INTRODUCTION
The directed extension of an axon towards its synaptic partner represents a critical step in establishing the complex wiring diagram of the brain. A wealth of cellular interactions, including adhesive signals between an axon and its correct target, repulsive signals from inappropriate partners, as well as interactions amongst afferent axons play important roles (Luo and Flanagan, 2007). However, how these interactions change growth cone morphology to direct axon extension toward specific targets remains unknown.
Growth cones are the sensory structures of the advancing axon. They form motile actin-rich protrusions, filopodia and lamellipodia, which probe the environment for guidance signals (Bentley and Toroian-Raymond, 1986; Chien et al., 1993; Dent et al., 2011). The effects of guidance and adhesive factors on growth cone shape and axon extension have been intensely studied in vitro (Vitriol and Zheng, 2012). Adhesion molecules propel growth cones by cross-linking the substrate with the actin cytoskeleton to increase traction, ultimately promoting formation of protrusions and growth cone advance (Bard et al., 2008; Giannone et al., 2009; Mitchison and Kirschner, 1988; Thoumine et al., 2006). However, little is known about how growth cones navigate the more complex environments encountered in vivo. Previous work correlated in vivo growth cone shape with axon extension, revealing that simple, highly polarized growth cones advance rapidly, while complex, less polarized growth cones frequently pause (Mason and Wang, 1997). However, how adhesion molecules might regulate these polarity changes remains incompletely understood, and it is unknown whether growth cone polarity can determine target specificity.
To address these issues, we studied photoreceptor (R cell) axon targeting in the Drosophila visual system (Hadjieconomou et al., 2011). The compound eye of the fruit fly comprises approximately 800 facets, called ommatidia, each of which contains eight photoreceptors (R1-R8). Due to the curvature of the eye and the arrangement of light sensing organs, each of the outer six R cells (R1-R6) within a single ommatidium receives light from a different point in space, and must therefore connect to a different target column in the brain. At the same time, specific groups of R1-R6 cells distributed amongst neighboring ommatidia receive light from the same point in space, and converge on the same target column (Figure 1A). This wiring principle is called neural superposition, and results in the formation of a retinotopic map (Clandinin and Zipursky, 2000).
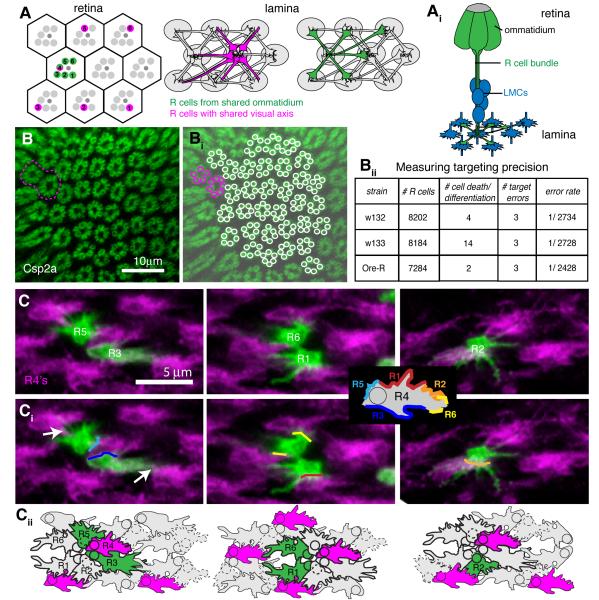
Figure 1. R cell growth cones find their targets with extremely high fidelity.
(A) Schemata of the Drosophila retina and lamina. Axons of R cells that sense light from the same point in space (magenta) innervate a common cartridge (grey ovals), while axons of R cells from the same ommatidium (green) innervate neighboring cartridges. (Ai) Side view of R cells from a single ommatidium with axonal projections in the lamina (green), and their synaptic targets, the lamina monopolar cells (LMC, blue), in mid pupae. (B) Wild-type adult lamina labeled with the pre-synaptic marker Csp2a. One targeting error is highlighted in magenta. (Bi) Reconstruction of R cell terminals. (Bii) Quantification of error measurements in three wild-type strains; n=15-18 laminas. (C) R4 growth cones contact both R3 and R5 from the same ommatidium, as well as R1, R2 and R6 from neighboring ommatidia. Single growth cones of different subtypes labeled with GFP-myr at 28% apf (green), co-labeled with R4 marked by mδ-Gal4 UAS-mtdTomato (magenta); ventral hemisphere, anterior up, equator to the right. (Ci) Major contact sites were marked with colored lines. Growth cones also made small filopodial contacts with other neighbors (white arrows). Inset shows model of putative adhesion sites of an R4 growth cone with its neighbors. (Cii) Schematic outline of growth cones, based on ultrastructural images by Meinertzhagen and Hanson (1993). Growth cones from within a bundle are outlined in bold. See also Figure S1.
Ultrastructural and genetic studies have provided insight into the mechanisms that direct R1-R6 axons to their targets (Hadjieconomou et al., 2011; Meinertzhagen and Hanson, 1993). R cell axons from each ommatidium are bundled together in a fascicle as they extend into the brain, following the path of the R8 axon. Upon reaching the lamina, R1-R6 axons defasciculate and each extends a lateral process that innervates a single column of five post-synaptic targets, the lamina monopolar cells (LMCs; Figure 1Ai). All R cells that “see the same point in visual space” innervate the same target column, and assemble a fascicle, called a cartridge, that contains both R cell axons and LMC dendrites. Remarkably, all targeting steps occur within a highly patterned structure, as every neurite and growth cone is precisely positioned with respect to its neighbors (Meinertzhagen and Hanson, 1993; Meinertzhagen and O’Neil, 1991). Furthermore, R cell targeting specificity is genetically hardwired (Hiesinger et al., 2006), and it is instructed by interactions amongst afferent R cell axons, while target derived cues are largely permissive (Clandinin and Zipursky, 2000).
The classical cadherin Ncad, the receptor tyrosine phosphatase LAR and the adaptor protein Liprin-α are critically involved in the extension of R cell growth cones toward their target column (Choe et al., 2006; Lee et al., 2001; Prakash et al., 2005; Prakash et al., 2009). Ncad is expressed both in R cells and LMCs and mediates interactions between these two cell types; R cells that lack Ncad, LAR or Liprin-α frequently fail to extend. In contrast, the atypical cadherin Fmi, together with its partner Golden Goal, regulates target choice by mediating interactions amongst R cell axons (Chen and Clandinin, 2008; Hakeda-Suzuki et al., 2011; Lee et al., 2003; Tomasi et al., 2008). Intriguingly, while removal of fmi from all R cells results in highly penetrant targeting phenotypes, loss of fmi in single R cells has only negligible effects, suggesting that at least one redundant pathway must exist (Chen and Clandinin, 2008; Lee et al., 2003). Here we provide new insight into the mechanisms by which R1-R6 cell axons find their targets using cadherin-mediated interactions. Our results demonstrate that a network of interactions between multiple partners provides redundant cues to orient growth cones.
RESULTS
R1-R6 cells choose synaptic partners with remarkable fidelity
Invertebrate nervous systems can display tremendous wiring precision. Microscopic reconstruction of the lamina of the Dipteran fly Calliphora failed to detect any targeting errors made by 650 photoreceptor axons (Horridge and Meinertzhagen, 1970). To measure targeting fidelity in Drosophila, we stained adult brains of three wild-type strains with the synaptic marker Cysteine String Protein 2a (Csp2a), which specifically labels R cell terminals in the lamina (Zinsmaier et al., 1990). Excluding equatorial and peripheral regions, if all R cells differentiate normally and target correctly, all cartridges should contain exactly six profiles. If one R cell fails to differentiate, or dies, a single cartridge will have five terminals, while all of its neighbors will have six. In total, we observed 20 errors of this type in 3,945 cartridges. In addition, there are two categories of true targeting errors that can be distinguished. First, if one axon innervates an incorrect target, one cartridge will contain seven terminals while a neighbor will contain five. Second, if an axon innervates two targets, one cartridge will contain seven terminals, while all neighbors will contain six (Figure 1B). Across the three wild type strains, we observed only 9 targeting errors in 3,945 cartridges scored, corresponding to an average error rate of only 1 in 2630 terminals (99.96% fidelity; Figure 1Bii). As each retina comprises approximately 4800 R1-R6 axons, this corresponds to fewer than 2 targeting errors per eye. Thus, the mechanisms that regulate R cell targeting must be very robust.
R cell growth cones polarize towards their targets prior to contacting them
To identify the cellular and molecular mechanisms that guide R cell targeting, we first examined the development of R cell growth cone morphology. After reaching the lamina, during late larval and early pupal development, R1-R6 growth cones expand laterally, with R cells from within a bundle forming an open “ring-like” configuration (Figure 1C). Growth cones begin to extend to their targets around 32% after puparium formation (apf), and appear to have little contact with their postsynaptic partners prior to this time (Meinertzhagen and Hanson, 1993). To examine individual R cell growth cones, we genetically labeled R4 axons using mδ-Gal4 driven expression of myristoylated tdTomato (Chen and Clandinin, 2008). In addition, we stochastically labeled single growth cones with myristoylated GFP, assigning R cell identity based on the morphology and position of the cell body within the retina (see Extended Experimental Procedures). Before they extended, R4 growth cones were closely apposed to a specific, invariant set of neighbors, both from the same ommatidium (R3 and R5), as well as from neighboring ommatidia (R1, R2 and R6; Figure 1C; Meinertzhagen and Hanson, 1993). Analogous, precise neighbor relationships were seen for all other growth cones as well (data not shown). Thus, each R cell can engage in adhesive interactions with several specific neighbors. Furthermore, we found that all R4 growth cones were of similar shape, aligned in parallel, and oriented toward the future target column (Figure S1).
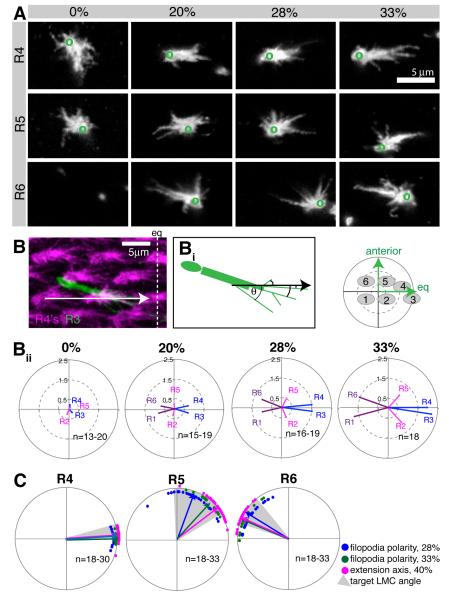
To understand how this polarity develops, we quantified the morphology of growth cones from the time they have expanded in the lamina, at the start of pupation, until they have extended towards their targets, around 33% apf (Figure 2A). Actin protrusions, such as filopodia and lamellipodia, are central to axon guidance (Bentley and Toroian-Raymond, 1986; Chien et al., 1993; Geraldo and Gordon-Weeks, 2009). Furthermore, growth cones in vivo make more filopodial contact with their targets as compared to neighboring non-targets (Raper et al., 1983). As R cell growth cones form no discernable lamellipodia, we therefore used filopodial distribution as a measure of polarity (Figure 2B). Each filopodium was represented as a vector with a defined length and angle relative to a landmark (Figure 2Bi); from these we calculated a mean polarity vector for each growth cone and each R cell sub-type (see Extended Experimental Procedures). At the start of pupation (0% apf), R1 and R6 growth cones had not expanded, and the growth cones of R2-R5 were of uniform shape and projected filopodia in all directions, producing short mean vectors that reflect a lack of polarization (Figure 2A,Bii; Figure S2). However, at 20% apf, mean polarity vectors had increased in magnitude as growth cones began to orient toward their future targets. Polarity increased progressively until 33% apf, when growth cones extended.
Figure 2. R cell growth cone polarity increases over time.
(A) Single R4, R5 and R6 growth cones labeled with GFP-myr, at 0, 20, 28 and 33% apf; ventral hemisphere, equator to the right, anterior up. Green circles indicate location of the axon shaft. Images for R1-R3 are in Figure S2. R6 growth cones have not expanded yet at 0% apf, and only the axons shaft is visible. (B) R4 growth cones were labeled by mδ-Gal4 UAS-mtdTomato (magenta), and a single R3 with GFPmyr (green); maximum intensity projection of 5μm thick stack, 33% apf; eq: equator. (Bi) Filopodia were measured from the growth cone base to their tip; the angle θ is the angle of the filopodium with respect to the alignment of R4 growth cones (black arrow). (Bii) Polar plots of the mean R cell polarity vectors between 0 and 33% apf. The small plot denotes the spatial orientation and location of the R cell targets. (C) Growth cone polarity at 28% apf (blue) and 33% apf (green) correlates with the angle of extension at 40% apf (magenta). Shown are mean polarity angles; arrows indicate mean polarity vector for the population; each dot represents a single R cell. Grey areas show the angles of the target LMCs. Plots for R1-R3 are in Figure S2.
To relate growth cone polarity to target choice, we plotted the mean polarity angles at 28% and 33% apf, as well as the angles at which growth cones have extended at 40% apf (Figure 2C, Figure S2). For R1, R3, R4, and R6, growth cone polarity and targeting angles almost completely overlapped. For R2 and R5 there was a consistent 30° difference between the polarity at 28% and extension angles at 40% apf, which decreased to around 10° by 33% apf (Figure 2C). We also plotted the angular size and position of all target cartridges, relative to the R cell bundle, and observed extensive overlap between R cell growth cone polarization and target position (Figure 2C). In addition, we observed an inverse relationship between the angular target size and the degree of R cell polarity, creating the following ranked order of polarization: R3> R4 > R1= R6 > R2= R5. These observations suggest that R cell growth cones “pre-select” their post-synaptic partners prior to interacting with them, through a process of directed polarization.
Fmi and Ncad have redundant roles in R cell targeting
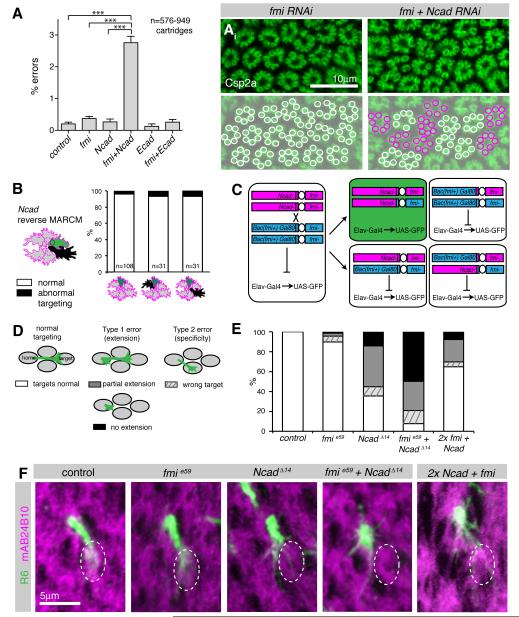
We next sought molecules that mediate interactions between R cells, and might direct growth cone polarization. The atypical cadherin Fmi was a strong candidate, as its loss in all R cells results in severe targeting defects (Lee et al., 2003). However, when single cells are made mutant for fmi, they invariably target normally (Chen and Clandinin, 2008), suggesting the existence of additional adhesive factors. We therefore examined two cadherins expressed in R cell growth cones at this stage, namely Ncad and E-cadherin (Ecad; Lee et al., 2001; Prakash et al., 2005), and probed whether these genes interact genetically with fmi. We took advantage of our ability to quantitatively measure the fidelity of axon targeting, and generated a sensitized genetic background by expression of fmi RNAi using a late R cell driver (gmrFlp; actin-FRT-y(+)-FRT-Gal4). In this background, Fmi levels were only moderately reduced (Figure S3) and the error rate of targeting was not significantly increased relative to controls (Figure 3A). While knocking down Ecad in this background had no effect, co-depletion of Ncad and Fmi resulted in an approximately tenfold increase in the fraction of R cells that made targeting errors (Figure 3A). These data raise the possibility that Ncad might serve two functions in R cell growth cones, mediating both interactions between R cells and their targets, as well as amongst R cell growth cones, where it could act redundantly with Fmi.
Figure 3. Fmi and Ncad genetically interact to mediate R cell growth cone extension.
(A) Single and double RNAi against fmi, Ncad, Ecad and LAR under control of gmr-Flp: actin>y>Gal4. Shown are % of errors by single R cells. Mean values + SEM. ***p<0.001, one-way ANOVA with Newman-Keuls post hoc test. (Ai) Lamina and its reconstruction in fmi single and Ncad fmi double RNAi expressing flies, labeling Csp2a; cartridges with six terminals in white and with five or seven terminals in magenta. (B) Targeting defects of single Ncad homozygous wild-type R cell growth cones (green) with or without Ncad mutant neighbors (black) in a heterozygous background (grey), at 38% apf; left bar: all neighbors are wild-type, center bar: one direct neighbor is mutant, right bar: one indirect neighbor is mutant. None of the groups are significantly different from each other (Fisher’s exact test). R1-6 cells were pooled. (C) Schemata of MARCM strategy to generate single cells double mutant for Ncad and fmi. Flies were mutant for fmie59/fmi192 and rescued with a Bac construct (Bac(fmi+)), see Figure S3), which sits in cis to Gal80 and in trans to Ncadδ14. After mitotic recombination, only the double mutant daughter cell will have lost the Gal80 containing chromosome arm, resulting in de-repression of GFP (green). The wild-type sister clone (top right) contains two wild-type copies of both Ncad and fmi, while two other outcomes of recombination result in cells heterozygous wild-type for both genes. (D) Schemata illustrating types of targeting defects. (E) Quantification of targeting phenotypes of fmi and Ncad single and double mutant cells, as well as cells homozygous wild-type for both fmi and Ncad (2x) using reverse MARCM (see text for description) at 38% apf. R1-R6 cells were pooled. All groups were significantly different from control with p<0.001, or p<0.05 for control vs. fmi; Fisher’s exact test, adjusted for multiple comparisons; n=71-123. (F) Single R6 growth cones at 38% apf labeled with CD8GFP (green), counterstained with mAb24B10 (magenta) to visualize cartridges. The wild-type R6 target cartridge is marked with a dotted white line; projected stacks of 3-5.5 μm; dorsal lamina hemisphere, equator down. Growth cones mutant for Ncad, for Ncad and fmi, as well as growth cones with elevated cadherin levels (2x) showed reduced or no target interactions.
Given this synergy between Ncad and Fmi, we first tested whether manipulating Ncad altered interactions amongst R cells, using reverse MARCM (Lee et al., 2000). This experiment generated cells mutant for Ncad that were labeled by the absence of gmr:RFPmyr, while their homozygous wild-type sister cells were labeled by mCD8GFP and analyzed for targeting defects (Figure 3B). All other cells were heterozygous wild-type. As clonally related R cells could be either sorted into the same ommatidium, a separate ommatidium, or die, a wild-type mCD8GFP positive cell can be in an ommatidium containing only wild-type cells, or it can sit directly next to a mutant cell, or it can be separated from the mutant cell by two or three wild-type cells. Surprisingly, targeting defects were both very infrequent, and independent of the presence or position of an Ncad mutant cell (Figure 3B). As analogous experiments performed with Fmi did reveal targeting phenotypes (Chen and Clandinin, 2008), these data demonstrate that Ncad is not necessary for these interactions.
Ncad and Fmi act redundantly to mediate growth cone extension
To test how the simultaneous loss of both Fmi and Ncad might affect R cell targeting, we sought to generate single cell somatic mosaic clones mutant for both genes. However, the Ncad and fmi are located on different arms of the same chromosome, complicating standard experiments (Lee and Luo, 1999). We took two approaches to circumvent this problem. First, we generated fmi mutants using Mosaic Analysis with a repressible cell marker (MARCM), while simultaneously knocking down Ncad in the same cells using Gal4-mediated expression of Ncad RNAi. Second, we moved the fmi locus onto the same chromosome arm as Ncad, by inserting a bacterial artificial chromosome (Bac) containing all fmi coding and regulatory sequences (CH321-66D09, Figure S3) onto 2L and placed it in an fmi trans-heterozygous null mutant background. This Bac completely rescued both lethality and planar cell polarity defects associated with fmi mutants (Figure S3). To induce Ncad and fmi double mutant clones, we then placed this fmi rescue construct in trans to an Ncad mutant chromosome (Figure 3C).
We divided growth cone targeting defects into two categories (Figure 3D). Type 1 targeting errors describe growth cones that either completely failed to extend to their targets, remaining in the home cartridge, or partially failed to extend, innervating both the home and the correct target cartridge. Type 2 targeting errors describe growth cones that extended away from the home cartridge, but innervated the wrong target. While a type 1 error emerges from defects in growth cone extension, a type 2 error reflects an error in target choice.
As previously reported, single cells mutant for fmi almost always targeted normally, while approximately 55% of cells mutant for Ncad displayed type 1 errors, and only a few cells displayed type 2 errors (Figure 3D, E; Chen and Clandinin, 2008; Prakash et al., 2005). Remarkably, R cells homozygous mutant for both fmi and Ncad displayed significant phenotypic enhancement, with 79% showing type1 and 13% showing type 2 errors. We obtained comparable results when we expressed Ncad RNAi in fmi MARCM clones (Figure S3). Thus, Fmi acts redundantly with Ncad in single R cells to mediate growth cone extension.
Relative, but not absolute, levels of cadherin proteins regulate extension
Next, we examined whether cadherin expression levels are important for targeting by generating single cells with two copies of Ncad and/or Fmi in a background where all other cells have only a single copy of each gene, using reverse MARCM. No appreciable targeting defects were observed when cells had two copies of either fmi (Chen and Clandinin, 2008) or Ncad alone (Figure 3B, 3.7%), relative to neighbors with only one copy. In contrast, single cells that had the wild type copy number of both genes, relative to neighbors that had only one copy of both, frequently displayed Type 1 errors (Figure 3E,F). As these mistargeting cells expressed normal levels of cadherin, these data demonstrate that relative, rather than absolute levels of cadherin expression are important for axon extension. Finally, these data, together with the fact that Fmi is neither expressed nor required in LMCs (Chen and Clandinin, 2008), argue strongly that Fmi and Ncad mediate interactions amongst R cells, rather than interactions between R cells and LMCs.
Ncad and Fmi act together to direct R cell targeting specificity
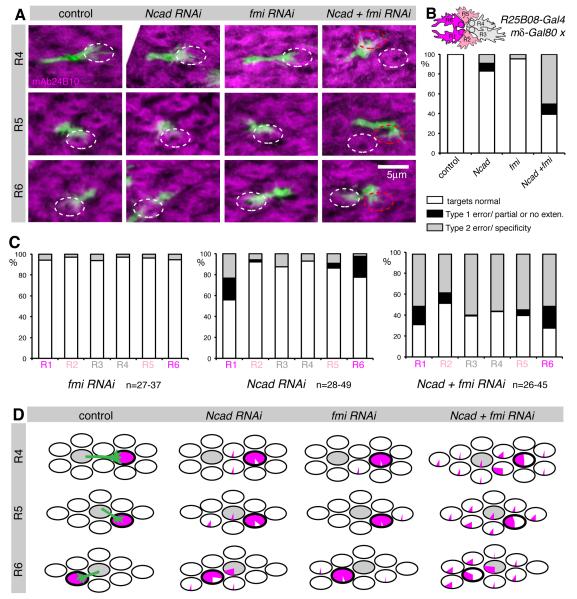
We were surprised that most Ncad and fmi double mutant growth cones did not display defects in target specificity, but rather failed to extend. Furthermore, we found only minor growth cone polarity defects in these mutant cells (Figure S4). One explanation could be that Ncad and Fmi are used in multiple growth cone interactions, such that wild-type cells can compensate for a single mutant cell in their midst. According to this hypothesis, the mutant cell is passively “molded” into the polarization pattern established by its wild-type neighbors. If this hypothesis is correct, axons should frequently extend to the wrong targets if more cells are made mutant. Indeed, large Ncad or fmi mutant clones display severe targeting phenotypes (Lee et al., 2003; Prakash et al., 2005). However, these effects are complicated by the fact that Fmi is required in R3 and R4 cells to specify ommatidial polarity, as well as in R8 to establish retinotopy (Lee et al., 2003; Senti et al., 2003; Usui et al., 1999). To circumvent these requirements, we generated an R cell subset-specific driver line that was not expressed in R3, R4 or R8, by combining R25B08-Gal4 (Pfeiffer et al., 2010) with mδ-Gal80 (Figure S4). This compound driver was specifically expressed in R1 and R6 at 28% apf, and also at lower levels in a subset of R2 and R5 cells at 0% apf. Using this driver, we expressed RNAi against Ncad and/or fmi, while stochastically labeling single growth cones with GFPmyr (Figure S4). In line with R25B08-Gal4 mδGal80 not expressing in R3, R4 and R8, planar cell polarity and topographic mapping defects were very rare when this driver was used to knockdown fmi (Figure S4, data not shown). Using this driver, knockdown of Ncad in R1 and R6 growth cones resulted in 20% of these cells displaying Type 1 extension defects, consistent with the RNAi construct moderately reducing Ncad activity (Figure 4A-C). Type 1 errors were rare in R2 and R5 (2-5%) and absent for R3 and R4, in line with the Gal4 expression pattern (Figure 4C). Similarly, knocking down fmi produced few defects in R cell target choice (Figure 4A-C). In contrast, when both Ncad and Fmi protein levels were knocked down, we observed a strong synergistic interaction, with approximately 50% of growth cones displaying Type 2 errors, innervating the wrong cartridge (Figure 4A-C). Notably, there was no significant increase in Type 1 errors, the phenotype observed when Ncad and Fmi were simultaneously removed from single R cells, likely due to the incomplete loss of both proteins (Figure 4). Moreover, despite different levels of Ncad and Fmi knockdown in different R cell subtypes, Type 2 targeting errors were equally prevalent across all R cells (Figure 4C), suggesting that reciprocal interactions between growth cones of different subtypes are required for target selection. We infer that balanced adhesion amongst many R cells via Ncad and Fmi is required for growth cone targeting specificity and both genes need to be disrupted in multiple R cells to give phenotypes that affect target specificity, rather than extension.
Figure 4. Ncad and Fmi direct targeting specificity through R cell- R cell interactions.
Ncad and fmi single or double knockdown using R25B08-Gal4 mδ-Gal80. The Gal4 line generates strong hypomorphs in R1 and R6, weaker hypomorphs in R2 and R5, while R3 and R4 are wild-type (see schemata in B). Single R cells were labeled with GFPmyr and counterstained with mAb24B10 at 40% apf. (A) Loss of both Ncad and fmi resulted in targeting defects of R4, R5 and R6. All R4 from ventral and R5 + R6 from dorsal hemisphere, confocal stacks of 4-6.5μm; white oval: wild-type target, red oval: incorrect target. (B) Quantification of targeting defects. All groups were significantly different from control with p<0.001, or p<0.05 for control vs. fmi; Fisher’s exact test, adjusted for multiple comparisons; n=190-239. Schema indicates driver expression: magenta (high), light pink (medium), grey (low-none). (C) Targeting defects distributed over different R cell types. (D) Graphical summary of R cell targeting. Each oval represents one cartridge. Pie charts display the percentage of R4-R6 cells that targeted each cartridge. The home cartridge is shown in grey; the correct target cartridge is outlined in bold. Plots for R1-R3 cells are in Figure S4.
Subset-specific reduction of Ncad and Fmi affects growth cone polarity
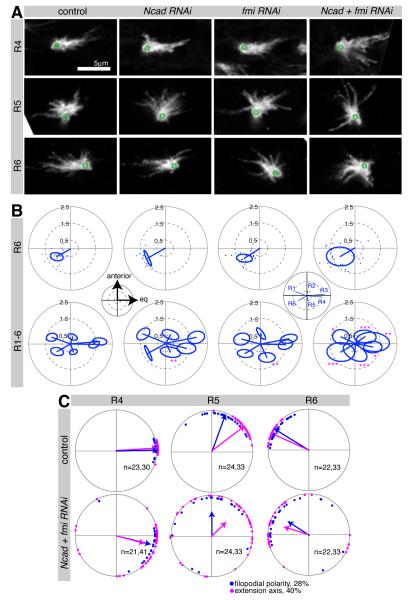
To understand the mechanistic underpinning of these target specificity defects, we examined the spatial distribution of targeting errors made when Ncad and fmi were knocked down in R1, R2, R5 and R6, and found that the majority of mistargeted axons innervated columns close to their correct target (Figure 4D, Figure S4). We next tested whether these targeting defects were preceded by changes in growth cone polarity by examining wild-type and mutant growth cones before they extended to their targets at 28% apf. We observed striking defects in growth cone morphology in all R cells, despite the fact that each had different levels of RNAi-mediated knockdown caused by the expression pattern of the driver (Figure 5A, and data not shown). We then quantified filopodial distribution, and observed significantly increased variability in polarity when both proteins were knocked down (Figure 5B), while knockdown of Ncad and fmi alone had little or no effect. Notably, these polarity defects were not associated with substantial changes in the number or length of filopodia (Figure S5). If growth cone polarization towards the correct target is a necessary prelude to targeting specificity, there should be a tight correlation between growth cone polarization and targeting in this subset-specific loss of N-cadherin and Flamingo. We found that mean growth cone polarity angles at 28% apf and extension angles at 40% apf had similar distributions, except for R2 and R5, where the difference in angles was the same as in wild type (Figures 5C and S5). Thus, our data strongly support the notion that Ncad and Fmi shape R cell targeting by directing growth cone polarization, and that the angle and degree of polarization directs the trajectory of the growth cone towards its target.
Figure 5. R cell growth cones in Ncad fmi subset knockdown have polarity defects.
Ncad and fmi single or double knockdown in an R cell subset using R25B08-Gal4 mδ-Gal80. Single R cells were labeled by GFPmyr at 28% apf. (A) R4-R6 growth cones displayed polarity defects in Ncad + fmi RNAi; ventral hemisphere of lamina, anterior up, equator to the right; green circles denote position of axon shaft. (B) Polar plots display the mean polarity vectors for each growth cone subtype, with the standard ellipse (see Extended Experimental Procedures). For R6 growth cones, polarity vector endpoints are shown as one dot per growth cone. Subset loss of Ncad and fmi resulted in a larger spread of polarity angles and thus an increased standard ellipse for the population. Insets show spatial coordinates and identity of growth cones. *p<0.05; **p<0.01; ***p<0.001 compared to control; testing for differences in the long axis of the standard ellipse using Bootstrap with Z-test and adjustment for multiple comparisons; n=17-27. (C) Polar plots of wild-type and Ncad fmi double mutants. Plots for R4-R6 with growth cone polarity angles at 28% (blue) and growth cone extension angle at 40% apf (magenta), are shown. For plots of R1-R3 see Figure S5.
Fmi, but not Ncad, is differentially expressed and localized to a specific growth cone subdomain
While both Ncad and Fmi mediate adhesive interactions between R cell growth cones, we wanted to examine whether these interactions depended on differences in either protein expression or localization. While Fmi is strongly expressed only in R cells at 28% apf (Lee et al., 2003), Ncad is expressed at higher levels in LMCs, and lower levels in R cells (Lee et al., 2001; Figure S6). As processes of both cell types were closely apposed, we could not directly distinguish Ncad protein localized in LMCs from Ncad protein within R cell growth cones. We therefore reduced Ncad protein levels using LMC-specific expression of an Ncad RNAi construct (Figure S6). In this background, R cells were stochastically labeled with myristoylated GFP and stained with Fmi and Ncad antibodies. We then determined the colocalization of the growth cone label with both Fmi and Ncad (see Experimental Procedures). Both proteins displayed discrete, partially overlapping localization (Figure S6). In addition, we found that Fmi, but not Ncad, was expressed differentially, with the highest levels in R2 and R5, intermediate levels in R1 and R6, and low levels in R3 and R4, even as growth cone sizes did not differ (Figure S6).
As R cell growth cones are only a few microns in size, these studies are insufficient to precisely define the sub-cellular localization of these proteins. We therefore used structured illumination super-resolution microscopy (SIM) to examine cadherin distributions with approximately 100nm spatial resolution, more than twice the resolution of confocal microscopy (Gustafsson, 2005; Schermelleh et al., 2008). As SIM can only be performed on thin tissue sections, precluding the use of retinal landmarks to identify R cell subtypes, we generated the intersectional driver R49A06-Gal4 mδ-Gal80 that sparsely labeled R2, R5 and R8 growth cones at 28% apf with mCD8GFP (Figure S6). Growth cones were co-stained with Fmi and SIM imaged in various orientations (see Extended Experimental Procedures). We found that, in contrast to imaging using confocal microscopy, both mCD8GFP and Fmi formed small, discrete patches at the cell membrane that rarely overlapped, suggesting that these two proteins formed distinct membrane domains (Figure 6A-D). 3D reconstructions of single growth cones show that Fmi was strongly enriched in the central domain of the growth cone, while being largely absent from filopodia (Figure 6A-D). To confirm that Fmi was predominantly present on the growth cone surface, we also stained R cell growth cones for Fmi in the absence of detergent, and found that this did not change the pattern of Fmi protein (Figure S6). Taken together, these data demonstrate that Fmi is distributed in a punctate pattern on the surface of the central domain of R cell growth cones.
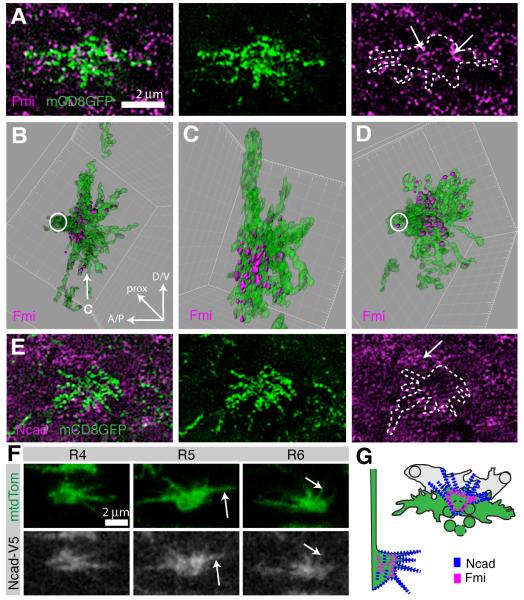
Figure 6. Fmi protein in enriched in the central domain of the growth cone, while Ncad protein is more broadly localized.
(A-E) R2 and R5 growth cones were sparsely labeled using R49A06-Gal4, mδ-Gal80 driving UAS-mCD8GFP (green) expression at 28% apf, co-stained with Fmi (magenta, A-D) or Ncad (magenta, E) and imaged using SIM. (A,E) Single optical sections of 125nm. (B,C) 3D reconstruction of the growth cone shown in (A) and Fmi contained within it (see Experimental Procedures). Fmi forms prominent patches at the base of the growth cone and is largely excluded from filopodia. (D) Second example of reconstructed growth cone. (B,D) en face view and (C) lateral view as indicated by arrow in (B). (E) Similar to Fmi, Ncad forms small patches at the membrane, but Ncad levels are low in the growth cone as compared to surrounding LMCs (arrow). (F) Single confocal sections at 28% apf, labeled with mtdTomato (green) and Ncad-V5 (white). Ncad localizes broadly within the growth cone, including filopodia (arrows). Growth cones from ventral hemisphere, equator to right. (G) Schemata of Ncad (blue) and Fmi (magenta) localization, en face view at left top and lateral view at right bottom. See also Figure S6.
We also imaged Ncad expression in wild-type animals using SIM. Single image sections show that, as with Fmi, Ncad localized to discrete patches (Figure 6E). However, as Ncad is more densely distributed in the lamina, we did not attempt to reconstruct growth cones across sections. Instead we made use of a conditional fusion of an epitope tag to the endogenous Ncad protein that allows labeling of native Ncad in single cells. In this approach, when Flipase is expressed under heat-shock control, an FRT-flanked stop cassette is excised to generate an Ncad-V5 fusion protein, as well as the LexA transcription factor, which activates expression of lexAop-mtdTomato (Pecot et al., 2013). Using this approach, we found that Ncad, in contrast to Fmi, localized broadly within the growth cone, where it occupied both the central domain and filopodia (Figure 6F). In summary, while Ncad is expressed at uniform levels and localized broadly within R cell growth cones, Fmi is highly enriched in the central domain and expressed at different levels in different R cell subtypes, consistent with the notion that Ncad and Fmi have different functions (Figure 6G).
Asymmetric increases in cadherin expression induce errors in target choice
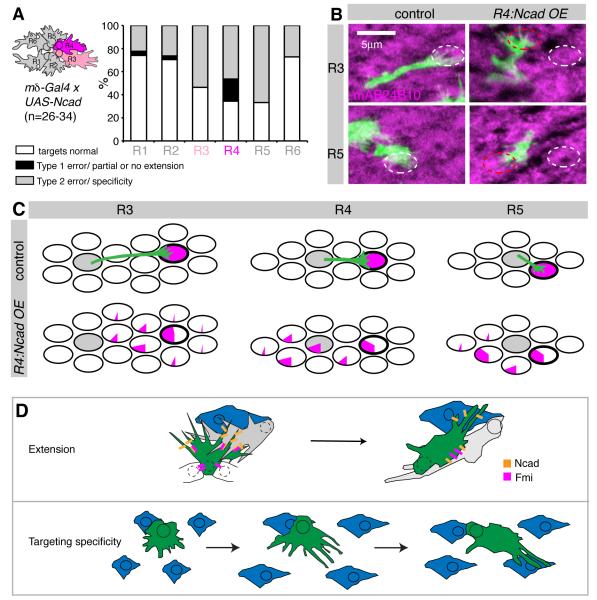
Each growth cone is closely apposed and potentially adhering to either four or five other growth cones, two from the same ommatidial bundle and either two or three from distinct, neighboring bundles (Figure 1, and data not shown). We wanted to investigate whether changing only one such interaction affects targeting and whether both interactions within and across bundles are required for target specificity. Previous data suggested that balanced interactions amongst growth cones within a bundle are necessary for targeting (Chen and Clandinin, 2008). However, targeting defects were infrequent in these studies. Thus, we decided to overexpress a cadherin using mδ-Gal4, the most specific R cell driver available, driving strong expression in R4 and weak expression in R3 (Chen and Clandinin, 2008). Fmi overexpression using mδ-Gal4 induces strong targeting defects, but also causes planar cell polarity defects, affecting targeting specificity indirectly (Chen and Clandinin, 2008). We therefore overexpressed Ncad together with mtdTomato to label R4, as well as gmr-FRT-white-FRT-GFPmyr to stochastically label single growth cones of all subtypes. Each R cell subtype forms at most one adhesive contact with R4. If interactions between neighbors within the bundle were instructive for targeting, we would expect that R4’s primary neighbors R3 and R5 to show strong targeting defects. Conversely if Ncad mediates R cell interactions between bundles, we expect that R1, R2 and R6 should show targeting defects, as these cells are contacted by R4 across bundles (Figure 1C). While R4 cells overexpressing Ncad displayed both Type 1 and Type 2 errors, other R cells almost never showed Type 1 errors but rather displayed many Type 2 errors in specificity (Figure 7A,B). Thus, elevating Ncad levels in R4 affected targeting specificity non-autonomously in other R cells, suggesting that manipulation of a single interaction by overexpression can dominantly change target selection. Furthermore, Ncad overexpression did not merely cause R cells to stick to and innervate the same targets as R4. Instead R3 and R5, for example, innervated targets close to their correct target, demonstrating that increasing adhesion with R4 broadly reduced targeting fidelity (Figure 7C). Finally, as all R cells showed Type 2 errors, this experiment revealed that both intra- and inter-bundle adhesions affect target specificity. However, cells with defective intra- but normal inter-bundles interactions (R3, R5) had twice as many targeting defects as growth cones with normal intra-, but defective inter-bundle interactions (R1, R2, R6; Figure 7A). Thus, interactions between neighboring growth cones within a bundle may be more important for regulating targeting specificity than interactions between neighbors of separate bundles.
Figure 7. Ncad is sufficient to mediate R cell interactions.
(A-C) Ncad overexpression (OE) in R4 and weaker and early in R3 (see cartoon) using mδ-Gal4, analyzed at 40% apf. (A) Quantification of targeting defects across different R cell subtypes. (B) Examples of R3 and R5 growth cones in both wild-type and R4:Ncad OE (green), counterstained with mAb24B10 to visualize cartridges. Correct targets are shown in white ovals, incorrect targets in red ovals; z-stacks of 4.5-6μm, dorsal hemisphere, equator to right. (C) Graphical summary of R cell targeting. Each oval represents one cartridge. Pie charts display the percentage of R3-R5 cells that targeted each cartridge. The home cartridge is in grey; the correct target cartridge outlined in bold. (E) Models of how cadherin-mediated adhesion controls axon extension and targeting specificity. LMC processes: blue, Ncad: orange, Fmi: magenta.
DISCUSSION
Here we demonstrate how a novel mode of afferent-afferent interactions can robustly instruct axon targeting, creating a complex, yet essentially error-free wiring diagram. We found that R cell growth cones reliably polarize to orient their filopodia towards their synaptic partners prior to interacting with them; this polarization then strongly predicts target choice. We further demonstrated that both target specificity and axon extension are critically dependent on redundant functions of two cadherins: Fmi and Ncad. Surprisingly, both processes require these molecules in different ways. Axon extension displays conventional molecular redundancy: single cells lacking both molecules polarize normally, but almost invariably fail to extend to the target. Conversely, if the activities of both molecules are moderately reduced in a broader subset of cells, most growth cones extend, but frequently polarize abnormally, and select inappropriate targets. Thus target specificity depends on the redundant functions of Ncad and Fmi across a distributed network of growth cone interactions. As Fmi is expressed at different levels in specific growth cones, and as Ncad and Fmi show distinct localization patterns within the growth cone, axons could distinguish their neighbors by measuring the relative amounts of Ncad and Fmi across their surface. These differences in cadherin activity could then polarize growth cones, orienting them to their target. Thus, redundant utilization of a small number of adhesion molecules, combined with quantitative expression differences, and subcellular localization, can direct the formation of highly complex neuronal connections with remarkable fidelity.
Specificity and Redundancy in Cadherin Function
Cadherins play central roles in directing axons toward their appropriate synaptic partners (Hirano and Takeichi, 2012). However, a surprising result has been the relative specificity of their functions, given the apparent breadth of the expression of many cadherins, such as Ncad in Drosophila (Hummel and Zipursky, 2004; Lee et al., 2001; Nern et al., 2008; Prakash et al., 2005; Zhu and Luo, 2004). As we demonstrate, some of this apparent specificity can emerge through multiple forms of redundancy. In particular, Ncad and Fmi act together and have two separable functions in R cell growth cones: axon extension and targeting specificity. Strikingly, the conventional approach to exploring redundant functions, examining the phenotypes of single cells homozygous mutant for both genes, would overlook the role of these proteins in determining growth cone polarity. It seems likely then, that redundancy both within and between families of adhesion molecules could be a prominent feature in the developing brain.
Ncad and Fmi act together to direct axon extension
While Ncad, together with Lar and Liprin-α, mediates interactions between R cells and their LMC targets that are necessary for axon extension (Choe et al, 2006, Prakash et al., 2005), Fmi specifically mediates interactions amongst R cells, and cells lacking Fmi extend normally (Chen and Clandinin, 2008). Thus, the fact that single R cells mutant for both Ncad and fmi display severe defects in axon extension argues that both LMCs, as well as neighboring R cells, act as substrates to facilitate growth cone extension through homophilic adhesion. While growth cones are extending to their target, they are passed by growth cones extending in the opposite direction toward a different target (Meinertzhagen and Hanson, 1993), suggesting that an en passant interaction promotes extension with each growth cone using the other as a substrate (Figure 7D). In addition, single growth cones with higher levels of Ncad and Fmi relative to their neighbors often fail to extend to their target, suggesting that growth cones also need to reduce adhesions with some of their neighbors in order to extend. Similarly, both reduced and increased levels of focal adhesions inhibit growth cone advance in vitro (Myers and Gomez, 2011; Woo et al., 2009). Thus, growth cone extension occurs through progressive shifts in cell-surface interactions, first amongst R cell growth cones, and then between R cells and their target LMCs, with site-specific loss and gain of adhesion.
Cadherin-mediated growth cone polarization shapes R cell target selection
Previous work demonstrated that interactions amongst afferents play a central role in directing R cell axons towards appropriate post-synaptic targets, but the mechanisms by which they might do so were unknown (Chen and Clandinin, 2008; Clandinin and Zipursky, 2000; Lee et al., 2003). Here we show that afferent interactions mediated by Ncad and Fmi polarize the formation or stabilization of filopodia on each growth cone, such that filopodia become preferentially aligned towards their target (Figure 7D). In wild-type animals, this polarization emerges before R cell axons contact their targets, and tightly correlates with the angle of growth cone extension. Moreover, we observed that growth cones still polarized in Ncad and fmi subset-specific mutants, but that the angle of polarization was less precise across the population of each R cell type. Importantly, even under these conditions, the distribution of growth cone polarization angles closely matched the distribution of targets chosen. Taken together, these correlative studies argue strongly that growth cone polarization is a critical step in determining target specificity. These data shed new light on previous studies that demonstrated that the behavior of retinal ganglion cell growth cones correlates with their shape and polarity, such that highly polarized growth cones advance rapidly, while complex, un-polarized growth cones are often stationary and occupy choice points (Godement et al., 1994; Mason and Wang, 1997). Similarly, we found that R cell growth cones early in development are stationary and relatively un-polarized; once they become sufficiently polarized so as to choose one target over another, they extend. Thus growth cone polarization is likely a critical, predictive step in target selection in many systems.
How do Ncad and Fmi determine targeting specificity?
Previous results demonstrated that growth cones compare levels of Fmi between their neighbors, and it was proposed that differential adhesion might instruct axon targeting (Chen and Clandinin, 2008). Indeed, we found that different growth cone subtypes expressed different levels of Fmi, and that Fmi localization is restricted to the central domain of the growth cone. To our knowledge, this is the first demonstration that a cadherin can be localized to a growth cone specific compartment, creating new possibilities for diversifying adhesive interactions. Each growth cone forms contacts with either four or five of its neighbors. For example, within the bundle, R4 adheres to one neighbor with low (R3) and one neighbor with high (R5) Fmi levels. Furthermore, R4 forms contacts across bundles with one growth cone with high Fmi (R2) and two growth cones with lower Fmi (R1, R6). Analogous patterns are evident for all other growth cones. Thus, based on levels of Fmi protein, adhesive strength might vary systematically across the surface of each growth cone, depending on the identity of each neighbor, resulting in a regular mosaic tiling of all growth cones.
Ncad is localized broadly within the growth cone, and is not necessary to mediate interactions between R cells within the bundle. This suggests that Ncad might primarily mediate adhesive interactions between growth cones across bundles, in addition to mediating adhesive interactions between R cells and LMCs. However, when overexpressed in R4, Ncad can dominantly alter both inter- and intra-bundle interactions. Notably, R cells neighboring R4, such as R5, do not merely stick to R4 to innervate a common target. Instead, the abnormal adhesive contact with R4 likely alters the balance of adhesive contacts each R cell makes with its other neighbors. Furthermore, intra-bundle interactions are more sensitive to cadherin levels and more crucial for target specificity. This is in line with previous observations that R cells in bundles that are flipped by 180° in certain mutants, or R cells that project across the equator in wild-type almost invariably target normally, despite sitting adjacent to a different set of growth cones from neighboring bundles (Clandinin and Zipursky, 2000, Horridge and Meinertzhagen, 1970). In aggregate, our results suggest that growth cones form distinct adhesive contacts with their neighbors, and that the relative adhesiveness of each of these contacts is integrated to determine growth cone polarization and ultimately target specificity.
A distributed network of adhesive interactions directs target selection
Here we propose a new model of how neuronal targeting can be achieved through a series of adhesive interactions that orient growth cones within a sheet. Each growth cone engages in local, cadherin-mediated interactions with multiple neighbors, effectively constructing a large network of interconnected cells. We hypothesize that growth cone polarity evolves as a result of balancing the adhesive forces generated by contact with neighboring growth cones of different adhesivity. Analogous adhesion networks have been described in various epithelia, where all cells are equally adhesive, and equilibrium is reached when most cells have acquired a hexagonal shape (Classen et al., 2005; Lecuit and Lenne, 2007). By contrast, R cell growth cones are more complex in shape, they form domains containing different levels of different cadherins and they are spatially constrained by their association with an axon bundle. As a result, the adhesive forces acting on growth cones change their shapes, and orientations within the lamina as a function of R cell subtype. As many interactions act in concert to orient each growth cone, the system becomes highly robust. This can be observed experimentally when single cells within the network are made mutant for critical adhesion factors, yet orient correctly towards their targets. Furthermore, this redundant network of interactions achieves extremely high targeting fidelity in wild-type animals. Analogous targeting strategies could be widely used, as many sensory systems, both in insects and vertebrates, are organized into topographic maps, whose development often relies on afferent-afferent interactions (Imai et al., 2009; Millard et al., 2007; Millard et al., 2010; Ting et al., 2005). Thus, quantitative differences in relative adhesion, and distributed networks of adhesive interactions are likely to be central in patterning many systems in the brain.
EXPERIMENTAL PROCEDURES
Fly Stocks
A detailed description of all fly stocks, crosses, transgenes and imaging studies used can be found in the Extended Experimental Procedures, and in Supplemental Tables 1 and 2.
Generation of Transgenic lines
Transgenic flies carrying mδ-Gal80, the fmi Bac rescue construct and UAS-myrEGFP were generated using standard procedures. For details, please refer to Extended Experimental Procedures.
Immunohistochemistry and Confocal Imaging
Immunohistochemistry was performed using standard protocols. For details, see Extended Experimental Procedures. Images were acquired on a Leica TCS SP2 AOBS confocal microscope, using a 100x N.A. 1.4 lens and were rendered and analyzed using Bitplane Imaris and Image J. Figures were prepared using Adobe Photoshop and Illustrator. Statistics were calculated using Graph Pad Prism and Matlab.
Cryosectioning, SIM imaging and Image Analysis
See Extended Experimental Procedures for details.
Supplementary Material
HIGHLIGHTS.
- Cadherins determine targeting specificity by polarizing growth cones
- Cadherins are targeted to specific growth cone subdomains
- Cellular and molecular redundancies achieve targeting with high fidelity
- A complex interaction network polarizes growth cones towards their targets
ACKNOWLEDGEMENTS
We thank P. Garrity, L. Luo, G. Rubin, I. Salecker, G. Struhl, L. Zipursky, the Bloomington and San Diego Stock Centers, as well as the Developmental Studies Hybridoma Bank, BacPac Resources and the Vienna Drosophila RNAi Center for fly strains, antibodies and plasmids. We are grateful to D. Clark for help with quantification and statistics, to M. Silies for help with fly husbandry, and also to K. Shen, A. Huberman, and to members of the Clandinin lab for helpful comments on the manuscript. This work was supported by a postdoctoral fellowship from Walter V. and Idun Berry (T.S.), by a travel fellowship of the Heinrich Hertz-Stiftung (H.N.), and by funding from the National Eye Institute, R01 EY015231 (T.R.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION Supplemental Information that includes Extended Experimental Procedures, six Figures, and two tables can be found with this article online at ….
REFERENCES
- Bard L, Boscher C, Lambert M, Mege RM, Choquet D, Thoumine O. A molecular clutch between the actin flow and N-cadherin adhesions drives growth cone migration. J Neurosci. 2008;28:5879–5890. doi: 10.1523/JNEUROSCI.5331-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D, Toroian-Raymond A. Disoriented pathfinding by pioneer neurone growth cones deprived of filopodia by cytochalasin treatment. Nature. 1986;323:712–715. doi: 10.1038/323712a0. [DOI] [PubMed] [Google Scholar]
- Chen PL, Clandinin TR. The cadherin Flamingo mediates level-dependent interactions that guide photoreceptor target choice in Drosophila. Neuron. 2008;58:26–33. doi: 10.1016/j.neuron.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CB, Rosenthal DE, Harris WA, Holt CE. Navigational errors made by growth cones without filopodia in the embryonic Xenopus brain. Neuron. 1993;11:237–251. doi: 10.1016/0896-6273(93)90181-p. [DOI] [PubMed] [Google Scholar]
- Choe KM, Prakash S, Bright A, Clandinin TR. Liprin-alpha is required for photoreceptor target selection in Drosophila. Proc Natl Acad Sci U S A. 2006;103:11601–11606. doi: 10.1073/pnas.0601185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin TR, Zipursky SL. Afferent growth cone interactions control synaptic specificity in the Drosophila visual system. Neuron. 2000;28:427–436. doi: 10.1016/s0896-6273(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Classen AK, Anderson KI, Marois E, Eaton S. Hexagonal packing of Drosophila wing epithelial cells by the planar cell polarity pathway. Dev Cell. 2005;9:805–817. doi: 10.1016/j.devcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Dent EW, Gupton SL, Gertler FB. The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S, Gordon-Weeks PR. Cytoskeletal dynamics in growth-cone steering. J Cell Sci. 2009;122:3595–3604. doi: 10.1242/jcs.042309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Mege RM, Thoumine O. Multi-level molecular clutches in motile cell processes. Trends Cell Biol. 2009;19:475–486. doi: 10.1016/j.tcb.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Godement P, Wang LC, Mason CA. Retinal axon divergence in the optic chiasm: dynamics of growth cone behavior at the midline. J Neurosci. 1994;14:7024–7039. doi: 10.1523/JNEUROSCI.14-11-07024.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MG. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci U S A. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjieconomou D, Timofeev K, Salecker I. A step-by-step guide to visual circuit assembly in Drosophila. Curr Opin Neurobiol. 2011;21:76–84. doi: 10.1016/j.conb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Hakeda-Suzuki S, Berger-Muller S, Tomasi T, Usui T, Horiuchi SY, Uemura T, Suzuki T. Golden Goal collaborates with Flamingo in conferring synaptic-layer specificity in the visual system. Nat Neurosci. 2011;14:314–323. doi: 10.1038/nn.2756. [DOI] [PubMed] [Google Scholar]
- Hiesinger PR, Zhai RG, Zhou Y, Koh TW, Mehta SQ, Schulze KL, Cao Y, Verstreken P, Clandinin TR, Fischbach KF, et al. Activity-independent prespecification of synaptic partners in the visual map of Drosophila. Curr Biol. 2006;16:1835–1843. doi: 10.1016/j.cub.2006.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Takeichi M. Cadherins in brain morphogenesis and wiring. Physiol Rev. 2012;92:597–634. doi: 10.1152/physrev.00014.2011. [DOI] [PubMed] [Google Scholar]
- Horridge GA, Meinertzhagen IA. The accuracy of the patterns of connexions of the first- and second-order neurons of the visual system of Calliphora. Proc R Soc Lond B Biol Sci. 1970;175:69–82. doi: 10.1098/rspb.1970.0012. [DOI] [PubMed] [Google Scholar]
- Hummel T, Zipursky SL. Afferent induction of olfactory glomeruli requires N-cadherin. Neuron. 2004;42:77–88. doi: 10.1016/s0896-6273(04)00158-8. [DOI] [PubMed] [Google Scholar]
- Imai T, Yamazaki T, Kobayakawa R, Kobayakawa K, Abe T, Suzuki M, Sakano H. Pre-target axon sorting establishes the neural map topography. Science. 2009;325:585–590. doi: 10.1126/science.1173596. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Lenne PF. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol. 2007;8:633–644. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- Lee CH, Herman T, Clandinin TR, Lee R, Zipursky SL. N-cadherin regulates target specificity in the Drosophila visual system. Neuron. 2001;30:437–450. doi: 10.1016/s0896-6273(01)00291-4. [DOI] [PubMed] [Google Scholar]
- Lee RC, Clandinin TR, Lee CH, Chen PL, Meinertzhagen IA, Zipursky SL. The protocadherin Flamingo is required for axon target selection in the Drosophila visual system. Nat Neurosci. 2003;6:557–563. doi: 10.1038/nn1063. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Luo L, Flanagan JG. Development of continuous and discrete neural maps. Neuron. 2007;56:284–300. doi: 10.1016/j.neuron.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Mason CA, Wang LC. Growth cone form is behavior-specific and, consequently, position-specific along the retinal axon pathway. J Neurosci. 1997;17:1086–1100. doi: 10.1523/JNEUROSCI.17-03-01086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinertzhagen IA, Hanson TE. In: The Development of the optic lobe. Bate M, Martinez-Arias A, editors. Cold Spring Harbor Laboratory Press; 1993. pp. 1363–1491. [Google Scholar]
- Meinertzhagen IA, O’Neil SD. Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J Comp Neurol. 1991;305:232–263. doi: 10.1002/cne.903050206. [DOI] [PubMed] [Google Scholar]
- Millard SS, Flanagan JJ, Pappu KS, Wu W, Zipursky SL. Dscam2 mediates axonal tiling in the Drosophila visual system. Nature. 2007;447:720–724. doi: 10.1038/nature05855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard SS, Lu Z, Zipursky SL, Meinertzhagen IA. Drosophila dscam proteins regulate postsynaptic specificity at multiple-contact synapses. Neuron. 2010;67:761–768. doi: 10.1016/j.neuron.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Cytoskeletal dynamics and nerve growth. Neuron. 1988;1:761–772. doi: 10.1016/0896-6273(88)90124-9. [DOI] [PubMed] [Google Scholar]
- Myers JP, Gomez TM. Focal adhesion kinase promotes integrin adhesion dynamics necessary for chemotropic turning of nerve growth cones. J Neurosci. 2011;31:13585–13595. doi: 10.1523/JNEUROSCI.2381-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Zhu Y, Zipursky SL. Local N-cadherin interactions mediate distinct steps in the targeting of lamina neurons. Neuron. 2008;58:34–41. doi: 10.1016/j.neuron.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot MY, Tadros W, Nern A, Bader M, Chen Y, Zipursky SL. Multiple interactions control synaptic layer specificity in the Drosophila visual system. Neuron. 2013;77:299–310. doi: 10.1016/j.neuron.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Caldwell JC, Eberl DF, Clandinin TR. Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nat Neurosci. 2005;8:443–450. doi: 10.1038/nn1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, McLendon HM, Dubreuil CI, Ghose A, Hwa J, Dennehy KA, Tomalty KM, Clark KL, Van Vactor D, Clandinin TR. Complex interactions amongst N-cadherin, DLAR, and Liprin-alpha regulate Drosophila photoreceptor axon targeting. Dev Biol. 2009;336:10–19. doi: 10.1016/j.ydbio.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper JA, Bastiani MJ, Goodman CS. Guidance of neuronal growth cones: selective fasciculation in the grasshopper embryo. Cold Spring Harb Symp Quant Biol. 1983;48(Pt 2):587–598. doi: 10.1101/sqb.1983.048.01.063. [DOI] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Usui T, Boucke K, Greber U, Uemura T, Dickson BJ. Flamingo regulates R8 axon-axon and axon-target interactions in the Drosophila visual system. Curr Biol. 2003;13:828–832. doi: 10.1016/s0960-9822(03)00291-4. [DOI] [PubMed] [Google Scholar]
- Thoumine O, Lambert M, Mege RM, Choquet D. Regulation of N-cadherin dynamics at neuronal contacts by ligand binding and cytoskeletal coupling. Mol Biol Cell. 2006;17:862–875. doi: 10.1091/mbc.E05-04-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CY, Yonekura S, Chung P, Hsu SN, Robertson HM, Chiba A, Lee CH. Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development. 2005;132:953–963. doi: 10.1242/dev.01661. [DOI] [PubMed] [Google Scholar]
- Tomasi T, Hakeda-Suzuki S, Ohler S, Schleiffer A, Suzuki T. The transmembrane protein Golden goal regulates R8 photoreceptor axon-axon and axon-target interactions. Neuron. 2008;57:691–704. doi: 10.1016/j.neuron.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL, Takeichi M, Uemura T. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell. 1999;98:585–595. doi: 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- Vitriol EA, Zheng JQ. Growth cone travel in space and time: the cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron. 2012;73:1068–1081. doi: 10.1016/j.neuron.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S, Rowan DJ, Gomez TM. Retinotopic mapping requires focal adhesion kinase-mediated regulation of growth cone adhesion. J Neurosci. 2009;29:13981–13991. doi: 10.1523/JNEUROSCI.4028-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Luo L. Diverse functions of N-cadherin in dendritic and axonal terminal arborization of olfactory projection neurons. Neuron. 2004;42:63–75. doi: 10.1016/s0896-6273(04)00142-4. [DOI] [PubMed] [Google Scholar]
- Zinsmaier KE, Hofbauer A, Heimbeck G, Pflugfelder GO, Buchner S, Buchner E. A cysteine-string protein is expressed in retina and brain of Drosophila. J Neurogenet. 1990;7:15–29. doi: 10.3109/01677069009084150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.