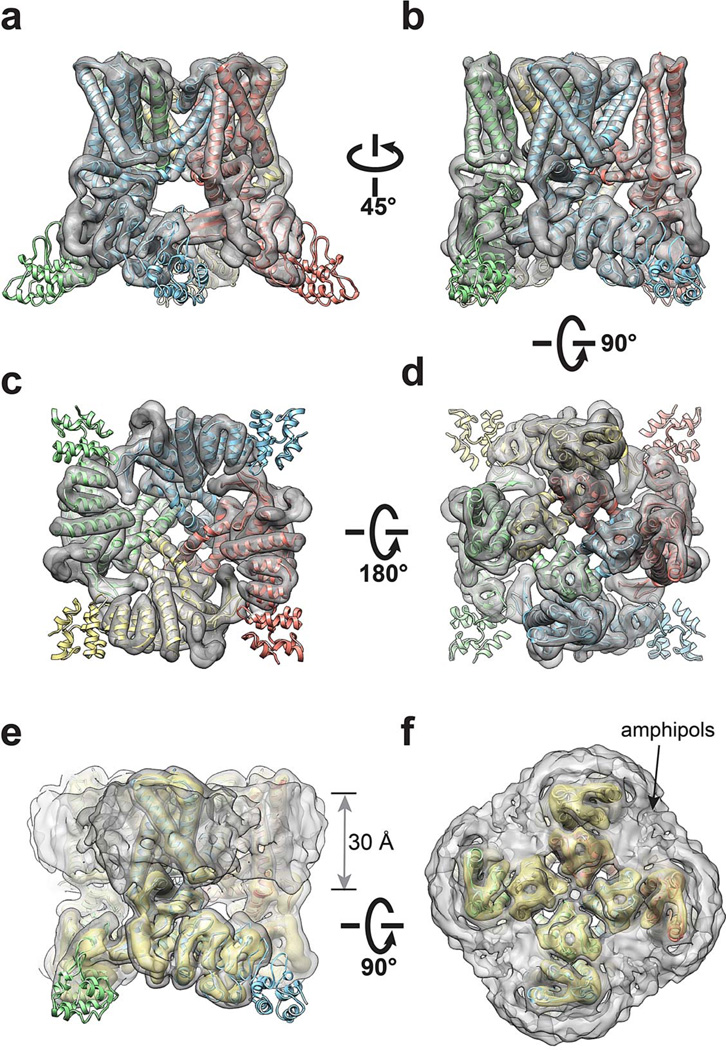
Extended Data Figure 4. 3D reconstruction of TRPV1-capsaicin complex low-pass filtered at 6Å resolution.
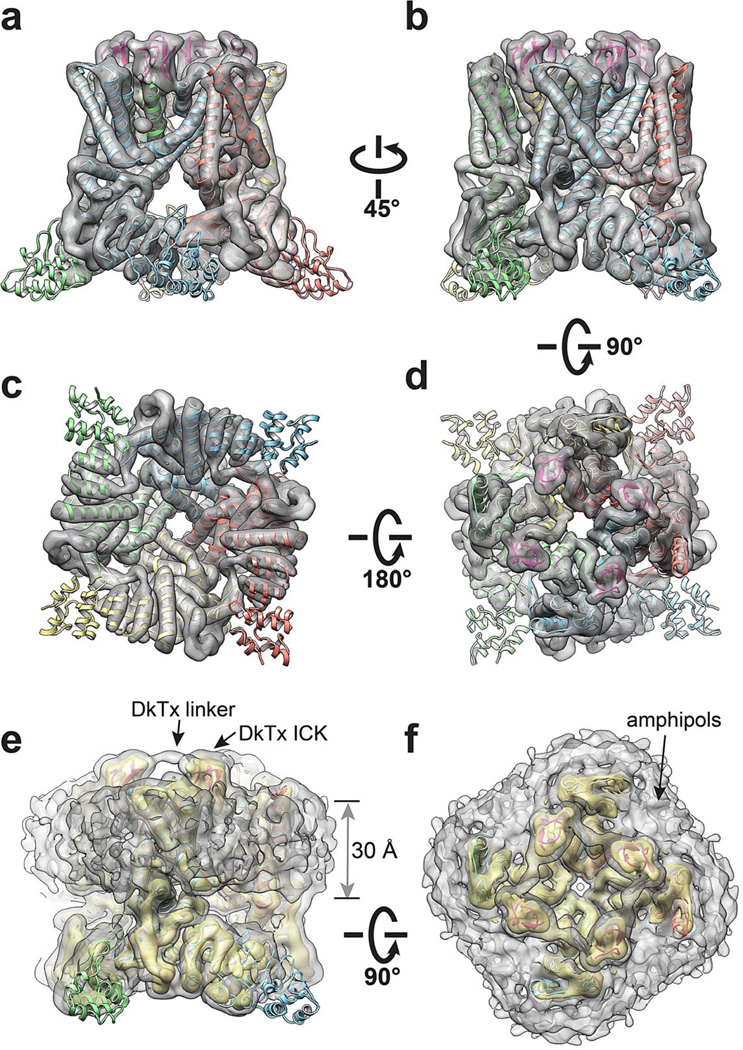
a-d, Four different views of the 3D reconstruction low-pass filtered at 6Å and amplified by a temperature factor of −100Å2, fitted with de novo atomic model of TRPV1-capsaicin complex built as described in Methods. e, f, Two views of the 3D reconstruction displayed at two different isosurface levels (high in yellow and low in gray). At the low isosurface level, the belt-shaped density of amphipols is visible with a thickness of ~30Å.
Extended Data Figure 5. 3D reconstruction of TRPV1-RTX/DkTx complex low-pass filtered at 3.8Å resolution.
a-d, Four different views of the 3D reconstruction low pass filtered at 3.8Å with a temperature factor of −100Å2, fitted with de novo atomic model of TRPV1-RTX/DkTx complex (toxin is shown in magenta and indicated by arrows) built as described in Methods.
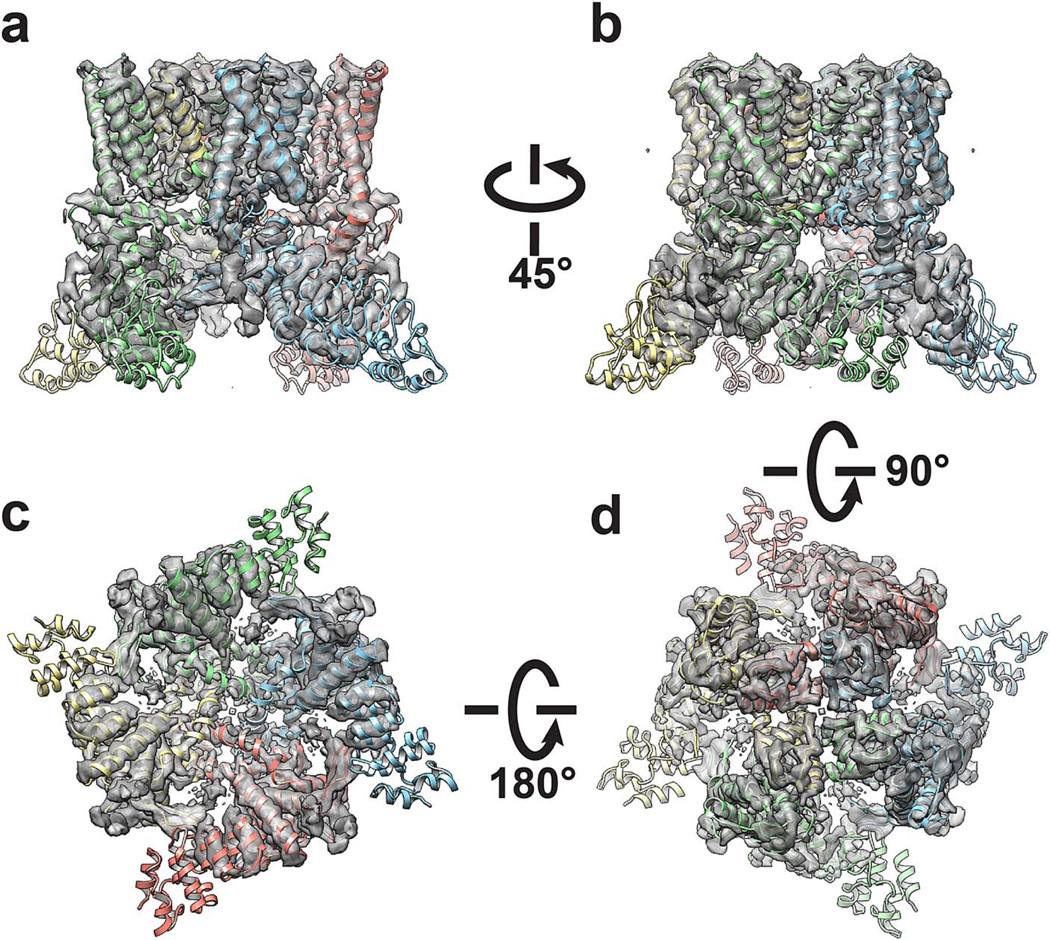
Extended Data Figure 6. 3D reconstruction of TRPV1-capsaicin complex low-pass filtered at 4.2Å resolution.
a-d, Four different views of the 3D reconstruction low pass filtered to 4.2Å with a temperature factor of −150Å2, fitted with de novo atomic model of TRPV1-capsaicin complex built as described in Methods.
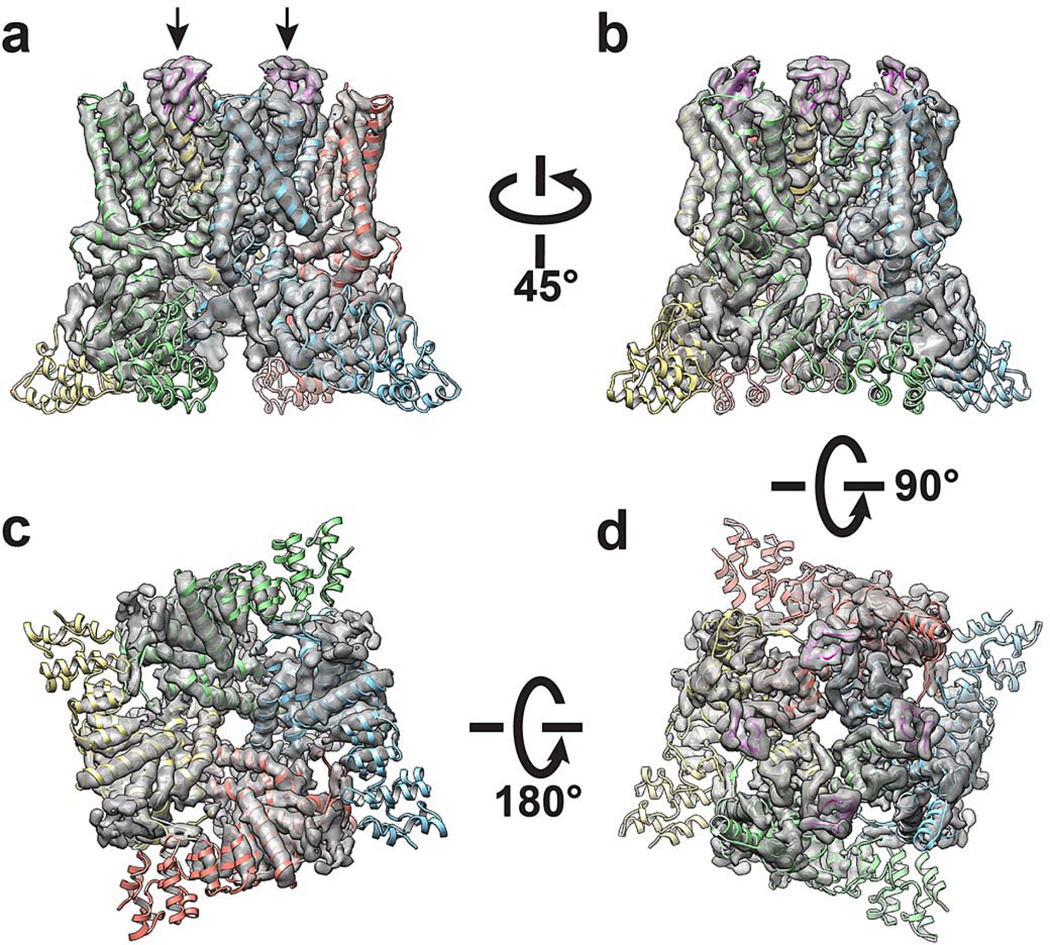
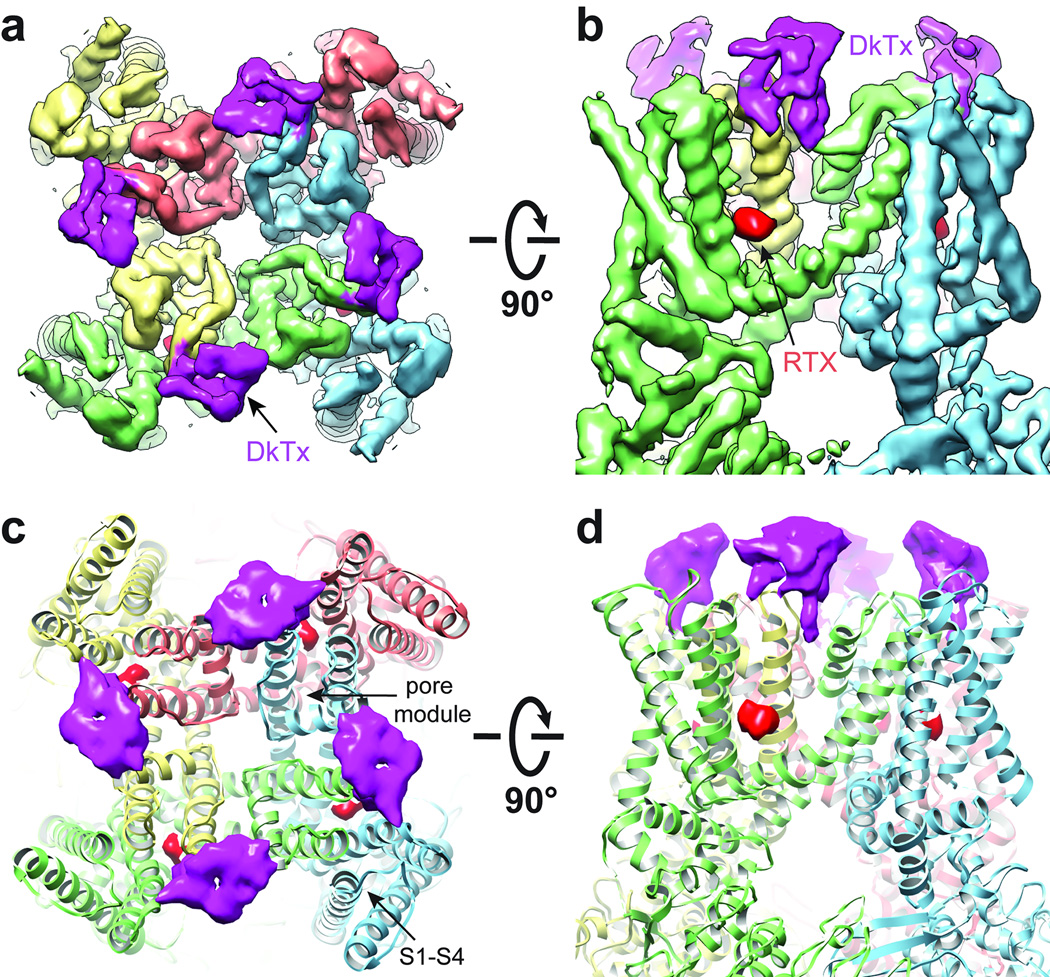
In the RTX/DkTx structure, density corresponding to two DkTx molecules (i.e. four toxin moieties) was observed atop each tetrameric channel complex (Fig. 1 and Extended Data Fig. 2). Mutations within extracellular loops of the TRPV1 outer pore region specifically abrogate DkTx-evoked responses 15, outlining a putative vanillotoxin site. Indeed, we see that DkTx is bound to the channel in precisely this region, but in an interesting and unexpected way: each toxin moiety sits at subunit interfaces by contacting residues at the top of the pore helix from one subunit and the outer pore loop proximal to S6 from the neighboring subunit (Figs. 1 and 2a). Thus, two bivalent toxin molecules ‘staple’ the channel together in its activated state, reminiscent of interactions between psalmotoxin and homo-trimeric ASIC1a channels, wherein the toxin bridges distinct regions on extracellular domains of adjacent subunits 2.
Figure 1. Structure of TRPV1 in complex with vanilloid ligand and spider toxin.
a, b, Cryo-EM density map of TRPV1 in complex with vanilloid agonist (resiniferatoxin, RTX; red sphere) and vanillotoxin (DkTx spider toxin; magenta and denoted by arrow) shown as top-down and side views. The map is filtered to 3.8Å and amplified with a temperature factor of −100Å2. c, d, Structures from a and b with TRPV1 channel rendered as ribbon diagram. Density corresponding to each ligand represents signal from difference map (4.7σ) generated by comparison with apo-TRPV1 structure.
Extended Data Figure 2. 3D reconstruction of TRPV1-ligand complexes filtered at 6Å resolution.
a-d, Four different views of the 3D reconstruction low-pass filtered at 6Å and amplified by a temperature factor of −100Å2, fitted with de novo atomic model of TRPV1-RTX/DkTx complex (toxin is shown in magenta and indicated by arrowhead) built as described in Methods. e, f, Two views of the 3D reconstruction displayed at two different isosurface levels (high in yellow and low in gray). At the low isosurface level, the belt-shaped density of amphipols is visible with a thickness of ~30Å. DkTx-related densities are also clearly visible, including the linker peptide that connects the toxin’s two inhibitor cysteine knot (ICK) moieties, as noted.
Figure 2. Binding sites for spider toxin and vanilloid agonists.
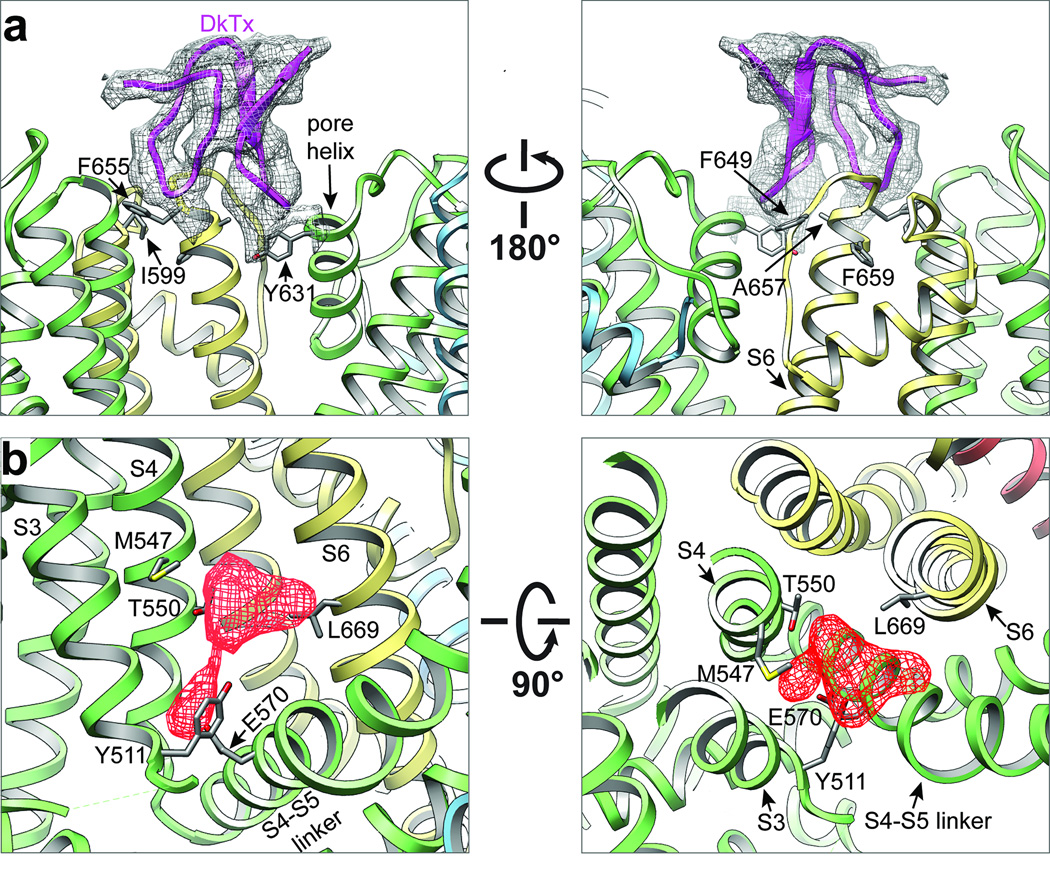
a, Detailed views of interaction between TRPV1 subunits (yellow and green) and one ICK knot from DkTx spider toxin (purple). TRPV1 resides in close proximity to the toxin are highlighted, including four (I599, F649, A657, F659) which, when mutated, render the channel specifically insensitive to DkTx. b, Vanilloid binding pocket defined by EM density of RTX (red, filtered at 4.5Å with a temperature factor of −200Å2, 8σ) viewed from the side (left) or top down (i.e. from the extracellular face; right). Residues in close proximity to observed densities are highlighted, including several (Y511, S512, M547, and T550) that have been previously implicated in vanilloid binding.
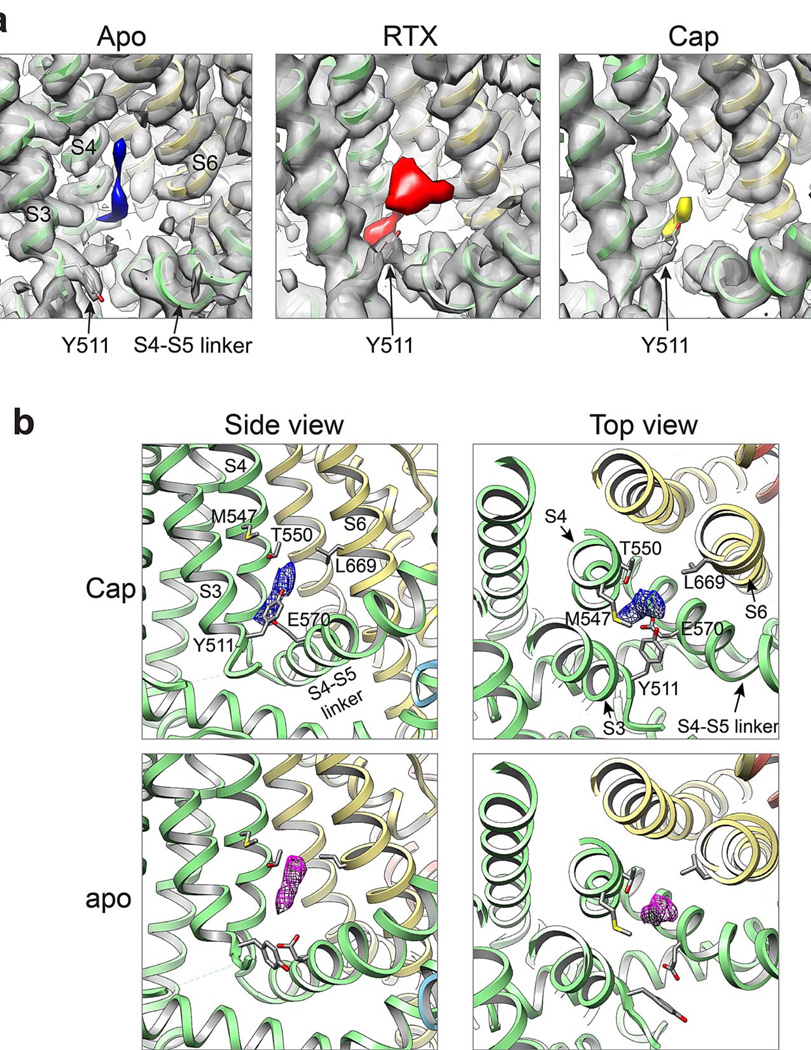
Mutagenesis studies have pinpointed residues that specify sensitivity to small vanilloid ligands, including Y511 and S512 in S3 and M547 and T550 in S4 22–25. Indeed, in the RTX/DkTx structure, we see robust density in close proximity to these residues, likely representing a molecule of RTX coordinated in a binding pocket just above E570 in the S4-S5 linker from the same subunit, and proximal to L669 in S6 of a neighboring subunit (Figs. 1 and 2b and Extended Data Fig. 7a). In VGICs, the S4-S5 linker plays a critical role in translating movement of the voltage sensor into gating of the pore 9,11. Thus, the putative vanilloid site is well positioned to affect channel gating by impinging on two regions (i.e. S4-S5 linker and S6) that likely influence structure of the pore domain (see below). This model is further supported by the capsaicin-bound TRPV1 structure, where we also see density in the vanilloid pocket (Extended Data Fig. 7). The weaker signal observed with capsaicin may reflect its substantially lower affinity for TRPV1 and smaller mass (MW 305) compared to RTX (MW 628). Interestingly, capsaicin and RTX densities are in close proximity, consistent with overlapping, but non-identical binding sites 26. Intriguingly, we observed density in this same hydrophobic pocket in the apo TRPV1 structure, perhaps representing a lipid or other small hydrophobic molecule associated with the channel in the absence of a competing exogenous ligand (Extended Data Fig. 7). Although our model lacks sufficient detail to reveal precisely how vanilloids bind, we see that Y511 assumes two distinct rotamers in apo versus liganded structures wherein its side chain points away from or into the binding pocket, respectively (Extended Data Fig. 7), suggesting that vanilloid binding involves an ‘induced fit’ mechanism akin to some enzyme/substrate interactions. Taken together, these densities define an important allosteric regulatory site for inflammatory agents, such as anandamide and other ‘endovanilloids’, or inverse agonists such as capsazepine.
Extended Data Figure 7. Observed densities in vanilloid pocket.
a, Non-protein associated densities in the region adjacent to S3-S4 transmembrane helices observed in 3D density maps of apo TRPV1 structure (blue, 3.4Å, −100Å2) or TRPV1 in complex with RTX/DkTx (red, 3.8Å, −150Å2) or capsaicin (yellow, 3.9Å, −150Å2), as indicated. b, Density of bound capsaicin (blue) viewed from the side (left) and top down (i.e. from the extracellular face; right). Density is also observed in the apo-channel structure (purple), possibly representing an endogenous lipid or other small hydrophobic molecule. All maps were low-pass filtered to 4.5Å with a temperature factor of −200Å2, normalized and displayed at the same sigma level (8Å).
Pore profiles reveal a dual gate
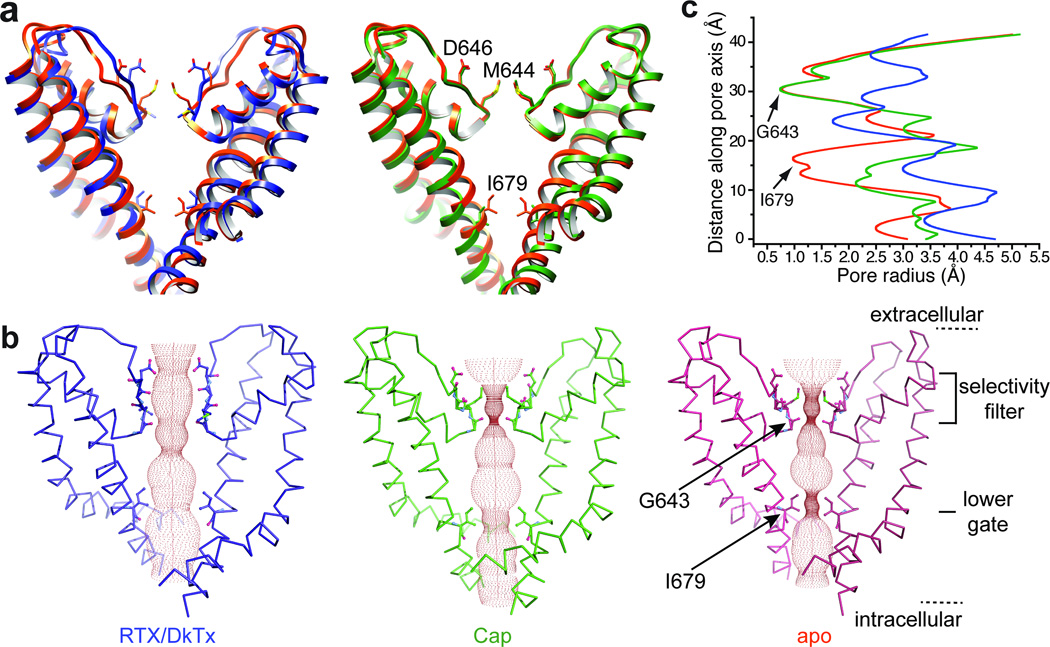
The three TRPV1 structures described here and in the accompanying study 8 represent the channel in distinct states that are readily discernible by comparing diameters of their ion conduction pathways (Fig. 3 and Extended Data Fig. 8a–c). In the apo state, the pathway is constricted at the selectivity filter, as well as the lower gate. In the capsaicin bound structure, there is no change in the selectivity filter, whereas the lower gate is markedly expanded. In the RTX/DkTx structure, the channel is fully open with the ion conduction pathway relieved of any constrictions. These profiles argue for a dual gating mechanism involving substantial conformational changes at both the selectivity filter and lower gate. TRPV1 has been shown to undergo pore dilation, enabling conduction of organic cations following prolonged activation 27. We do not believe that any of the configurations described here correspond to a dilated state because their pores are likely too narrow to accommodate such large cations. Nonetheless, the plasticity that we observe at both upper and lower gates may allow TRPV1, and possibly other TRPs, to assume a pore dilated state.
Figure 3. Comparison of ion permeation pathway in apo- versus liganded channels.
a, Superimposition of S5-P-S6 pore module from apo- (orange) versus RTX/DkTx (blue; left) or capsaicin (green; right) bound structures. In each case, only two diagonally opposed subunits are shown for clarity. Key residues in the selectivity filter and lower gate are highlighted to display side chain movements associated with gating. b, Solvent-accessible pathway along the pore mapped using the HOLE program for RTX/DkTx bound, capsaicin bound, and apo TRPV1 structures. Residues located at the selective filter and lower gate are rendered as sticks. c, Comparison of pore radii (calculated with the program HOLE) for RTX/DkTx bound (blue), capsaicin bound (green), and apo (orange) TRPV1 structures.
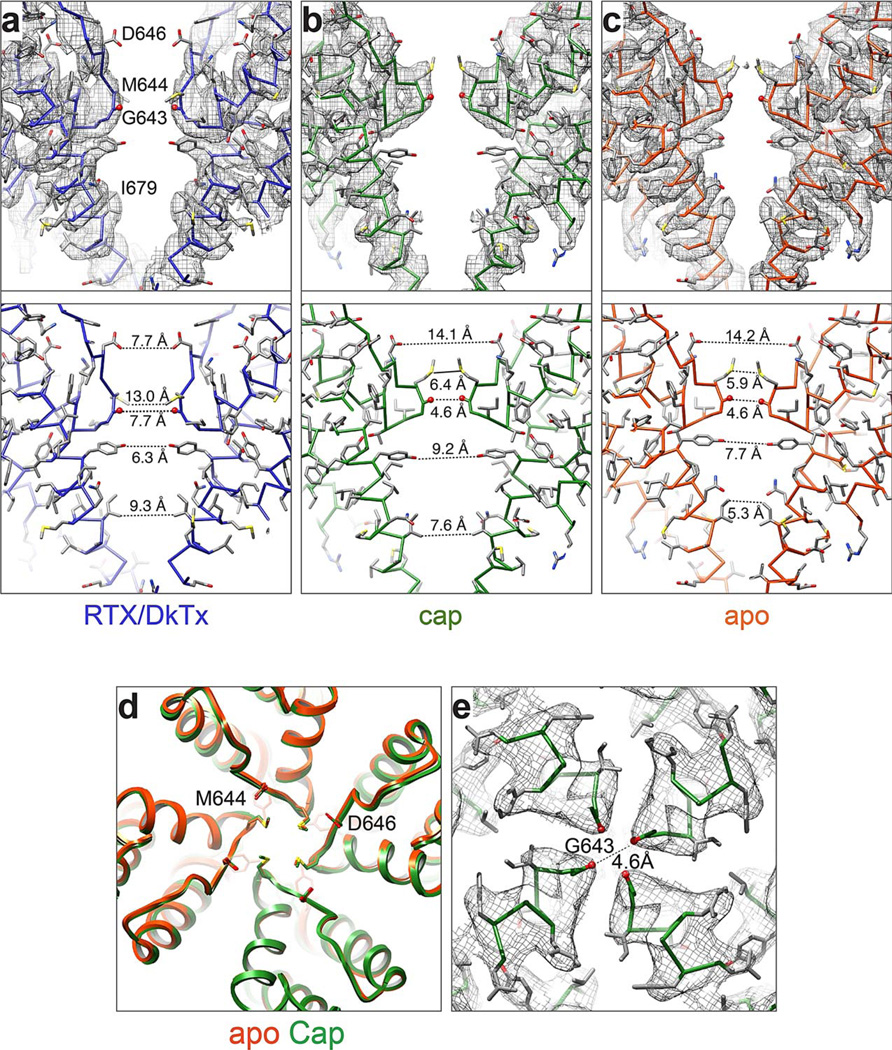
Extended Data Figure 8. Structural details of the TRPV1 pore with and without ligands.
a–c Density maps for pore region for two diagonally opposed monomers superimposed onto their atomic models (top). Distances between specific side chain atoms along the pore are also indicated (bottom). d, Superimposed top-down view of apo- and capsaicin bound TRPV1 outer pore regions (orange and green, respectively). e, Density map of selectivity filter in capsaicin-bound TRPV1 structure. The distance between carbonyl oxygens of diagonally opposed G643 residues (4.6Å) does not differ from that of the apo structure (4.6Å).
Rearrangements in the outer pore region
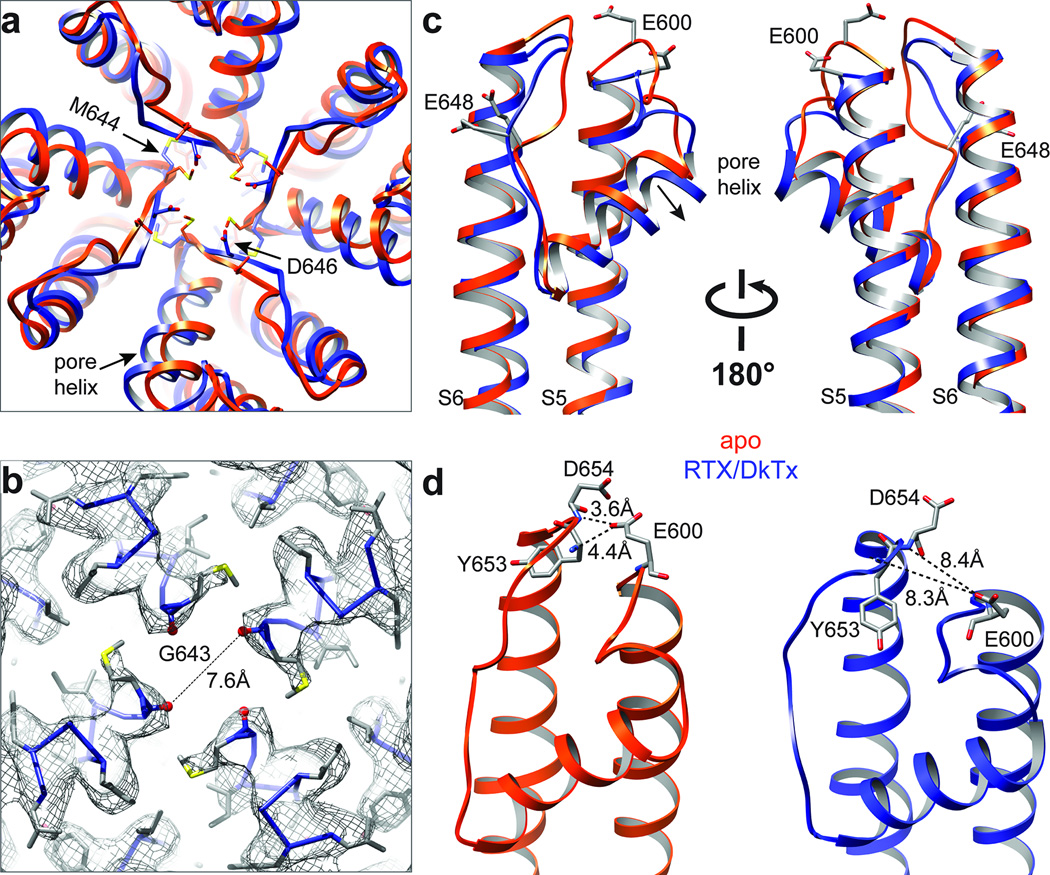
Superimposition of TRPV1 structures in apo versus RTX/DkTx bound states shows a pronounced shift in the relative position of the pore helix. This rearrangement is characterized by a rigid body tilt of the helix away from the central axis of the channel by ~1.9Å and widening of the selectivity filter at its narrowest point, increasing the distance between G643 carbonyl oxygens from 4.6 to 7.6Å (Fig. 4a–c). The downward movement of M644 also contributes to widening of the filter, where the distance between side chains increases from 5.9 to 13.0Å (Extended Data Fig. 8a–c). Moreover, conformational changes in the selectivity filter shorten the distance between Cα atoms of diagonally apposed D646 residues from 15.6 to 13.0Å, placing their side chains in a better geometry to coordinate and partially dehydrate cations for efficient conduction. The pore helix shift is also associated with rearrangements in outer pore loops that follow S5 and precede S6 (Fig. 4c), where movements have been suggested to play a role in the activation of TRPV channels by chemical or thermal stimuli 19,28–31. For example, two glutamates (E600 and E648) within these loops of TRPV1 are known to be important for acid-evoked potentiation or activation. In the apo TRPV1 structure, these loops are bridged through potential hydrogen bonding between the side chain of E600 and main chain nitrogen atoms of Y653 and D654 (Fig. 4d). These interactions, which likely maintain the outer pore in a non-conductive conformation in the resting state, are broken in the RTX/DkTx structure due to rearrangements that increase distance between these loops. We suggest that protonation at E600 similarly disrupts these hydrogen bonds to facilitate gating-associated movements of the pore helix and widening of the selectivity filter as a mechanism by which extracellular protons sensitize the channel to thermal or chemical stimuli.
Figure 4. Structural rearrangements in the outer pore region.
a, Superimposed top-down views of outer pore regions from apo- and RTX/DkTx bound TRPV1 structures (orange and blue, respectively). Note shift in relative positions of pore helix and selectivity filter. b, Density map with atomic model showing distance between diagonally opposed G643 residues, which represents the narrowest point in the outer pore region. c, Superimposed side views of the outer pore domains of apo- and RTX/DkTx bound TRPV1 structures (orange and blue, respectively). Residues important for proton-mediated sensitization (E600) or activation (E648) are labeled. Arrow indicates downward rigid body tilt of pore helix in RTX/DkTx structure. d, Proximity of E600 to neighboring residues in the apo structure (left) allows for hydrogen bonding, which is disrupted by rearrangements in the RTX/DkTx bound structure (right).
Generally speaking, the movements we observe in the outer pore and pore helix of TRPV1 stand in contrast to what is seen with potassium channels, where these regions are believed to remain relatively stationary during normal gating 10,20. However, there are instances, such as C-type inactivation, ‘flicker transitions’, or multiple subconductance states, that likely result from rearrangements within the K+ channel selectivity filter 32–34. Indeed, activation of TRPV1 by most stimuli (chemical or thermal) produces flickery behavior in which channels make frequent excursions to the closed state 35–37, perhaps reflecting the dynamic nature of the outer pore. Such flickers are substantially reduced when open channels bind DkTx, presumably reflecting toxin-mediated stabilization of the outer pore (including pore helix and selectivity filter) in fully conducing conformations 15.
Structure of the TRPV1 outer pore in the presence of capsaicin is superimposable with that of the apo, closed channel (Extended Data Fig. 8d, e). In the absence of DkTx, this structure is unlikely to be trapped in its fully open state and may, instead, represent the channel during one of its frequent excursions to partially or transiently closed states. Nonetheless, the channel has undergone conformational changes associated with gating, including movements of the S4-S5 linker and lower gate, as discussed below. Thus, we conclude that our capsaicin-TRPV1 structure represents a partially activated state.
Opening of the lower gate
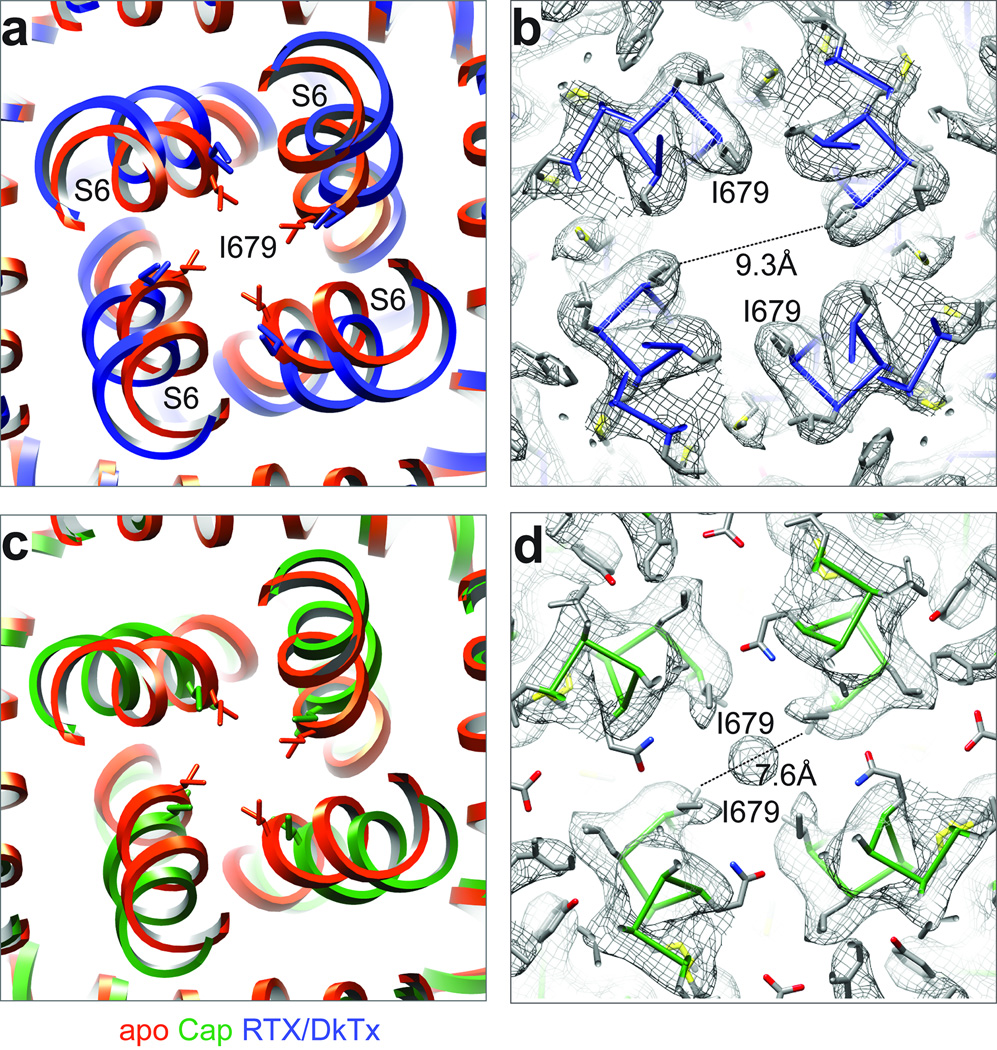
As we show in the accompanying study 8, I679 in S6 forms a hydrophobic seal sufficiently narrow (5.3Å) to block permeation by hydrated ions when TRPV1 is in its closed conformation. In the RTX/DkTx bound structure, we see a remarkable disruption of this seal with consequent expansion of the lower gate to 9.3Å in diameter (Fig. 5a, b). This is driven by conformational changes in the lower half of the S6 helix, resulting in ~30° rotation of I679 side chains away from the central canal. With capsaicin, the lower gate expands to 7.6Å (Fig. 5c, d). These changes in pore diameter are associated with a slight bowing out of S6 away from the vertical axis, which occurs in nearly opposite directions for capsaicin versus RTX/DkTx (Fig. 6a). Consequently, rotation of I679 away from the central axis is more pronounced for RTX/DkTx, accounting for wider opening of the lower gate compared to capsaicin (Extended Data Fig. 8).
Figure 5. Opening of the lower gate.
a, Superimposition of inner pore region from apo- and RTX/DkTx bound TRPV1 structures (orange and blue, respectively). The residue (I679) that forms the hydrophobic seal of the lower gate is highlighted, and its side chain shown in stick format. b, Density map with atomic model showing distances between I679 in the RTX/DkTx bound channel. Note substantial expansion of the lower gate relative to apo-structure. c, Superimposition of apo- and capsaicin bound TRPV1 inner pore regions (orange and green, respectively). d, Density map with atomic model showing distances between I679 in the capsaicin bound channel. Note expansion of the lower gate relative to apo-structure, but to a lesser extent than seen in RTX/DkTx bound channel. The density at the central cavity may represent noise amplified by applying symmetry, or a trapped hydrated ion. Interestingly, this density is not observed in apo or RTX/DkTx structures.
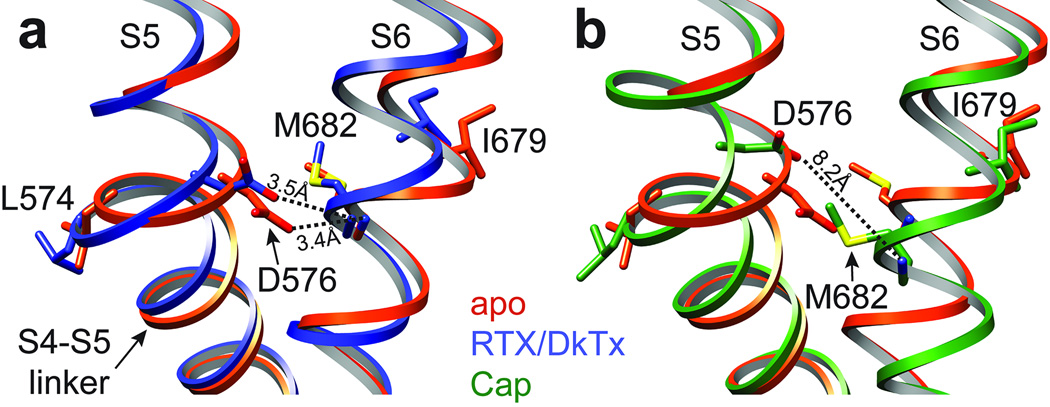
Figure 6. Coupling of S4-S5 linker and S6 helix.
a, b, Superimposition of apo (orange) with RTX/DkTx (blue) or capsaicin (green) bound structures highlighting interactions between residues in the S4-S5 linker and S6 helix. Through these interactions, movement of the S4-S5 linker is translated to rotation or displacement of I679 away from the central axis, breaking the hydrophobic seal to open the lower gate.
In VGICs, movement within the S1-S4 voltage sensor is coupled to channel opening through the S4-S5 linker helix 9,10. In Kv channels, lateral movement of the S4-S5 linker leads to splaying of the lower S6 segment below a kink formed by a conserved -Pro-X-Pro- or -Gly- signature, resulting in widening of the activation gate at the apex of the inverted teepee 10,38. In TRPV1, the S4-S5 linker and S6 also interact, but in a way that suggests a distinct gating mechanism. Whereas in VGICs these contacts take place well above the activation gate, in TRPV1 they occur just slightly beneath I679, placing them in direct alignment with the lower gate (Fig. 6). Thus, in both apo and RTX/DkTx structures, hydrogen bonding is observed between two residues that are invariant in all TRPV subtypes, namely D576 in the S4-S5 linker and M682 in S6. Consequently, movement at the junction between S5 and the S4-S5 linker (~2Å) produces a concomitant shift in S6 and associated movement of I679 away from the central canal. In the capsaicin bound structure, we see a somewhat larger shift (~2.7Å) of the S4-S5 linker (with loss of hydrogen bonding between D576 and M682), and outward movement of I679 (Fig. 6b). Thus, we propose that opening of the TRPV1 lower gate involves more subtle rearrangements in S6 compared to VGICs, and occurs closer to the point of interaction with the S4-S5 linker. Indeed, the S6 helix in TRPV1 lacks a proline or glycine kink that underlies the more pronounced outward splaying mechanism of Kv gate opening 10. Movement of the S4-S5 linker and S6 is, however, accompanied by lateral displacement of the TRP domain at the helical ‘elbow’ proximal to S6 (Extended Data Fig. 9c, d), where a 3.5Å displacement between Cα atoms of I696 is readily seen when comparing apo versus RTX/DkTx bound structures. These observations are consistent with the idea that the TRP domain functions as an allosteric modulatory site 39.
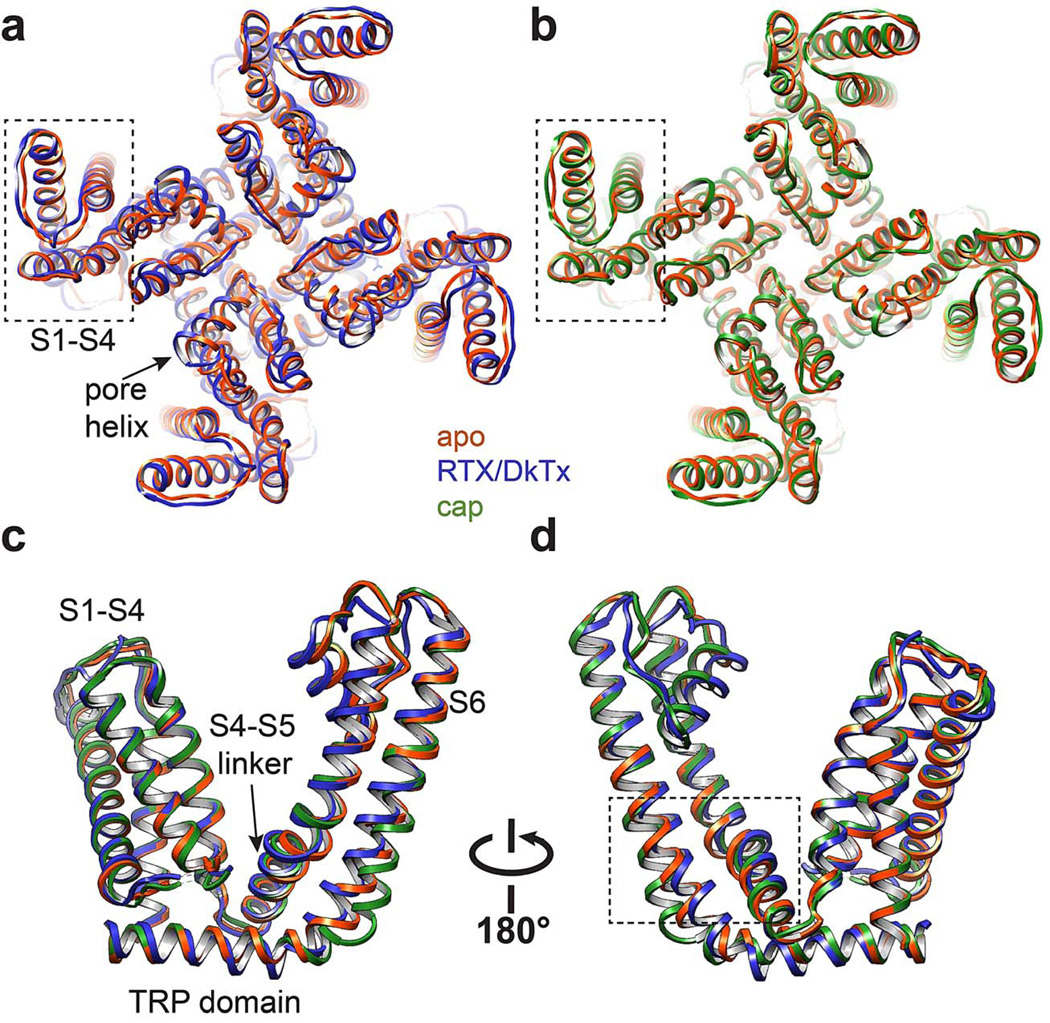
Extended Data Figure 9. S1-S4 as a stationary domain.
a, Superimposition of apo- and RTX/DkTx bound TRPV1 structures (orange and blue, respectively) from top-down view. S1-S4 domain (outlined in dashed box) shows near complete overlap. b, Same comparison for apo- and capsaicin bound channel structures (orange and green, respectively). c, d, Superimposition of transmembrane core of apo- versus RTX/DkTx- or capsaicin-bound TRPV1 structures (orange, blue, and green, respectively). Dashed box denotes region highlighted in Figure 6.
Static nature of the S1-S4 domain
In contrast to VGICs, we find that the S1-S4 domain of TRPV1 is virtually superimposable when comparing apo and activated structures (Extended Data Fig. 9), suggesting that this domain plays a less active role in TRP channel gating. This may explain why many gain-of-function mutations in TRPV subtypes are found within the S4-S5 linker, S5-P-S6 pore, and TRP domain, but not within the S1-S4 domain 18,40,41. Taken together, we posit that S1-S4 domain functions as a passive anchor upon which the S4-S5 linker moves in response to ligand binding. This is consistent with our observation that opening of the TRPV1 inner gate does not involve splaying of the lower half of S6, which would be constrained by the static S1-S4 domain.
TRPV1 and certain other TRP subtypes exhibit weak voltage dependence 42–44, for which the structural basis remains obscure. Our observations suggest that this is unlikely to involve movement within S1-S4, consistent with the fact that TRPV1 lacks positive charges in S4 that are characteristic of VGICs. However, as detailed above, the outer pore undergoes substantial downward movement during gating, away from the extracellular face of the bilayer. Because this helix forms a negative dipole at its C-terminal end, movement should be facilitated during membrane depolarization, enhancing open probability at positive membrane potentials, especially when the lower gate is open and membrane potential is focused on the upper gate. This conformational change may contribute to aspects of voltage sensitivity, such as outward rectification characteristic of many TRP channel subtypes 44. This idea (and that of dual gating) is consistent with TRPV4 mutagenesis studies suggesting that rectification is mediated by a voltage-dependent gating mechanism that operates in tandem with the primary intracellular gate 45.
Summary
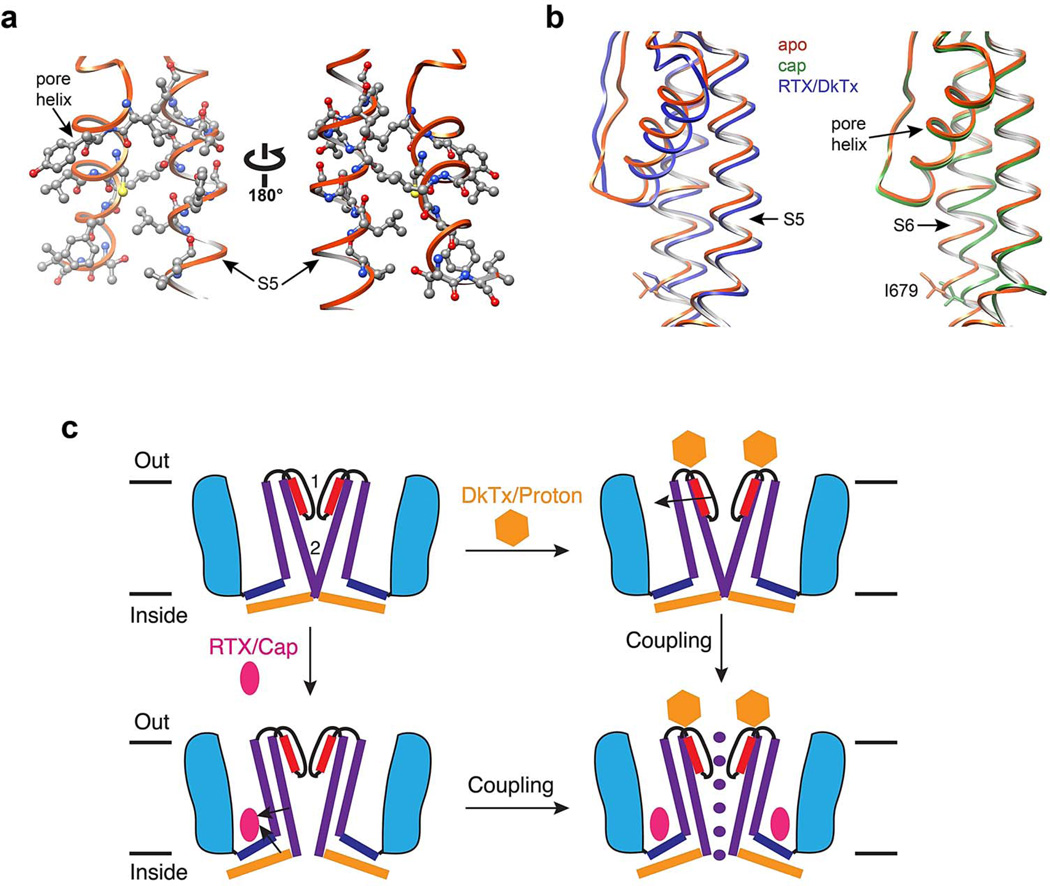
TRP channels are noted for their ability to function as polymodal signal integrators 37, which in the case of TRPV1 underlies its role in pain hypersensitivity 1. The structures presented here demonstrate that regions targeted by inflammatory agents induce or undergo substantial conformational changes associated with gating. These include the outer pore domain, which we show to be unusually dynamic compared to VGICs, as well as the hydrophobic pocket defined by the external surface of the S3-S4 helices, S4-S5 linker and S6 helix. Thus, pro-algesic agents can modulate channel activity by exerting immediate effects on either of two restriction points defined by the selectivity filter and lower gate. While these regions can be independently targeted, pharmacological observations suggest that they are allosterically coupled. For example, distinct stimuli (chemical or thermal) show cross sensitization, and the inverse vanilloid agonist, capsazepine, inhibits channel activation by capsaicin, protons, spider toxins, heat, or constitutive pore helix mutations 18,37,46,47. We suspect that the pore helix represents a critical structural element in coupling upper and lower gates; in the RTX/DkTx bound structure, downward tilt of the pore helix away from the central canal occurs concomitantly with movement of S5, likely through close side chain interactions that foster physical coupling between the two helices to facilitate opening of the lower gate (Extended Data Fig. 10a, b). Such dynamic communication between upper and lower gates could underlie integration of diverse physiologic signals (Extended Data Fig. 10c).
Extended Data Figure 10. Dual gate model for TRPV1 activation.
a, Pore helix and upper half of S5 helix are in close proximity and appear to be physically coupled, representing a potential mechanisms for allosteric coupling between upper and lower gates. Several residues on both helices are rendered as ball-and-stick to highlight close apposition. b, Downward tilt of pore helix away from the central pore is associated with movement of the S5 helix in RTX/DkTx structure (left). This structural arrangement is not observed in capsaicin bound structure (right). c, Model depicting two gate mechanism of TRPV1 activation. Two main constriction points at the selectivity filter (1) and lower gate (2) block ion permeation in the apo, closed state (upper left). Some pharmacological agents (e.g. protons or spider toxins; gold hexagon) target the outer pore region of the channel to open or stabilize the conductive conformation of the selectivity filter (upper right). Arrow denotes proposed coupling between the pore helix and S5. Small vanilloid ligands (e.g. RTX and capsaicin; red ellipse) bind within a hydrophobic pocket formed by the S3-S4 helices, the S4-S5 linker, and the pore module, eliciting conformational changes that expand the lower gate (lower left). Arrows indicate proposed coupling between S4-S5 helix, S6 and TRP domain. Full expansion of the ion permeation pathway and ion conduction is achieved when both upper and lower gates are opened (lower right). Pharmacological and mutagenesis data suggest that these gates are allosterically coupled.
Of course, a fascinating aspect of TRPV1 function relates to its robust thermosensitivity. Heat is ubiquitous and can affect the energy landscape of both the upper and lower gates. Indeed, structure-function and pharmacological studies have implicated various regions as determinants of thermosensitivity, including cytoplasmic and outer pore domains 17,19,48–50. Further insights into the mechanism of heat-evoked gating would be greatly facilitated by obtaining ‘snapshots’ of the channel at different temperatures. We believe that recent advances in cryo-EM make it possible to achieve this goal, which is likely out of reach for more established methods of protein crystallography.
Online Methods
Sample preparation and data acquisition for cryo-EM analysis
TRPV1 protein was purified as described in the accompanying study 8. For preparation of channel in complex with different pharmacological agents, TRPV1 protein (final concentration 5 µM) in amphipols was mixed with spider toxin, which has two toxin moieties and a total molecular weight of 8.5 kDa, (final concentration 10 µM) and RTX (50 µM; MW 628 Da), or with capsaicin (50 µM; MW 305 Da) for 30 min prior to grid preparation. Cryo-EM grids were prepared by the same procedure as described in the accompanying paper. Frozen hydrated TRPV1 complexes were imaged on TF30 Polara microscope operated at 300kV using K2 Summit in dose fractionation super-resolution counting mode. Imaging conditions were identical with those used for imaging channel alone, as described in the accompanying study.
Image processing
Particle picking, CTF determination and 2D classifications were carried out by the same procedure as described in the accompanying paper. After particle picking and 2D screening, 148,670 particles of RTX/DkTx bound TRPV1 and 95,897 particles of capsaicin bound TRPV1 were selected for 3D classification in RELION51, with the 3D reconstruction of apo TRPV1 filtered to a resolution of 60 Å as an initial model. Refinement of the final selected 3D classes was also carried out in RELION. 36,158 and 33,238 particles of RTX/DkTx and capsaicin bound TRPV1, respectively, were used to calculate the final maps. Resolution of final 3D reconstruction was estimated by gold standard FSC = 0.143 criteria, after applying a soft spherical mask on the two reconstructions independently refined from the half-data sets 21. The resolutions of the final 3D reconstructions were 3.8Å for RTX/DkTx bound TRPV1 and 4.2Å for capsaicin bound TRPV1. For model building and visualization, amplitude of the final 3D density map was amplified by a temperature factor of −150Å2. All maps deposited to the EMDB database are the raw maps without amplitude sharpening, masking or filtering.
Before calculating difference maps between 3D reconstructions of liganded and apo TRPV1, all maps were filtered to the same resolution of 6Å and applying the same temperature factor of −100Å2. Difference maps were calculated by subtracting the density of apo TRPV1 from the liganded TRPV1, using program “diffmap.exe” (written by Nikolaus Grigorieff). For visualizing ligand densities (Fig. 2b and Extended Data Fig. 7b), liganded and apo TRPV1 maps were filtered to 4.5Å with the same temperature factor (−200Å2) and normalized using mapman (Uppsala Software Factory), and visualized with the same threshold (8σ). UCSF Chimera 52 was used to visualize and segment the cryo-EM maps, and the rigid body fitting of atomic model was done using the fit-in-map function from Chimera.
Model building in Coot
The apo-structure of TRPV1 was docked into the 3D density maps of capsaicin- and RTX/DkTx-bound structures as a rigid body. The S1-S4 domain fits nicely into both maps, therefore only slight adjustments were performed at this part of the model. On the other hand, the S4-S5 linker, pore module (S5-P-S6) and TRP domain of the TRPV1 apo-structure exhibit substantial deviation from both maps, indicating structural rearrangements during ligand binding. This part of the model was rebuilt in Coot 53,54 using the apo-structure as a reference. The density map for the pore module of capsaicin bound structure is of sufficient quality to assign most side chains in this region, allowing us to obtain an accurate pore profile by calculating pore radii using HOLE program55. However, density for the TRP domain, especially the part that immediately follows S6, does not allow for side chain resolution, likely reflecting the dynamic nature of this region during gating. Nonetheless, the invariant W697 in helical TRP domain shows excellent density and serves as a landmark to orient the entire helix. The map of RTX/DkTx bound structure is of sufficient quality to build the model without ambiguity. A polyalanine model (a total of 31 aa) based on the NMR structure of hanatoxin (pdb code: 1D1H) 56 was docked onto the DkTx density with slight adjustment.
Extended Data Figure 3. Cryo-EM of TRPV1 in complex with capsaicin.
a, b, Fourier transform (a) of a representative image (b). c, 2D class averages of cryo-EM particles. d, Enlarged view of three representative 2D class averages. e, Gold-standard FSC curve (red) of the final 3D reconstruction, marked with the resolutions that correspond to FSC = 0.5 and 0.143. The FSC curve between the final map and that calculated from the atomic model is shown in blue. f, Euler angle distribution of all particles used for calculating the final 3D reconstruction. The sizes of balls represent the number of particles. The accuracy of rotation is 4.989°, as reported by RELION.
Acknowledgements
We thank X. Li for assistant in data acquisition using TF30 Polara and K2 Summit camera, and J.P Armache, C. Bohlen, J. Cordero-Morales, and J. Osteen for discussion and critical reading of the manuscript. This work was supported by grants from the National Institute of Health (R01GM098672 and S10RR026814 to Y.C. and R01NS065071 and R01NS047723 to D. J.), the National Science Foundation (DBI-0960271 to D. Agard and Y.C.), and the UCSF Program for Breakthrough Biomedical Research (Y.C.). E.C. was a fellow of the Damon Runyon Cancer Research Foundation.
Footnotes
Author contributions
All authors designed experiments. E.C. expressed, purified all protein samples used in this work and performed all functional studies. M.L carried out all cryo-EM experiments, including data acquisition and processing. E.C. built atomic model based on the cryo-EM maps. All authors analyzed data and wrote the manuscript.
Author Information
3D cryo-EM density maps of TRPV1 complexes without low pass filter and amplitude modification have been deposited in the Electron Microscopy Data Bank under the accession number: EMD-5776 (TRPV1-RTX/DkTx) and EMD-5777 (TRPV1-capsaicin). The coordinates of atomic models of TRPV1 in these two states have been deposited in the Protein Data Bank under the accession number: 3J5Q and 3J5R. The authors declare no competing financial interests.
References
- 1.Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baconguis I, Gouaux E. Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature. 2012;489:400–405. doi: 10.1038/nature11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen SB, Tao X, MacKinnon R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 2011;477:495–498. doi: 10.1038/nature10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hattori M, Gouaux E. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature. 2012;485:207–212. doi: 10.1038/nature11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whorton MR, MacKinnon R. X-ray structure of the mammalian GIRK2-betagamma G-protein complex. Nature. 2013;498:190–197. doi: 10.1038/nature12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohlen CJ, Julius D. Receptor-targeting mechanisms of pain-causing toxins: How ow? Toxicon. 2012;60:254–264. doi: 10.1016/j.toxicon.2012.04.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vriens J, Appendino G, Nilius B. Pharmacology of vanilloid transient receptor potential cation channels. Mol Pharmacol. 2009;75:1262–1279. doi: 10.1124/mol.109.055624. [DOI] [PubMed] [Google Scholar]
- 8.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catterall WA. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron. 2010;67:915–928. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 11.Swartz KJ. Sensing voltage across lipid membranes. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu S, Darbon H, Dyason K, Verdonck F, Tytgat J. Evolutionary origin of inhibitor cystine knot peptides. Faseb J. 2003;17:1765–1767. doi: 10.1096/fj.02-1044fje. [DOI] [PubMed] [Google Scholar]
- 13.Phillips LR, et al. Voltage-sensor activation with a tarantula toxin as cargo. Nature. 2005;436:857–860. doi: 10.1038/nature03873. [DOI] [PubMed] [Google Scholar]
- 14.Swartz KJ, MacKinnon R. Hanatoxin modifies the gating of a voltage-dependent K+ channel through multiple binding sites. Neuron. 1997;18:665–673. doi: 10.1016/s0896-6273(00)80306-2. [DOI] [PubMed] [Google Scholar]
- 15.Bohlen CJ, et al. A bivalent tarantula toxin activates the capsaicin receptor, TRPV1, by targeting the outer pore domain. Cell. 2010;141:834–845. doi: 10.1016/j.cell.2010.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siemens J, et al. Spider toxins activate the capsaicin receptor to produce inflammatory pain. Nature. 2006;444:208–212. doi: 10.1038/nature05285. [DOI] [PubMed] [Google Scholar]
- 17.Grandl J, et al. Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat Neurosci. 2010;13:708–714. doi: 10.1038/nn.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers BR, Bohlen CJ, Julius D. A yeast genetic screen reveals a critical role for the pore helix domain in TRP channel gating. Neuron. 2008;58:362–373. doi: 10.1016/j.neuron.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F, Cui Y, Wang K, Zheng J. Thermosensitive TRP channel pore turret is part of the temperature activation pathway. Proc Natl Acad Sci U S A. 2010;107:7083–7088. doi: 10.1073/pnas.1000357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 21.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou MZ, Mtui T, Gao YD, Kohler M, Middleton RE. Resiniferatoxin binds to the capsaicin receptor (TRPV1) near the extracellular side of the S4 transmembrane domain. Biochemistry. 2004;43:2501–2511. doi: 10.1021/bi035981h. [DOI] [PubMed] [Google Scholar]
- 23.Gavva NR, et al. Molecular determinants of vanilloid sensitivity in TRPV1. J Biol Chem. 2004;279:20283–20295. doi: 10.1074/jbc.M312577200. [DOI] [PubMed] [Google Scholar]
- 24.Jordt SE, Julius D. Molecular basis for species-specific sensitivity to"hot" chili peppers. Cell. 2002;108:421–430. doi: 10.1016/s0092-8674(02)00637-2. [DOI] [PubMed] [Google Scholar]
- 25.Phillips E, Reeve A, Bevan S, McIntyre P. Identification of species-specific determinants of the action of the antagonist capsazepine and the agonist PPAHV on TRPV1. J Biol Chem. 2004;279:17165–17172. doi: 10.1074/jbc.M313328200. [DOI] [PubMed] [Google Scholar]
- 26.Szallasi A, Blumberg PM, Annicelli LL, Krause JE, Cortright DN. The cloned rat vanilloid receptor VR1 mediates both R-type binding and C-type calcium response in dorsal root ganglion neurons. Mol Pharmacol. 1999;56:581–587. doi: 10.1124/mol.56.3.581. [DOI] [PubMed] [Google Scholar]
- 27.Chung MK, Guler AD, Caterina MJ. TRPV1 shows dynamic ionic selectivity during agonist stimulation. Nature neuroscience. 2008;11:555–564. doi: 10.1038/nn.2102. [DOI] [PubMed] [Google Scholar]
- 28.Jordt SE, Tominaga M, Julius D. Acid potentiation of the capsaicin receptor determined by a key extracellular site. Proc Natl Acad Sci U S A. 2000;97:8134–8139. doi: 10.1073/pnas.100129497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SE, Patapoutian A, Grandl J. Single residues in the outer pore of TRPV1 and TRPV3 have temperature-dependent conformations. PLoS One. 2013;8:e59593. doi: 10.1371/journal.pone.0059593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryu S, Liu B, Yao J, Fu Q, Qin F. Uncoupling proton activation of vanilloid receptor TRPV1. J Neurosci. 2007;27:12797–12807. doi: 10.1523/JNEUROSCI.2324-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh BI, Kim YK, Jabbar W, Huang CL. Conformational changes of pore helix coupled to gating of TRPV5 by protons. Embo J. 2005;24:3224–3234. doi: 10.1038/sj.emboj.7600795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berneche S, Roux B. A gate in the selectivity filter of potassium channels. Structure. 2005;13:591–600. doi: 10.1016/j.str.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 33.Cuello LG, Jogini V, Cortes DM, Perozo E. Structural mechanism of C-type inactivation in K(+) channels. Nature. 2010;466:203–208. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoshi T, Armstrong CM. C-type inactivation of voltage-gated K+ channels: pore constriction or dilation? J Gen Physiol. 2013;141:151–160. doi: 10.1085/jgp.201210888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hui K, Liu B, Qin F. Capsaicin activation of the pain receptor, VR1: multiple open states from both partial and full binding. Biophys J. 2003;84:2957–2968. doi: 10.1016/S0006-3495(03)70022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B, Hui K, Qin F. Thermodynamics of heat activation of single capsaicin ion channels VR1. Biophys J. 2003;85:2988–3006. doi: 10.1016/S0006-3495(03)74719-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tominaga M, et al. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron. 1998;21:531–543. doi: 10.1016/s0896-6273(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 38.Jiang Y, et al. The open pore conformation of potassium channels. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 39.Latorre R, Zaelzer C, Brauchi S. Structure-functional intimacies of transient receptor potential channels. Q Rev Biophys. 2009;42:201–246. doi: 10.1017/S0033583509990072. [DOI] [PubMed] [Google Scholar]
- 40.Dai J, et al. TRPV4-pathy, a novel channelopathy affecting diverse systems. J Hum Genet. 2010;55:400–402. doi: 10.1038/jhg.2010.37. [DOI] [PubMed] [Google Scholar]
- 41.Lin Z, et al. Exome sequencing reveals mutations in TRPV3 as a cause of Olmsted syndrome. Am J Hum Genet. 2012;90:558–564. doi: 10.1016/j.ajhg.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Latorre R, Vargas G, Orta G, Brauchi S. In: TRP Ion Channel Function in Sensory Transduction and Cellular Signaling Cascades Frontiers in Neuroscience. Liedtke WB, Heller S, editors. 2007. [PubMed] [Google Scholar]
- 43.Matta JA, Ahern GP. Voltage is a partial activator of rat thermosensitive TRP channels. J Physiol. 2007;585:469–482. doi: 10.1113/jphysiol.2007.144287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nilius B, et al. Gating of TRP channels: a voltage connection? J Physiol. 2005;567:35–44. doi: 10.1113/jphysiol.2005.088377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loukin S, Su Z, Zhou X, Kung C. Forward genetic analysis reveals multiple gating mechanisms of TRPV4. J Biol Chem. 2010;285:19884–19890. doi: 10.1074/jbc.M110.113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao E, Cordero-Morales JF, Liu B, Qin F, Julius D. TRPV1 channels are intrinsically heat sensitive and negatively regulated by phosphoinositide lipids. Neuron. 2013;77:667–679. doi: 10.1016/j.neuron.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Der Stelt M, Di Marzo V. Endovanilloids. Putative endogenous ligands of transient receptor potential vanilloid 1 channels. Eur J Biochem. 2004;271:1827–1834. doi: 10.1111/j.1432-1033.2004.04081.x. [DOI] [PubMed] [Google Scholar]
- 48.Brauchi S, Orta G, Salazar M, Rosenmann E, Latorre R. A hot-sensing cold receptor: C-terminal domain determines thermosensation in transient receptor potential channels. J Neurosci. 2006;26:4835–4840. doi: 10.1523/JNEUROSCI.5080-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papakosta M, et al. The chimeric approach reveals that differences in the TRPV1 pore domain determine species-specific sensitivity to block of heat activation. J Biol Chem. 2011;286:39663–39672. doi: 10.1074/jbc.M111.273581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao J, Liu B, Qin F. Modular thermal sensors in temperature-gated transient receptor potential (TRP) channels. Proc Natl Acad Sci U S A. 2011;108:11109–11114. doi: 10.1073/pnas.1105196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi H, et al. Solution structure of hanatoxin1, a gating modifier of voltage-dependent K(+) channels: common surface features of gating modifier toxins. J Mol Biol. 2000;297:771–780. doi: 10.1006/jmbi.2000.3609. [DOI] [PubMed] [Google Scholar]