Abstract
In Spain, rates of ciprofloxacin resistance in pneumococci were low during the last decade (2.6% in 2002 and 2.3% in 2006). In 2012, the rate remained at 2.3%, equivalent to 83 of 3,621 isolates. Of the 83 resistant isolates, 15 showed a low level (MIC of 4 to 8 μg/ml) and 68 a high level (MIC of 16 to 128 μg/ml) of ciprofloxacin resistance. Thirteen low-level-resistant isolates had single changes in ParC, one had a single ParE change, and one did not present any mutations. High-level-resistant isolates had GyrA changes plus additional ParC and/or ParE changes: 51, 15, and 2 isolates had 2, 3, or 4 mutations, respectively. Although 24 different serotypes were observed, 6 serotypes accounted for 51.8% of ciprofloxacin-resistant isolates: 8 (14.5%), 19A (10.8%), 11A (7.2%), 23A (7.2%), 15A (6.0%), and 6B (6.0%). A decrease in pneumococcal 7-valent conjugate vaccine (PCV7) serotypes was observed from 2006 (35.7%) to 2012 (16.9%), especially of serotype 14 (from 16.3% to 2.4%; P < 0.001). In comparison with findings in 2006, multidrug resistance was greater in 2012 (P = 0.296), mainly due to the increased presence and/or emergence of clonal complexes associated with non-PCV7 serotypes: CC63 expressing serotypes 8, 15A, and 19A; CC320 (with serotype 19A); and CC42 (with serotype 23A). Although rates of ciprofloxacin resistance remained low and stable throughout the last decade, changes in serotype and genotype distributions were observed in 2012, notably the expansion of a preexisting multidrug-resistant clone, CC63, and the emergence of the CC156 clone expressing serotype 11A.
INTRODUCTION
Streptococcus pneumoniae is an important cause of morbidity and mortality worldwide, and it is a major etiological agent of community-acquired pneumonia, meningitis, and acute otitis media (1). Following the introduction of the pneumococcal 7-valent conjugate vaccine (PCV7) in 2000 in the United States, the incidence of invasive pneumococcal disease declined drastically, coinciding with a decrease in penicillin resistance (2–4). In Spain, where PCV7 was introduced in 2001, a decrease in the incidence of invasive disease due to PCV7 serotypes was also observed (5). However, shortly after introduction of PCV7, an emergence of nonvaccine serotypes was observed worldwide (6, 7).
Fluoroquinolones (FQs) target type II DNA topoisomerases. Despite the functional similarities between topoisomerase IV (topo IV) and gyrase, their susceptibilities to FQs vary across bacterial species (8). Isolates of S. pneumoniae that are resistant to FQs have been shown to present mutations at specific regions (quinolone resistance determining regions [QRDRs]) of the topoisomerase IV (parC and parE) and DNA gyrase (gyrA) genes. In recent decades, the new-generation FQs levofloxacin (LVX) and moxifloxacin (MOX), which have enhanced activity against pneumococci and other respiratory pathogens, have become therapeutic alternatives in the treatment of community-acquired pneumonia. In S. pneumoniae, the primary target for ciprofloxacin (CIP) and LVX is topo IV (9–12), whereas gyrase is the primary target for MOX (13). Although CIP has low activity against S. pneumoniae and is not recommended for treatment, it has proved to be useful for detection of first-step mutations. In the present study, FQ resistance was considered when the CIP MIC was ≥4 μg/ml, following the criteria established by Chen et al. (14), which coincides with the current (>2 μg/ml) EUCAST breakpoints (15). The differences observed in the rates of susceptibility to CIP compared with those to LVX and MOX are due to isolates with first-step QRDR mutations. These isolates (CIP resistant but LVX or MOX susceptible) could become highly resistant under selective FQ pressure and are associated with treatment failure when FQs are used (16). By using a CIP resistance breakpoint MIC of ≥4 μg/ml, we have detected first-step mutations in isolates susceptible to LVX by the CLSI criteria (LVX MIC of 1 to 2 μg/ml). In addition, among isolates with a CIP MIC of 2 μg/ml, no first-step mutations were detected in our previous studies (17, 18). The killing effect of FQs has been related to the resolution of reaction intermediates of DNA-FQ-topoisomerase complexes, which subsequently generates irreparable double-stranded DNA breaks (19).
CIP resistance in S. pneumoniae continues to show a low prevalence (<3%) in Europe (18, 20), although higher rates have been detected in Asia (10.5%) (21) and Canada (7.3%) (22). Resistance to FQs can evolve during treatment, and there are numerous reports of treatment failures with the use of FQs in pneumococcal infections caused by strains with first-step mutations (14, 23). These cases tend to involve elderly patients with chronic respiratory diseases, such as chronic obstructive pulmonary disease (COPD), in which higher rates of FQ resistance have been detected (24). Although the development of FQ resistance has been associated with FQ consumption (14, 22, 25), the dissemination of pneumococcal FQ-resistant clones has rarely been observed so far (26). However, previous epidemiological studies (17, 18) have revealed a low genetic diversity of pneumococcal clones among FQ-resistant pneumococci in Spain.
The present study investigates the prevalence of FQ-resistant pneumococci in Spain during 2012. Resistance mutations in the QRDRs of parC, parE, and gyrA were studied, as were resistance associations with other antibiotics and the characteristics of the clones harboring this resistance. In order to assess changes in the epidemiology of FQ resistance, the results of the present study were compared with those of two similar studies conducted in 2002 and 2006.
MATERIALS AND METHODS
Bacterial isolates, serotyping, and susceptibility tests.
A total of 3,621 S. pneumoniae isolates from 112 hospitals nationwide were sent to the Spanish Pneumococcus Reference Laboratory during 2012: 2,926 isolates were from adults, and 695 were from children. In terms of their origins, 2,252 (62.2%) isolates were from blood or other sterile sites, while the remaining 1,369 (37.8%) were from respiratory samples. Isolates were confirmed as S. pneumoniae by standard methods, with serotypes being determined by the Quellung reaction. Antimicrobial susceptibility was tested by agar dilution at the Spanish Reference Laboratory. The MICs of CIP, LEV, and MOX of 83 isolates with CIP MICs of ≥4 μg/ml were confirmed by Etest and broth microdilution methods, according to the Clinical and Laboratory Standards Institute guidelines (27). S. pneumoniae ATCC 49619 was included as a quality control.
Molecular typing.
Clonal complexes (CCs) were characterized by means of pulsed-field gel electrophoresis (PFGE). Briefly, genomic DNA embedded in agarose plugs was restricted with SmaI or ApaI, and fragments were separated by PFGE in a Chef-DRIII apparatus (Bio-Rad). PFGE patterns were visually compared with those of representative international pneumococcal clones of the Pneumococcal Molecular Epidemiology Network (28), and isolates with patterns that varied by three or fewer bands were considered to represent the same PFGE type. Major clusters, which share the same PFGE pattern/serotype combination, were defined as those that included three or more pneumococcal isolates. In order to assess identity with global pneumococcal clones, at least one isolate of each cluster (n = 42) was analyzed by multilocus sequence typing (MLST), as described previously (29). Allele numbers and sequence types (ST) were assigned using the MLST web site (http://www.mlst.net).
PCR amplification and DNA sequence determination.
parE and parC QRDRs were amplified by using the oligonucleotides parE398 (30) and parC152 (9). To amplify and sequence the gyrA QRDRs, the oligonucleotides gyrA44 and gyrA170 (30) were used. These PCR fragments were sequenced as previously described (17). To detect the presence of the ant gene, the oligonucleotides used in PCR amplifications were antUP and antDOWN (31).
Statistical analysis.
The χ2 test or Fisher's exact test were used as appropriate. Two-sided P values of <0.05 were considered statistically significant.
RESULTS
Ciprofloxacin resistance and multidrug resistance.
The rate of CIP resistance in 2012 was 2.3% (83/3,621). Among these 83 CIP-resistant (Cipr) isolates, 15 (18.1%) with MICs of 4 to 8 μg/ml were classified as low-level resistant (LL-Cipr), while the remaining 68 (81.9%), with MICs of ≥16 μg/ml, were classified as high-level resistant (HL-Cipr) (Table 1). Global Cipr rates remained stable across the three time periods studied (2002, 2006, and 2012), and no statistically significant variations were found in the rates of LL- and HL-Cipr isolates (Table 1). In addition, there was no difference between the three sets of results in relation to age groups except for a decrease in the prevalence of Cipr among pneumococci isolated from patients >64 years old (7.2% in 2002 versus 3.9% in 2012). In 2012, Cipr rates among pneumococci isolated from noninvasive disease (3.7%; 51/1,369) were higher than those for pneumococci isolated from invasive disease (1.4%; 32/2,252; P < 0.001), this being consistent with the previous two reports (17, 18).
TABLE 1.
Comparison of three surveillance studies of ciprofloxacin-resistant Streptococcus pneumoniae isolates in Spain: data for 2002, 2006, and 2012a
| Characteristic | No. of Cipr isolates/total no. of isolates (%)c |
P valued |
|||
|---|---|---|---|---|---|
| 2002 | 2006 | 2012 | 2002 vs 2012 | 2006 vs 2012 | |
| Ciprofloxacin resistance | |||||
| Global | 75/2,882 (2.6) | 98/4,215 (2.3) | 83/3,621 (2.3) | 0.419 | 0.940 |
| Low level (MICs of 4–8 μg/ml) | 14/75 (18.7) | 30/98 (30.6) | 15/83 (18.1) | 1.000 | 0.059 |
| High level (MICs ≥ 16 μg/ml) | 61/75 (81.3) | 68/98 (69.3) | 68/83 (/81.9) | 1.000 | 0.059 |
| In persons <15 years of age | 0/978 (0) | 2/1,446 (0.14) | 2/695 (0.4) | 0.172 | 0.600 |
| In persons 15–64 years of age | 22/1,166 (1.9) | 34/1,455 (2.3) | 19/1,336 (1.4) | 0.431 | 0.100 |
| In persons >64 years of age | 53/738 (7.2) | 61/1,314 (4.7) | 62/1,590 (3.9) | <0.001 | 0.355 |
| PCV7 serotypes | 49/75 (65.3) | 35/98 (37.5) | 14/83 (16.9) | <0.001 | 0.014 |
| PCV13 serotypes | 56/75 (74.7) | 47/98 (48.0) | 31/83 (37.3) | <0.001 | 0.176 |
| Serotype 8 | 0/75 (0) | 7/98 (7.1) | 12/83 (14.5) | <0.001 | 0.145 |
| Serotype 11A | 1/75 (1.3) | 4/98 (4.1) | 6/83 (7.2) | 0.120 | 0.516 |
| Serotype 14 | 14/75 (18.7) | 16/98 (16.3) | 2/83 (2.4) | <0.001 | <0.001 |
| Serotype 19A | 1/75 (1.3) | 8/98 (8.2) | 9/83 (10.8) | 0.019 | 0.613 |
| Other antimicrobial drug resistance | |||||
| Penicillin MIC ≥ 0.12 μg/ml | 55/75 (73.3) | 44/98 (44.9) | 37/83 (44.5) | <0.001 | 1.000 |
| Chloramphenicol MIC ≥ 8 μg/ml | 33/75 (44.0) | 11/98 (11.2) | 4/83 (4.8) | <0.001 | 0.176 |
| Multidrug resistanceb | 55/75 (73.3) | 48/98 (49.0) | 48/83 (57.8) | 0.046 | 0.296 |
Cipr, resistant to ciprofloxacin, defined by Chen et al. as ≥4 μg/ml (22); low-level resistant, MICs of 4 to 8 μg/ml; high-level resistant, MICs ≥ 16 μg/ml.
Multidrug resistance, resistance to CIP and to at least two other antimicrobial groups.
Values for “Other antimicrobial drug resistance” are no. of resistant isolates/no. of ciprofloxacin-resistant isolates (%).
Values in bold indicate statistically significant differences (P < 0.05) between time periods.
Forty-eight (57.8%) Cipr pneumococci were considered multidrug resistant (MDR), defined as resistance to CIP plus at least two other antimicrobial groups. The MDR rate showed a slight increase from 2006 to 2012 (Table 1).
Amino acid substitutions in QRDRs of pneumococcal isolates.
The parC, parE, and gyrA QRDRs of the 83 Cipr (MIC ≥ 4 μg/ml) isolates were characterized. In addition, 15 randomized isolates with CIP MICs of 2 μg/ml were analyzed, with their QRDRs showing susceptible sequences, in agreement with previous findings of our group (17, 18). QRDRs of parE and parC were amplified in a single PCR with the oligonucleotides parE398 and parC152. All isolates yielded 1.6-kb PCR products, with the exception of a recombinant strain, which yielded a bigger fragment (see below). Although most Cipr isolates (79/83) showed low nucleotide sequence variation (≤1%), four isolates exhibited high variation (>4%), suggesting a recombinant origin for these genes (see below). The different patterns of amino acid substitutions in the QRDRs of all Cipr pneumococci, as well as their MICs to FQs, are shown in Table 2. Among the 15 LL-Cipr isolates, all but 1 had mutations producing amino acid changes in topoisomerase IV subunits: 13 produced changes in ParC, and 1 did so in ParE. The remaining isolate, with a CIP MIC of 4 μg/ml, presented no changes in its QRDRs.
TABLE 2.
Fluoroquinolone MICs of 83 isolates and amino acid changes in their DNA topoisomerase genesa
| No. of isolates | Amino acid substitution for: |
MIC (μg/ml) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ParC |
ParE |
GyrA |
||||||||||
| D78 | S79 | D83 | K426 | D435 | E474 | G79 | S81 | E85 | CIP | LVX | MOX | |
| 1 | — | — | — | — | — | — | — | — | — | 4 | 2 | 0.25 |
| 3 | — | F | — | — | — | — | — | — | — | 4–8 | 1–4 | 0.25–0.5 |
| 6 | — | Y | — | — | — | — | — | — | — | 4–8 | 1–2 | 0.12–8 |
| 3 | — | — | N | — | — | — | — | — | — | 4–8 | 1–4 | 0.25–0.5 |
| 1 | N | — | — | —‡ | —‡ | —‡ | —‡ | —‡ | —‡ | 8 | 2 | 0.5 |
| 1 | — | — | — | — | — | K | — | — | — | 8 | 2 | 1 |
| 1 | N | — | — | — | — | — | F | — | 32 | 8 | 4 | |
| 1 | — | F | — | — | — | — | A | — | — | 32 | 16 | 8 |
| 1 | — | F | — | — | — | — | — | A | — | 64 | 32 | 8 |
| 31 | — | F | — | — | — | — | — | F | — | 32–64 | 16–32 | 2–8 |
| 1 | — | F | — | — | — | — | — | Y | — | 32 | 16 | 2 |
| 1 | —‡ | Y | —‡ | —‡ | —‡ | —‡ | —‡ | F‡ | —‡ | 32 | 16 | 4 |
| 5 | — | Y | — | — | — | — | — | F | — | 32–64 | 16–32 | 2–4 |
| 1 | — | Y | — | — | — | — | — | V | — | 32 | 16 | 4 |
| 2 | — | Y | — | — | — | — | — | Y | — | 32 | 16 | 4 |
| 2 | — | F | — | — | — | — | — | — | K | 32 | 16 | 4 |
| 2 | — | — | Y | — | — | — | — | F | — | 32 | 16 | 2–4 |
| 1 | — | — | — | — | N | — | — | A | — | 64 | 32 | 1 |
| 1 | — | — | — | — | N | — | — | F | — | 32 | 16 | 2 |
| 1 | — | — | — | — | N | — | —‡ | F‡ | —‡ | 32 | 16 | 8 |
| 2 | — | F | N | — | — | — | — | F | — | 64 | 32 | 8 |
| 1 | — | F | Y | — | — | — | — | Y | — | 64 | 32 | 8 |
| 1 | — | Y | — | — | — | — | — | F | K | 64 | 32 | 1 |
| 1 | — | F | — | — | — | — | — | Y | K | 64 | 32 | 4 |
| 4 | — | F | — | — | — | — | — | F | K | 64 | 32 | 4–8 |
| 1 | — | Y | — | — | — | — | — | F | Q | 64 | 32 | 16 |
| 2 | — | F | — | — | N | — | — | F | — | 128 | 64 | 8 |
| 1 | — | Y | — | — | N | — | — | F | — | 128 | 64 | 4 |
| 1 | — | Y | — | N | — | — | — | F | — | 64 | 32 | 64 |
| 1 | — | — | N | — | — | K | — | Y | — | 32 | 16 | 2 |
| 1 | — | F | Y | N | — | — | — | V | 128 | 64 | 8 | |
| 1 | — | F | — | — | N | — | — | F | K | 128 | 64 | 64 |
Only changes involved in resistance are shown. —, no change; ‡, the residue is located in a recombinant gene. Additional amino acid changes, not involved in resistance, were ParC N91D (the isolate with mosaic parC gene), ParE I460V (28 isolates), and GyrA S114G (the three isolates with mosaic gyrA genes). CIP, ciprofloxacin; LVX, levofloxacin; MOX, moxifloxacin.
All HL-Cipr isolates had at least one amino acid change in topoisomerase IV genes, as well as mutations producing changes in the gyrase A subunit (Table 2). Among the 68 HL-Cipr isolates, 51 had double changes (70.6% at ParC and GyrA and 4.4% at ParE and GyrA), 15 had triple changes (either 1 or 2 changes at ParC and 1 or 2 changes at GyrA or 1 change each at ParC, ParE, and GyrA), and the remaining 2 isolates had four changes (one isolate had 2 ParC, 1 ParE, and 1 GyrA changes, while the other had 1 ParC, 1 ParE, and 2 GyrA changes). Mutations found were classical mutations involved in resistance, which have been previously found in clinical isolates and shown to be involved in resistance by genetic transformation (10, 12, 17, 18, 32–35).
Three isolates carried recombinant genes, one in parE, parC, and gyrA, one in parE and gyrA, and one in gyrA. These isolates probably acquired these genes from resistant Streptococcus mitis group isolates given the presence of ParC N91D in their ParC recombinant proteins and of GyrA S114G in their GyrA proteins (31). In addition, amplification of the isolate with the parE plus parC mosaic genes using oligonucleotides parE398 and parC152 rendered a fragment of 3.5 kb, which is longer than the 1.6 kb observed in the remaining isolates. This characteristic is typical of S. mitis group isolates. In addition, an ant gene was detected by PCR amplification (31) in the intergenic parE-parC region of this isolate (data not shown).
Dynamics of pneumococcal serotypes and genotypes.
A total of 24 different serotypes were detected among the Cipr pneumococci, but 6 of them accounted for 51.8% (43 of 83) of the isolates (Fig. 1): 8 (14.5%), 19A (10.8%), 11A (7.2%), 23A (7.2%), 6B (6.0%), and 15A (6.0%). A gradual decrease in PCV7 serotypes was found over the years from 2002 (65.3%) to 2006 (35.7%) to 2012 (16.9%), the most important concerning serotype 14 (Table 1). In Spain, PCV13 was licensed for adults in 2012, with the percentage of Cipr pneumococci belonging to PCV13 serotypes in that year being 37.3%, lower than the figure for 2006 (48.0%; P = 0.176). However, if one considers only the PCV13 serotypes not included in the PCV7 (1, 3, 5, 6A, 7F, and 19A), then a slight increase was observed (16.3% [16/98] in 2006, versus 20.5% [17/83] in 2012; P = 0.563); this increase was due mainly to the appearance and spread of serotype 19A, which was ranked second in 2012. It should also be noted that two non-PCV13 serotypes were detected in 21.7% of the overall Cipr episodes: serotype 8, which emerged in 2006 and whose frequency increased again in 2012 (Table 1), and serotype 11A, which showed a stepwise increase across the three sets of data.
FIG 1.
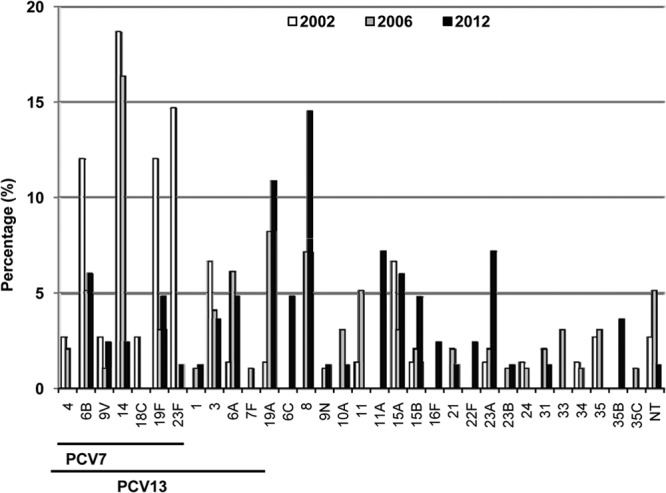
Serotype distribution of ciprofloxacin-resistant pneumococci isolated in Spain in 2002, 2006, and 2012. A total of 75 isolates from 2002 (white columns), 98 from 2006 (gray columns), and 83 from 2012 (black columns) were compared. “PCV7” and “PCV13” indicate serotypes included in the respective conjugate pneumococcal vaccines.
Genetic relatedness among resistant isolates was determined primarily by PFGE in order to make comparisons with global clones, while representative isolates were further studied in terms of their allelic profiles by MLST (n = 42/83). Although 32 different PFGE clonal complexes (CCs) were detected among the Cipr isolates, 3 of them (CC63, CC156, and CC42) accounted for nearly 40% of isolates (Fig. 2). The CC63 clone, expressing serotypes 8, 15A, 19A, and 19F, was found in 25.3% (21/83) of Cipr isolates (Table 3); CC156, with serotypes 9V and 11A, was found in 8.4% (7/83), while CC42, with serotype 23A, accounted for 6.0% (5/83) (Fig. 2). In addition, there were four CCs, with three isolates each, which had not been detected in the previous studies: CC1662 with serotype 15B, CC558 with serotype 35B, CC320 with serotype 19A, and CC460 with serotype 6A.
FIG 2.
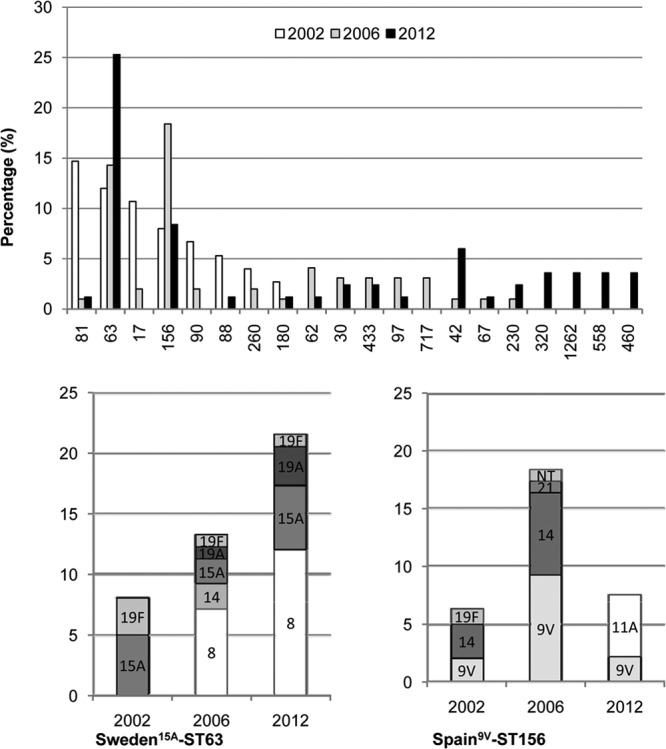
Genotype of ciprofloxacin-resistant pneumococci isolated in Spain in 2002, 2006, and 2012 (top panel) or serotypes expressed in the indicated clones (bottom panels).
TABLE 3.
Phenotypic characteristics and changes in QRDRs among 21 isolates of the CC63 clone found in 2012a
| Strain IDb | Serotype | Amino acid changes in QRDR of: |
No. of resistant mutations | MIC (μg/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ParC |
ParE |
GyrA |
|||||||||
| S79 | D83 | K426 | I460 | S81 | E85 | CIP | LVX | MOX | |||
| 14 | 8 | F | — | — | V | A | — | 2 | 64 | 32 | 8 |
| 3–6, 19, 20, 96 | 8 | F | — | — | V | F | — | 2 | 64 | 32 | 4–8 |
| 47 | 8 | Y | — | — | V | F | — | 2 | 64 | 32 | 4 |
| 55 | 8 | Y | — | — | — | F | — | 2 | 64 | 32 | 2 |
| 24, 56 | 8 | F | N | — | V | F | — | 3 | 64 | 32 | 4 |
| 91 | 15A | — | Y | — | V | F | — | 2 | 32 | 16 | 2 |
| 53 | 15A | Y | — | — | V | F | — | 2 | 32 | 16 | 2 |
| 71 | 15A | F | — | — | V | F | — | 2 | 64 | 32 | 4 |
| 58 | 15A | Y | — | — | V | F | Q | 3 | 64 | 32 | 16 |
| 26 | 15A | F | Y | N | V | V | — | 4 | 128 | 64 | 8 |
| 81 | 19A | Y | — | — | V | — | — | 1 | 4 | 1 | 4 |
| 12 | 19A | F | — | — | V | F | — | 2 | 64 | 32 | 4 |
| 9 | 19A | Y | — | — | — | F | K | 3 | 64 | 32 | 8 |
| 51 | 19F | F | — | — | V | F | — | 2 | 64 | 32 | 2 |
Changes involved in resistance are shown in bold type. —, no change. CIP, ciprofloxacin; LVX, levofloxacin; MOX, moxifloxacin.
Strain identification no.
The two most frequent CCs identified in 2012 (CC63 and CC156) had isolates with different serotypes, suggesting capsular switch events (Fig. 2). The CC63 clone showed a dramatic increase after 2002, and it was detected in 25.3% of Cipr pneumococci in 2012, associated mainly with serotype 8 (12 out of 21 isolates). Likewise, CC156 was maintained in 2012 and was associated with serotype 11A (5 out of 7 isolates).
DISCUSSION
Current rates of fluoroquinolone resistance in Spain (2.3% for CIP) are similar to those reported previously in 2002 (2.6%) and 2006 (2.3%). The prevalence of FQ resistance has been directly correlated with the consumption of FQs, especially CIP (14, 25). Data from the Spanish Medicines Agency (http//agemed.es) indicate that CIP consumption in Spain has remained essentially stable since 2002, namely, at 1.1 defined daily dose per 1,000 inhabitants per day (DDD). Over the same period, MOX consumption showed only a slight variation, from 0.3 (2002) to 0.4 (2006) and back to 0.3 DDD (2012). However, LVX consumption has increased from 0.2 DDD in 2002 to 0.4 DDD in 2006 to 0.6 DDD in 2012.
In line with a previous study from Canada (36), this increase in FQ consumption has not led to increased FQ resistance rates. One explanation for this could be the greater efficacy of the new FQs, which may reduce selective pressure in relation to the pneumococcal QRDR mutations involved in FQ resistance (37). In addition, due to their toxicity, FQs are not used to treat children, and therefore they do not produce selective pressure on the pneumococcal population colonizing the nasopharynx of children (the main pneumococcal reservoir). In fact, in the present series, only two Cipr pneumococci were isolated from children (ages 1 and 3), and these cases were probably due to family cross-transmission.
The introduction of PCV7 in June 2001 and likely also that of PCV13 in June 2010 had an impact on the ecology of pneumococci causing disease in children and adults, but it has also substantially reduced the incidence of antibiotic resistance (2); in fact, the majority (65.3%) of Cipr isolates in 2002 belonged to serotypes included in PCV7, compared with only 35.7% of Cipr isolates in 2006 and 16.9% in 2012. In line with the changes found in the serotype distribution, the MDR rate also decreased in 2006 as a consequence of the decreased number of MDR clones associated with these PCV7 serotypes: CC81 (Spain23F-ST81), CC90 (Spain6B-ST90), CC17 (Spain14-ST17), and CC88 of serotype 19F. However, although these clones have almost disappeared in 2012, MDR rates increased in this period (P = 0.296), mainly due to the increase and/or emergence of CCs associated with non-PCV7 MDR serotypes: CC63 (serotypes 8 and 15A), CC320 (serotype 19A), and CC42 (serotype 23A). These changes in clone and serotype distribution reflect changes in the pneumococcal population isolated from the nasopharynx of children, in which serotypes 15A, 15B, 19A, 6C, and 11A have increased in recent years (38, 39), and also in respiratory samples from acute exacerbations in COPD patients (40).
In comparison with the data for 2002 and 2006, the most notorious Cipr isolates in 2012 were those expressing serotypes 8 and 11A, not included in PCV13. All serotype 8 isolates were associated with genotype CC63, suggesting a capsular switching event. This clone expressing serotype 8 was first detected in the 2006 study (18), and it was disseminated in the Madrid area, mainly among HIV-positive patients, with one isolate showing FQ resistance and a ParC S79F amino acid substitution (26, 41). In the present series, eight CC63-serotype 8 isolates were also isolated from patients at hospitals in Madrid, a fact that could explain the frequency of this serotype among Cipr pneumococci in 2012.
CC63 was the most frequently detected CC (21 of 83 Cipr isolates) in this study and expressed four serotypes (8, 15A, 19A, and 19F). Of these, only 19A and 19F are included in PCV13. These 21 isolates had either one (1 isolate), two (15 isolates), three (4 isolates), or four (1 isolate) mutations at parC, parE, or gyrA. Heterogeneity was observed in terms of both the amino acid (S79 or D83) affected at ParC and the change producing resistance. There was also heterogeneity in GyrA mutations, which were found at either S81 or E85. These results suggest that although some of the isolates could have a clonal origin, the majority of these Cipr isolates are likely to be the result of spontaneous mutations in a CC63 isolate, which became predominant among Cipr pneumococci in 2012. In agreement with this, the CC63 genotype ranked first (9.1%) among 206 noninvasive pneumococci collected from chronic obstructive pulmonary disease patients during 2009 to 2012, and 5/18 (27.8%) of them showed resistance to FQs (42). In contrast, although we have no data about the genotypes of all pneumococci sent to the Reference Laboratory in 2012, data from Barcelona reveal that the overall frequency of CC63 among invasive isolates was low: 3.3% (37/1121) in adults in the years 1997 to 2008 (6) and 1.5% (3/198) in children in the 1997-to-2006 period (42).
Regarding serotype 11A, the two previous studies reported that pneumococci expressing this serotype were related to the ST62 clone. In the present series, however, five of six Cipr pneumococci expressing serotype 11A belonged to genotype CC156, suggesting another capsular switching event. This is a cause of concern because it allows the persistence of a well-established clone that usually expresses PCV7 serotypes (9V and 14) through a vaccine escape phenomenon.
ACKNOWLEDGMENTS
We acknowledge the use of the Streptococcus pneumoniae MLST website at Imperial College London, funded by the Wellcome Trust.
This study was supported by grant BIO2011-25343 from the Ministerio de Ciencia y Tecnología, by grant PI11/0763 from the Fondo de Investigaciones Sanitarias, and by Ciber de Enfermedades Respiratorias, an initiative of the Instituto de Salud Carlos III. A.D. was supported by an Agustí Pumarola grant from the Societat Catalana de Malaties Infecciones i Microbiologia Clínica and the Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica.
Footnotes
Published ahead of print 10 February 2014
REFERENCES
- 1.World Health Organization. 2007. Pneumococcal conjugate vaccine for childhood immunization—WHO position paper. Wkly. Epidemiol. Rec. 82:93–104 [PubMed] [Google Scholar]
- 2.Whitney CG, Farley MM, Hadler J, Harrison LH, Bennett NM, Lynfield R, Reingold A, Cieslak PR, Pilishvili T, Jackson D, Facklam RR, Jorgensen JH, Schuchat A. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737–1746. 10.1056/NEJMoa022823 [DOI] [PubMed] [Google Scholar]
- 3.Kyaw MH, Lynfield R, Schaffner W, Craig AS, Hadler J, Reingold A, Thomas AR, Harrison LH, Bennett NM, Farley MM, Facklam RR, Jorgensen JH, Besser J, Zell ER, Schuchat A, Whitney CG. 2006. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N. Engl. J. Med. 354:1455–1463. 10.1056/NEJMoa051642 [DOI] [PubMed] [Google Scholar]
- 4.Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, Bennett NM, Reingold A, Thomas A, Schaffner W, Craig AS, Smith PJ, Beall BW, Whitney CG, Moore MR. 2010. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J. Infect. Dis. 201:32–41. 10.1086/648593 [DOI] [PubMed] [Google Scholar]
- 5.Fenoll A, Granizo JJ, Aguilar L, Giménez MJ, Aragoneses-Fenoll L, Hanquet G, Casal J, Tarragó D. 2009. Temporal trends of invasive Streptococcus pneumoniae serotypes and antimicrobial resistance patterns in Spain from 1979 to 2007. J. Clin. Microbiol. 47:1012–1020. 10.1128/JCM.01454-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ardanuy C, Pallarés R, Calatayud L, Domínguez MA, Rolo D, Grau I, Martín R, Liñares J. 2009. Epidemiology of invasive pneumococcal disease among adult patients in Barcelona before and after pediatric 7-valent pneumococcal conjugate vaccine introduction, 1997–2007. Clin. Infect. Dis. 48:57–64. 10.1086/594125 [DOI] [PubMed] [Google Scholar]
- 7.Moore MR, Gertz JRE, Woodbury RL, Barkocy-Gallagher GA, Schaffner W, Lexau C, Gershman K, Reingold A, Farley M, Harrison LH, Hadler JL, Bennett NM, Thomas AR, McGee L, Pilishvili T, Brueggemann AB, Whitney CG, Jorgensen JH, Beall B. 2008. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J. Infect. Dis. 197:1016–1027. 10.1086/528996 [DOI] [PubMed] [Google Scholar]
- 8.Drlica K, Zhao X. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muñoz R, de la Campa AG. 1996. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 40:2252–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janoir C, Zeller V, Kitzis M-D, Moreau NJ, Gutmann L. 1996. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob. Agents Chemother. 40:2760–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández-Moreira E, Balas D, González I, de la Campa AG. 2000. Fluoroquinolones inhibit preferentially Streptococcus pneumoniae DNA topoisomerase IV than DNA gyrase native proteins. Microb. Drug Resist. 6:259–267. 10.1089/mdr.2000.6.259 [DOI] [PubMed] [Google Scholar]
- 12.Tankovic J, Perichon B, Duval J, Courvalin P. 1996. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob. Agents Chemother. 40:2505–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houssaye S, Gutmann L, Varon E. 2002. Topoisomerase mutations associated with in vitro selection of resistance to moxifloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2712–2715. 10.1128/AAC.46.8.2712-2715.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen DK, McGeer A, de Azavedo JC, Low DE. 1999. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N. Engl. J. Med. 341:233–239. 10.1056/NEJM199907223410403 [DOI] [PubMed] [Google Scholar]
- 15.The European Committee on Antimicrobial Susceptibility Testing. 2014. Breakpoint tables for interpretation of MICs and zone diameters, version 4.0. http://www.eucast.org/clinical breakpoints/
- 16.Fuller JD, Low DE. 2005. A review of Streptococcus pneumoniae infection treatment failures associated with fluoroquinolone resistance. Clin. Infect. Dis. 41:118–121. 10.1086/430829 [DOI] [PubMed] [Google Scholar]
- 17.de la Campa AG, Balsalobre L, Ardanuy C, Fenoll A, Pérez-Trallero E, Liñares J. 2004. Fluoroquinolone resistance in penicillin-resistant Streptococcus pneumoniae clones, Spain. Emerg. Infect. Dis. 10:1751–1759. 10.3201/eid1010.040382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de la Campa AG, Ardanuy C, Balsalobre L, Pérez-Trallero E, Marimón JM, Fenoll A, Liñares J. 2009. Changes in fluoroquinolone-resistant Streptococcus pneumoniae after 7-valent conjugate vaccination, Spain. Emerg. Infect. Dis. 15:905–911. 10.3201/eid1506.080684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drlica K, Malik M, Kerns RJ, Zhao X. 2008. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 52:385–392. 10.1128/AAC.01617-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riedel S, Beekmann SE, Heilmann KP, Richter SS, García-de-Lomas J, Ferech M, Goosens H, Doern GV. 2007. Antimicrobial use in Europe and antimicrobial resistance in Streptococcus pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 26:485–490. 10.1007/s10096-007-0321-5 [DOI] [PubMed] [Google Scholar]
- 21.Ip M, Chau SSL, Chi F, Cheuk ECS, Ma H, Lai RWM, Chan PK. 2007. Longitudinally tracking fluoroquinolone resistance and its determinants in penicillin-susceptible and -nonsusceptible Streptococcus pneumoniae isolates in Hong Kong, 2000 to 2005. Antimicrob. Agents Chemother. 51:2192–2194. 10.1128/AAC.00139-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adam HJ, Hoban DJ, Gin AS, Zhanel GG. 2009. Association between fluoroquinolone usage and a dramatic rise in ciprofloxacin-resistant Streptococcus pneumoniae in Canada, 1997-2006. Int. J. Antimicrob. Agents 34:82–85. 10.1016/j.ijantimicag.2009.02.002 [DOI] [PubMed] [Google Scholar]
- 23.Fuller JD, McGeer A, Low DE. 2005. Drug-resistant pneumococcal pneumonia: clinical relevance and approach to management. Eur. J. Clin. Microbiol. Infect. Dis. 24:780–788. 10.1007/s10096-005-0059-x [DOI] [PubMed] [Google Scholar]
- 24.Domenech A, Ardanuy C, Calatayud L, Santos S, Tubau F, Grau I, Verdaguer R, Dorca J, Pallares R, Martín R, Liñares J. 2011. Serotypes and genotypes of Streptococcus pneumoniae causing pneumonia and acute exacerbations in patients with chronic obstructive pulmonary disease. J. Antimicrob. Chemother. 66:487–493. 10.1093/jac/dkq480 [DOI] [PubMed] [Google Scholar]
- 25.Liñares J, de la Campa AG, Pallarés R. 1999. Fluoroquinolone resistance in Streptococcus pneumoniae. N. Engl. J. Med. 341:1546–1547. 10.1056/NEJM199911113412013, [DOI] [PubMed] [Google Scholar]
- 26.Rodríguez-Avial I, Ramos B, Rios E, Cercenado E, Ordobas M, Sanz JC. 2011. Clonal spread of levofloxacin-resistant Streptococcus pneumoniae invasive isolates in Madrid, Spain, 2007 to 2009. Antimicrob. Agents Chemother. 55:2469–2471. 10.1128/AAC.01380-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. CLSI document M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 28.McGee L, McDougal L, Zhou J, Spratt BG, Tenover FC, George R, Hakenbeck R, Hryniewicz W, Lefevre JC, Tomasz A, Klugman KP. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565–2571. 10.1128/JCM.39.7.2565-2571.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enright MC, Spratt BG. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049–3060. 10.1099/00221287-144-11-3049 [DOI] [PubMed] [Google Scholar]
- 30.González I, Georgiou M, Alcaide F, Balas D, Liñares J, de la Campa AG. 1998. Fluoroquinolone resistance mutations in the parC, parE, and gyrA genes of clinical isolates of viridans group streptococci. Antimicrob. Agents Chemother. 42:2792–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balsalobre L, Ferrándiz MJ, Liñares J, Tubau F, de la Campa AG. 2003. Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47:2072–2081. 10.1128/AAC.47.7.2072-2081.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jorgensen JH, Weigel LM, Swenson JM, Whitney CG, Ferraro MJ, Tenover FC. 2000. Activities of clinafloxacin, gatifloxacin, gemifloxacin, and trovafloxacin against recent clinical isolates of levofloxacin-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2962–2968. 10.1128/AAC.44.11.2962-2968.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varon E, Janoir C, Kitzis M-D, Gutmann L. 1999. ParC and GyrA may be interchangeable initial targets of some fluoroquinolones in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:302–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weigel LM, Anderson GJ, Facklam RR, Tenover FC. 2001. Genetic analyses of mutations contributing to fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:3517–3523. 10.1128/AAC.45.12.3517-3523.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martín-Galiano AJ, de la Campa AG. 2003. High-efficiency generation of antibiotic-resistant strains of Streptococcus pneumoniae by PCR and transformation. Antimicrob. Agents Chemother. 47:1257–1261. 10.1128/AAC.47.4.1257-1261.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel SN, McGeer A, Melano R, Tyrrell GJ, Green K, Pillai DR, Low DE. 2011. Susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. Antimicrob. Agents Chemother. 55:3703–3708. 10.1128/AAC.00237-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ambrose PG, Grasela DM, Grasela TH, Passarell J, Mayer HB, Pierce PF. 2001. Pharmacodynamics of fluoroquinolones against Streptococcus pneumoniae in patients with community-acquired respiratory tract infections. Antimicrob. Agents Chemother. 45:2793–2797. 10.1128/AAC.45.10.2793-2797.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simoes AS, Pereira L, Nunes S, Brito-Avo A, de Lencastre H, Sa-Leao R. 2011. Clonal evolution leading to maintenance of antibiotic resistance rates among colonizing pneumococci in the PCV7 era in Portugal. J. Clin. Microbiol. 49:2810–2817. 10.1128/JCM.00517-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grivea IN, Tsantouli AG, Michoula AN, Syrogiannopoulos GA. 2011. Dynamics of Streptococcus pneumoniae nasopharyngeal carriage with high heptavalent pneumococcal conjugate vaccine coverage in central Greece. Vaccine 29:8882–8887. 10.1016/j.vaccine.2011.09.074 [DOI] [PubMed] [Google Scholar]
- 40.Domenech A, Ardanuy C, Tercero A, García-Somoza D, Santos S, Liñares J. 8 December 2013. Dynamics of the pneumococcal population causing acute exacerbations in COPD patients in a Barcelona hospital (2009-12): comparison with 2001-04 and 2005-08 periods. J. Antimicrob. Chemother. [Epub ahead of print.] 10.1093/jac/dkt476 [DOI] [PubMed] [Google Scholar]
- 41.Sanz JC, Cercenado E, Marín M, Ramos B, Ardanuy C, Rodríguez-Avial I, Bouza E. 2011. Multidrug-resistant pneumococci (serotype 8) causing invasive disease in HIV+ patients. Clin. Microbiol. Infect. 17:1094–1098. 10.1111/j.1469-0691.2011.03495.x [DOI] [PubMed] [Google Scholar]
- 42.Muñoz-Almagro C, Jordan I, Gene A, Latorre C, García-García JJ, Pallarés R. 2008. Emergence of invasive pneumococcal disease caused by nonvaccine serotypes in the era of 7-valent conjugate vaccine. Clin. Infect. Dis. 46:174–182. 10.1086/524660 [DOI] [PubMed] [Google Scholar]


