Abstract
Three invasive Streptococcus pneumoniae strains nonsusceptible to linezolid were isolated in the United States between 2001 and 2012 from the CDC's Active Bacterial Core surveillance. Linezolid binds ribosomal proteins where structural changes within its target site may confer resistance. Our study identified mutations and deletions near the linezolid binding pocket of two of these strains within the rplD gene, which encodes ribosomal protein L4. Mutations in the 23S rRNA alleles or the rplV gene were not detected.
TEXT
Linezolid was the first oxazolidinone to be licensed in the United States (in 2000) and marketed worldwide (1–3). Linezolid is approved by the U.S. Food and Drug Administration (FDA) for the treatment of complicated skin infections, meningitis, nosocomial pneumonia, endocarditis, sepsis, osteomyelitis, concurrent bacteremia, and bacteremia associated with community-acquired pneumonia (1, 2).
Linezolid blocks the assembly of a functional initiation complex for protein synthesis, thereby preventing mRNA translation. Other antibiotics that prevent mRNA translation include chloramphenicol, tetracycline, macrolides, and lincosamides. They allow the formation of an initiation complex but inhibit subsequent peptide elongation (3, 4).
The LEADER (Linezolid Experience and Accurate Determination of Resistance) program, which monitors linezolid-resistant clinical isolates, reports that, in the United States, linezolid-sensitive Streptococcus pneumoniae isolates have an MIC90 of 1 μg/ml (5–9). Therefore, S. pneumoniae clinical strains with linezolid MICs of >1 μg/ml should be monitored and investigated for potential mechanisms of resistance. This is consistent with the Clinical and Laboratory Standards Institute (CLSI) breakpoint of 2 μg/ml (10).
The mechanisms of resistance to linezolid that have been described to date include target modification and use of a mobile cfr element (2, 8, 11). The linezolid target (the 50S subunit) is composed of 5S and 23S rRNAs and 36 riboproteins (L1 through L36). Linezolid-resistant strains present mutations in one or more alleles of the 23S rRNA gene, decreasing the affinity of ribosomes for the drug (12). A clear correlation between the number of 23S rRNA alleles mutated and increased linezolid resistance has been demonstrated (13, 14). The most frequently reported mutation in linezolid-resistant clinical isolates of staphylococci and enterococci occur by G-to-U substitution in the peptidyl transferase center of 23S rRNA at position 2576 (2, 8). Additional mutations within the same 23S rRNA gene have also been described (e.g., A2059G, C2190T, and G2447T) (15–17).
The cfr mobile element includes the cfr gene, which encodes a methyltransferase that methylates the 23S rRNA at position A2503. This affects binding of linezolid to the 50S subunit (11, 18, 19). While carried by Staphylococcus aureus strains (20, 21) and recently described in Streptococcus suis (22), this mobile element has not been described in S. pneumoniae.
Only a few S. pneumoniae strains with reduced susceptibilities to linezolid have been isolated from disease cases (16, 23). For these strains, it was suggested that mutations in 23S rRNA genes and those encoding ribosomal proteins L4 and L22 confer linezolid resistance (16). However, direct evidence demonstrating deletions within the rplD gene of S. pneumoniae strain TN33388, encoding ribosomal protein L4, which is linked to reduced susceptibility to linezolid, was published by Wolter et al. (23). Strain TN33388 was identified through the Active Bacterial Core surveillance (ABCs), part of the Centers for Disease Control and Prevention's (CDC's) Emerging Infections Program.
In this study, the CDC Streptococcus laboratory identified two other additional S. pneumoniae strains (7828-04 and 2008227074) with reduced susceptibilities to linezolid. Overall, 3 of 45,099 pneumococci tested (<1%) were isolated from invasive disease in the United States between 2001 and 2012 through the ABCs, and they showed reduced susceptibilities to linezolid (Table 1). Mutations within demonstrated linezolid targets were investigated in these two isolates.
TABLE 1.
Phenotypic findings of S. pneumoniae strains with reduced susceptibilities to linezolid
| Strain (yr of isolation, state) | Serotype | L4 phenotype | MIC (μg/ml)a |
Reference or source | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LZD | VAN | PEN | AMO | ERY | CHL | CLI | TET | ||||
| 0566-02 (2001, GA) | 19A | S20N | 0.25 | 0.25 | 0.06 | 0.06 | 0.12 | ≤2 | 0.12 | ≤2 | This study |
| 3084-03 (2002, GA) | 19A | S20N | 1 | 0.25 | 0.12 | 0.06 | 0.25 | ≤2 | 0.5 | ≤2 | This study |
| 7828-04 (2004, CT) | 014 | ΔW65R66b | 4 | 0.5 | 2 | 2 | 2 | 8 | 0.12 | ≤2 | This study |
| 2008227074 (2007, NM) | 09N | Q67R, R72G | 4 | 0.25 | ≤0.03 | ≤0.03 | 1 | >8 | 0.06 | ≤2 | This study |
| TN33388 (2003, TN) | 33F | ΔK68G69 | 4 | 0.25 | ≤0.03 | ≤0.03 | 1 | 8 | 0.25 | ≤2 | 22 |
LZD, linezolid; VAN, vancomycin; PEN, penicillin; AMO, amoxicillin; ERY, erythromycin; CHL, chloramphenicol; CLI, clindamycin; TET, tetracycline. Current CLSI breakpoints: LZD susceptible (S), ≤2 μg/ml; VAN S, ≤1 μg/ml; PEN S, ≤2 μg/ml, PEN resistance (R), ≥8 μg/ml; AMO S, ≤2 μg/ml, and AMO R, ≥8 μg/ml; ERY S, ≤0.25 μg/ml, and ERY R, ≥1 μg/ml; CHL S, ≤4 μg/ml, and CHL R, ≥8 μg/ml; CLI S, ≤0.25 μg/ml, and CLI R, ≥1 μg/ml; and TET S, ≤1 μg/ml, and TET R, ≥4 μg/ml (10).
Δ, Deletion.
Strain TN33388 from the CDC (for whom its mechanism of resistance to linezolid had been investigated), two serotype 19A linezolid-susceptible strains, and the reference S. pneumoniae strain R6 were utilized as controls (23). The MICs for linezolid, vancomycin, penicillin, amoxicillin, erythromycin, chloramphenicol, clindamycin, and tetracycline were determined using the broth microdilution methodology according to the CLSI (24). The linezolid-susceptible strains shown in Table 1 had linezolid MICs of 0.25 or 1 μg/ml, whereas linezolid-nonsusceptible strains had MICs of 4 μg/ml. The strains were susceptible to penicillin, vancomycin, amoxicillin, and tetracycline. Except for one strain (3084-03), they were also susceptible to clindamycin. Linezolid-nonsusceptible strains were resistant to chloramphenicol and erythromycin (Table 1).
To investigate the molecular mechanism of reduced susceptibility to linezolid, we amplified, purified, and sequenced the rplD gene (encoding the ribosomal protein L4), the rplV gene (encoding the ribosomal protein L22), and all four 23S rRNA alleles. The presence of the cfr gene in these linezolid-nonsusceptible strains was also sought.
For DNA extraction, S. pneumoniae strains were cultured on Trypticase soy agar (TSA) supplemented with 5% sheep blood and incubated overnight at 37°C in 5% CO2. Chromosomal DNA was then extracted by using the QIAamp DNA minikit (Qiagen, Inc., Valencia, CA). An aliquot (100 ng) was used as the template in PCR mixtures containing 1× Platinum Taq DNA polymerase high fidelity (Life Technologies, Carlsbad, CA) and the primers L4F (AAATCAGCAGTTAAAGCTGG) and L4R (GAGCTTTCAGTGATGACAGG) for the rplD gene and primers L22F (GCAGACGACAAGAAAACACG) and L22R (ATTGGATGTACTTTTTGACC) to amplify rplV (23). Each of the four copies of the 23S rRNA alleles carried by the pneumococcus was amplified by using a previously published method (25). Briefly, the genes were initially amplified as overlapping contigs. Then, nested PCRs utilizing unique primers downstream of each 23S rRNA allowed the amplification of the peptidyl transferase region from each allele. The presence of the cfr gene was investigated with primers and conditions described previously (26–28).
PCR products were purified using a QIAquick PCR purification kit (Qiagen, Inc., Valencia, CA), and the concentrations were quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). Genes were sequenced at Eurofins MWG Operon (Huntsville, AL). Genes from strain R6 (29) were also sequenced and analyzed for comparison. Sequences were analyzed utilizing the software DNASTAR Lasergene 10 and aligned against the nucleotide sequence of a linezolid-susceptible S. pneumoniae R6 reference strain (GenBank accession number AE007317) using BLAST (23, 29).
Our sequence analysis found that, when compared to those genes carried by wild-type strain R6, linezolid-nonsusceptible strains had mutations and deletions within only the rplD gene (Table 1 and Fig. 1). Strain 2008227074 contained two mutations leading to the amino acid substitutions Q67R and R72G. These two mutations had not been described before in linezolid-nonsusceptible S. pneumoniae strains. Strain 7828-04 presented a 6-bp deletion (ΔW65R66) that was similar, but not identical, to that previously identified in strain TN33388 (23). The two linezolid-susceptible strains had a substitution (S20N) which was caused by a single-nucleotide change in position 59 (G59A) of the nucleotide sequence. S20N is apparently out of the linezolid binding pocket within L4 and has been reported in fully susceptible pneumococcal strains and in isolates resistant to macrolides (29).
FIG 1.
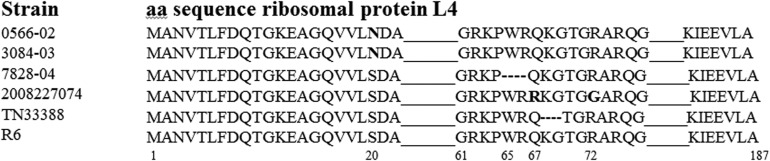
Alignment of ribosomal protein L4 of linezolid-nonsusceptible isolates of S. pneumoniae and linezolid-susceptible strains. Mutations are shown in bold type. Dashes represent deletions, and identical sequences, in comparison to strain R6, are indicated by a straight line. Numbers underneath the specific amino acids (aa) represent the position in the R6 sequence.
The sequences of the rplV gene of the linezolid-nonsusceptible and linezolid-susceptible strains were identical to that of strain R6. In contrast to the mechanism of linezolid resistance commonly found in staphylococcal isolates, these S. pneumoniae strains did not have mutations in any of the four copies of the 23S rRNA alleles. Moreover, the cfr gene could not be identified in any of these S. pneumoniae strains.
In conclusion, the 2 clinical isolates of S. pneumoniae with reduced susceptibilities to linezolid described in this study over a 12-year period have mutations only in the rplD gene, leading to changes in the amino acid sequence of the L4 protein. Part of ribosomal protein L4 is placed relatively close to the linezolid binding site on the ribosomes, suggesting that the mechanism for reduced susceptibility may include structural perturbation of the linezolid binding site.
Recently, mutations in 23S rRNA genes have been described in an in vitro-generated linezolid-resistant S. pneumoniae strain with an MIC of 32 μg/ml (30), which suggests another potential mechanism for resistance. However, these mutations have not been detected to date in clinical pneumococcal strains. Prudent use of linezolid in the United States may account for the low mutation rates in its target and therefore the continued activity against S. pneumoniae strains. Similarly, a global study that utilized strains (n = 636) isolated in 22 different countries showed susceptibility to linezolid in all S. pneumoniae strains (31). Despite many years of exposure to the drug, the very low rate of linezolid resistance in pneumococci suggests that the fitness cost of resistance (32) may be suppressing the successful dissemination of these strains in the pneumococcus.
ACKNOWLEDGMENTS
We are grateful for the contributions of members of the Centers for Disease Control and Prevention Active Bacterial Core surveillance/Emerging Infections Program Network. We also thank Magderie Klugman and Herbert P. Ludewick, Emory University, for their valuable assistance.
Footnotes
Published ahead of print 3 February 2014
REFERENCES
- 1.Vinh DC, Rubinstein E. 2009. Linezolid: a review of safety and tolerability. J. Infect. 59(Suppl 1):S59–S74. 10.1016/S0163-4453(09)60009-8 [DOI] [PubMed] [Google Scholar]
- 2.Bozdogan B, Appelbaum PC. 2004. Oxazolidinones: activity, mode of action, and mechanism of resistance. Int. J. Antimicrob. Agents 23:113–119. 10.1016/j.ijantimicag.2003.11.003 [DOI] [PubMed] [Google Scholar]
- 3.Livermore DM. 2003. Linezolid in vitro: mechanism and antibacterial spectrum. J. Antimicrob. Chemother. 51(Suppl 2):ii9–ii16. 10.1093/jac/dkg249 [DOI] [PubMed] [Google Scholar]
- 4.Swaney SM, Aoki H, Ganoza MC, Shinabarger DL. 1998. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob. Agents Chemother. 42:3251–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Draghi DC, Sheehan DJ, Hogan P, Sahm DF. 2005. In vitro activity of linezolid against key Gram-positive organisms isolated in the United States: results of the LEADER 2004 Surveillance Program. Antimicrob. Agents Chemother. 49:5024–5032. 10.1128/AAC.49.12.5024-5032.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell DJ, Mendes RE, Ross JE, Jones RN. 2009. Linezolid surveillance program results for 2008 (LEADER Program for 2008). Diagn. Microbiol. Infect. Dis. 65:392–403. 10.1016/j.diagmicrobio.2009.10.011 [DOI] [PubMed] [Google Scholar]
- 7.Jones RN, Fritsche TR, Sader HS, Ross JE. 2007. LEADER Surveillance Program results for 2006: an activity and spectrum analysis of linezolid using clinical isolates from the United States (50 medical centers). Diagn. Microbiol. Infect. Dis. 59:309–317. 10.1016/j.diagmicrobio.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 8.Jones RN, Ross JE, Castanheira M, Mendes RE. 2008. United States resistance surveillance results for linezolid (LEADER Program for 2007). Diagn. Microbiol. Infect. Dis. 62:416–426. 10.1016/j.diagmicrobio.2008.10.010 [DOI] [PubMed] [Google Scholar]
- 9.Flamm RK, Mendes RE, Ross JE, Sader HS, Jones RN. 2013. Linezolid surveillance results for the United States: LEADER Surveillance Program 2011. Antimicrob. Agents Chemother. 57:1077–1081. 10.1128/AAC.02112-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing—approved standard M100-S23, 23rd ed. CLSI, Wayne, PA [Google Scholar]
- 11.Mendes RE, Deshpande LM, Castanheira M, DiPersio J, Saubolle MA, Jones RN. 2008. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob. Agents Chemother. 52:2244–2246. 10.1128/AAC.00231-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long KS, Vester B. 2012. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 56:603–612. 10.1128/AAC.05702-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall SH, Donskey CJ, Hutton-Thomas R, Salata RA, Rice LB. 2002. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 46:3334–3336. 10.1128/AAC.46.10.3334-3336.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Besier S, Ludwig A, Zander J, Brade V, Wichelhaus TA. 2008. Linezolid resistance in Staphylococcus aureus: gene dosage effect, stability, fitness costs, and cross-resistances. Antimicrob. Agents Chemother. 52:1570–1572. 10.1128/AAC.01098-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorlozano A, Gutierrez J, Martinez T, Yuste ME, Perez-Lopez JA, Vindel A, Guillen J, Boquete T. 2010. Detection of new mutations conferring resistance to linezolid in glycopeptide-intermediate susceptibility Staphylococcus hominis subspecies hominis circulating in an intensive care unit. Eur. J. Clin. Microbiol. Infect. Dis. 29:73–80. 10.1007/s10096-009-0823-4 [DOI] [PubMed] [Google Scholar]
- 16.Farrell DJ, Morrissey I, Bakker S, Buckridge S, Felmingham D. 2004. In vitro activities of telithromycin, linezolid, and quinupristin-dalfopristin against Streptococcus pneumoniae with macrolide resistance due to ribosomal mutations. Antimicrob. Agents Chemother. 48:3169–3171. 10.1128/AAC.48.8.3169-3171.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong A, Reddy SP, Smyth DS, Aguero-Rosenfeld ME, Sakoulas G, Robinson DA. 2010. Polyphyletic emergence of linezolid-resistant staphylococci in the United States. Antimicrob. Agents Chemother. 54:742–748. 10.1128/AAC.00621-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57:1064–1073. 10.1111/j.1365-2958.2005.04754.x [DOI] [PubMed] [Google Scholar]
- 19.Toh SM, Xiong L, Arias CA, Villegas MV, Lolans K, Quinn J, Mankin AS. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506–1514. 10.1111/j.1365-2958.2007.05744.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales G, Picazo JJ, Baos E, Candel FJ, Arribi A, Pelaez B, Andrade R, de la Torre MA, Fereres J, Sanchez-Garcia M. 2010. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin. Infect. Dis. 50:821–825. 10.1086/650574 [DOI] [PubMed] [Google Scholar]
- 21.Kehrenberg C, Aarestrup FM, Schwarz S. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51:483–487. 10.1128/AAC.01340-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Li D, Song L, Liu Y, He T, Liu H, Wu C, Schwarz S, Shen J. 2013. First report of the multiresistance gene cfr in Streptococcus suis. Antimicrob. Agents Chemother. 57:4061–4063. 10.1128/AAC.00713-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolter N, Smith AM, Farrell DJ, Schaffner W, Moore M, Whitney CG, Jorgensen JH, Klugman KP. 2005. Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the pneumococcus. Antimicrob. Agents Chemother. 49:3554–3557. 10.1128/AAC.49.8.3554-3557.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically—approved standard M7-A8, 8th ed. CLSI, Wayne, PA [Google Scholar]
- 25.Tait-Kamradt A, Davies T, Cronan M, Jacobs MR, Appelbaum PC, Sutcliffe J. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118–2125. 10.1128/AAC.44.8.2118-2125.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendes RE, Deshpande LM, Farrell DJ, Spanu T, Fadda G, Jones RN. 2010. Assessment of linezolid resistance mechanisms among Staphylococcus epidermidis causing bacteraemia in Rome, Italy. J. Antimicrob. Chemother. 65:2329–2335. 10.1093/jac/dkq331 [DOI] [PubMed] [Google Scholar]
- 27.Miller K, Dunsmore CJ, Fishwick CW, Chopra I. 2008. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob. Agents Chemother. 52:1737–1742. 10.1128/AAC.01015-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156–1163. 10.1128/AAC.50.4.1156-1163.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoskins J, Alborn WE, Jr, Arnold J, Blaszczak LC, Burgett S, DeHoff BS, Estrem ST, Fritz L, Fu DJ, Fuller W, Geringer C, Gilmour R, Glass JS, Khoja H, Kraft AR, Lagace RE, LeBlanc DJ, Lee LN, Lefkowitz EJ, Lu J, Matsushima P, McAhren SM, McHenney M, McLeaster K, Mundy CW, Nicas TI, Norris FH, O'Gara M, Peery RB, Robertson GT, Rockey P, Sun PM, Winkler ME, Yang Y, Young-Bellido M, Zhao G, Zook CA, Baltz RH, Jaskunas SR, Rosteck PR, Jr, Skatrud PL, Glass JI. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709–5717. 10.1128/JB.183.19.5709-5717.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng J, Lupien A, Gingras H, Wasserscheid J, Dewar K, Legare D, Ouellette M. 2009. Genome sequencing of linezolid-resistant Streptococcus pneumoniae mutants reveals novel mechanisms of resistance. Genome Res. 19:1214–1223. 10.1101/gr.089342.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biedenbach DJ, Farrell DJ, Mendes RE, Ross JE, Jones RN. 2010. Stability of linezolid activity in an era of mobile oxazolidinone resistance determinants: results from the 2009 Zyvox Annual Appraisal of Potency and Spectrum program. Diagn. Microbiol. Infect. Dis. 68:459–467. 10.1016/j.diagmicrobio.2010.09.018 [DOI] [PubMed] [Google Scholar]
- 32.Klugman KP, Low DE, Metlay J, Pechere JC, Weiss K. 2004. Community-acquired pneumonia: new management strategies for evolving pathogens and antimicrobial susceptibilities. Int. J. Antimicrob. Agents 24:411–422. 10.1016/j.ijantimicag.2004.08.006 [DOI] [PubMed] [Google Scholar]


