Abstract
Although bacterial mechanisms involved in the resistance to inorganic arsenic are well understood, the molecular basis for organic arsenic resistance has not been described. Campylobacter jejuni, a major food-borne pathogen causing gastroenteritis in humans, is highly prevalent in poultry and is reportedly resistant to the arsenic compound roxarsone (4-hydroxy-3-nitrobenzenearsonic acid), which has been used as a feed additive in the poultry industry for growth promotion. In this study, we report the identification of a novel membrane transporter (named ArsP) that contributes to organic arsenic resistance in Campylobacter. ArsP is predicted to be a membrane permease containing eight transmembrane helices, distinct from other known arsenic transporters. Analysis of multiple C. jejuni isolates from various animal species revealed that the presence of an intact arsP gene is associated with elevated resistance to roxarsone. In addition, inactivation of arsP in C. jejuni resulted in 4- and 8-fold reductions in the MICs of roxarsone and nitarsone, respectively, compared to that for the wild-type strain. Furthermore, cloning of arsP into a C. jejuni strain lacking a functional arsP gene led to 16- and 64-fold increases in the MICs of roxarsone and nitarsone, respectively. Neither mutation nor overexpression of arsP affected the MICs of inorganic arsenic, including arsenite and arsenate, in Campylobacter. Moreover, acquisition of arsP in NCTC 11168 led to accumulation of less roxarsone than the wild-type strain lacking arsP. Together, these results indicate that ArsP functions as an efflux transporter specific for extrusion of organic arsenic and contributes to the resistance to these compounds in C. jejuni.
INTRODUCTION
Campylobacter, a Gram-negative microaerobic bacterium, is a leading bacterial cause of food-borne diseases, and Campylobacter infections account for 400 to 500 million cases of diarrhea each year in developed and developing countries (1). A recent estimate from the CDC indicated that campylobacteriosis accounts for 9% of food-borne illness (over 840,000 cases) every year in the United States (2). Campylobacter jejuni and Campylobacter coli are the two most important Campylobacter species that cause food-borne infections of humans, and raw poultry meat serves as the main source of infection (3). Roxarsone, an organoarsenic compound, has been extensively used as a feed additive in the poultry industry to control bacterial and coccidial infections and improve weight gain, feed utilization, and pigmentation (4). Although it was recently withdrawn from poultry use in the United States, it has been estimated that roxarsone was utilized in approximately 70% of the U.S. broiler production units (4). The concentrations of roxarsone used in feed formulations vary from 22.7 to 45.4 g/ton (5). In animals, roxarsone is excreted into fresh litter and then converted to inorganic arsenate [As(V)] in composted litter via the bioconversion processes (5, 6). The total arsenic concentration in the litter may range from 15 to 48 mg/kg (5–7).
In bacteria, several mechanisms for arsenic detoxification have been characterized, including reduction of As(V) to As(III) by arsenate reductases and extrusion or methylation of As(III) (8–10). The arsenic detoxification systems are encoded by various ars genes, including arsA, arsB, arsC, arsD, arsH, arsM, and arsR. These ars genes can be organized as operons, such as arsRBC, arsRABC, and arsRDABC, or exist singly on bacterial chromosomes (9, 11–17). ArsA functions as an ATPase, which is an energy source for ArsB (18, 19), while ArsB functions as an As(III)-specific transporter (8, 9). As arsenate reductase, ArsC converts As(V) to As(III) in the cytoplasm (8, 20), and then As(III) is extruded by other As(III)-specific transporters (8, 9). ArsD is an arsenic metallochaperone which transfers As(III) to ArsA and increases the rate of arsenic extrusion (10, 21–23). ArsH is an NADPH-flavin mononucleotide oxidoreductase, and the detoxification mechanism is probably through oxidation of arsenite to the less toxic arsenate or reduction of trivalent arsenicals to volatile arsines that escape from cells (24, 25). ArsM is an As(III) S-adenosylmethionine methyltransferase, which methylates As(III) to volatile trimethylarsine (10). ArsR functions as a transcription regulator which controls the expression of other ars genes (9, 26–29).
Campylobacter is well adapted in the poultry production system and must have means to overcome the toxic effects of arsenic compounds. Recently, a four-gene ars operon associated with high-level resistance to inorganic arsenic was identified in Campylobacter (9). This operon encodes a putative membrane permease (ArsP), a transcriptional repressor (ArsR), an arsenate reductase (ArsC), and an efflux protein (Acr3). The expression of this operon is directly regulated by ArsR, which binds to the ars promoter DNA and controls the expression of the ars genes. Insertional mutation of acr3 and arsC revealed that the two genes contribute to arsenite and arsenate resistance but had no effects on the MIC of roxarsone in Campylobacter (9). However, the function of ArsP has not been characterized. In general, the molecular mechanisms directly involved in organic arsenic (roxarsone) resistance have not been described for any bacteria. In this study, we identify ArsP as a specific mechanism for organic arsenic resistance in Campylobacter.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The key bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5α used for genetic manipulation was grown in Luria-Bertani (LB) broth or on LB agar supplemented with kanamycin (30 μg/ml), chloramphenicol (10 μg/ml), or ampicillin (100 μg/ml). C. jejuni strains were cultured on Mueller-Hinton (MH) agar or in MH broth at 42°C microaerobically (5% O2, 10% CO2, and 85% N2). When needed, kanamycin (30 μg/ml) or chloramphenicol (4 μg/ml) was used to supplement the media.
TABLE 1.
Key plasmids and bacterial strains used in this study
| Bacterial strain or plasmid | Description or relevant genotype | Source or reference |
|---|---|---|
| Plasmids | ||
| pUC19 | Cloning vector | 41 |
| pArsPC | pUC19+arsP+cat | This study |
| pRY112 | Shuttle vector | 42 |
| pRY112+arsP | pRY112+arsP | This study |
| pRY112+4ars | pRY112 containing the 4-gene ars operon | This study |
| Strains | ||
| DH5α | Plasmid propagation E. coli strain | Invitrogen |
| NCTC 11168 | Wild-type C. jejuni | 43 |
| 11168+arsP | NCTC 11168 derivative, pRY112+arsP | This study |
| 11168+4ars | NCTC 11168 derivative, pRY112 + 4ars | This study |
| CB5-28 | Wild-type C. jejuni | 9 |
| CB5-28 ΔarsC | CB5-28 derivative, ΔarsC::Kanr | 9 |
| CB5-28 ΔarsP | CB5-28 derivative, ΔarsP::cat | This study |
Chemical compounds and antibiotics.
The chemicals and antibiotics used in this study were purchased from Sigma-Aldrich Co. LLC (arsenite, arsenate, nitarsone, arsanilic acid, chloramphenicol, kanamycin, streptomycin, and ampicillin) and Fisher Scientific Inc. (roxarsone and carbarsone).
Bioinformatics.
Protein Basic Local Alignment Search Tool (BLASTP) was used to search ArsP homologues in databases of nonredundant protein sequences. Constraint-based Multiple Protein Alignment Tool (COBALT) (30) was used to carry out multiple-sequence alignment and construct the phylogenetic tree. A biological sequence alignment editor (BioEdit) was used to output the multiple-sequence alignment result. TopPred 0.01 (31), MEMSAT-SVM (32), and TMHMM 2.0 (33) were used to predict the transmembrane helices (TMH) of ArsP. The visual representation of ArsP transmembrane topology was demonstrated by TMRPres2D (34). The sequence logo was generated using WebLogo, version 2.8.2 (35).
Antimicrobial susceptibility tests.
The MICs of various arsenic compounds against C. jejuni strains were determined using the agar dilution or broth dilution antimicrobial susceptibility testing methods according to the protocol from the CLSI (36). MICs of azithromycin, ciprofloxacin, erythromycin, gentamicin, tetracycline, florfenicol, nalidixic acid, telithromycin, and clindamycin were determined using commercially prepared Sensititre broth microdilution plates (Trek Diagnostic Systems, Cleveland, OH). Every MIC test was repeated at least three times. C. jejuni ATCC 33560 was used as a quality control organism in this study.
Construction of the arsP mutant.
Primers AspT-F and ArsPT-R (Table 2) were used to amplify a 1.8-kb arsP fragment with two PsiI sites in the middle region of the fragment. The PCR fragment was cloned into the pUC19 in the SmaI sites, resulting in the construction of pArsP. The chloramphenicol resistance cat cassette amplified from pUOA18 using the Phusion HighFidelity DNA polymerase (NEB) (37) was ligated into the PsiI site of arsP in pArsP to yield the pArsPC plasmid. Suicide vector pArsPC was introduced into C. jejuni CB5-28 using electroporation. Transformants were selected on MH agar containing chloramphenicol at 4 μg/ml. The insertion of cat cassette into the arsP gene was confirmed by PCR analysis.
TABLE 2.
Key primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| S1730-F | TATTTACTGAGCTTTCTGTACTTT |
| S1730-R | TAGCCACGCCTAAAATAATC |
| Ars1-F | CATTCATTTTAGAGTTATTGCGTATAAAACATACT |
| ArsPN-R | CCATGTTTAAGAGCTCTTGTAGATCAC |
| ArsPT-F | CATCCATATACAAACTTCCAATACCCC |
| ArsPT-R | CCTGCGGAAAATACTTCGATATCTATGCC |
| ArsPW-F | ACATACTGGTTCTAAATTTCTGATCAAACG |
| ArsPW-R | AATCCAGAAAGCAAGGACTATCAACCT |
Cloning of the arsP and the ars operon into C. jejuni NCTC 11168.
The entire arsP gene, including its promoter region, was amplified from strain CB5-28 by PCR using primers Ars1-F and ArsPN-R (Table 2). The PCR product was cloned into the SmaI-digested pRY112 plasmid to construct pRY112+arsP. pRY112+arsP was sequenced, and it was confirmed that no mutations in the cloned sequence had occurred. The shuttle plasmid, pRY112+arsP, was transferred into NCTC 11168 by conjugation. The resulting strain was named 11168+arsP. The entire ars operon, including arsP, arsR, arsC, and acr3, was amplified from strain CB5-28 by PCR using primers ArsPW-F and ArsPW-R. The PCR product was cloned into the SmaI-digested pRY112 plasmid to construct pRY112+4ars. pRY112+4ars was sequenced, and it was confirmed that no mutations in the cloned sequence had occurred. The shuttle plasmid pRY112+4ars was transferred into NCTC 11168 by conjugation. The resulting strain was named 11168+4ars.
PCR and sequence analysis of arsP.
To detect the distribution of arsP with in various C. jejuni isolates, the S1730-F and S1730-R primers (Table 2) were used to amplify an intragenic region of the arsP gene. Since the arsP gene in some strains (e.g., C. jejuni NCTC 11168) is a pseudogene, if a positive PCR product was obtained from the strain with low MIC for roxarsone, the whole coding region of arsP was amplified using another pair of primers, ArsPW-F and ArsPW-R, and the product was then sequenced in the DNA facility at Iowa State University. PCR was performed in a volume of 50 μl containing 0.2 μM primers, 250 μM deoxynucleoside triphosphates (dNTPs), and 1.25 U of TaKaRa Ex Taq polymerase. An annealing temperature of 50°C and an elongation time of 1 min were used for the primer pair S1730-F and S1730-R, while an annealing temperature of 55°C and an elongation time of 4 min were used for the primer pair ArsPW-F and ArsPW-R.
Roxarsone accumulation assay.
To determine if ArsP contributes to reduce intracellular concentration of roxarsone, an accumulation assay was conducted using C. jejuni NCTC 11168 and 11168+arsP. NCTC 11168 and 11168+arsP were inoculated on MH agar plates and incubated microaerobically for 24 h. Bacterial cells were collected from the plates, inoculated into fresh MH broth (200 ml), and then incubated at 42°C with shaking at 160 rpm. After 4 to 5 h of incubation, the cells were harvested by centrifugation, washed once in phosphate-buffered saline (PBS), and then resuspended in PBS to 1010∼11 CFU/ml. Three different tubes were prepared for each sample. After incubation at 37°C for 10 min, roxarsone was added to a final concentration of 500 μg/ml. Aliquots (1.0 ml) were taken after 15 min and immediately mixed with 5.0 ml of ice-cold PBS. Bacterial cells were harvested by centrifugation at 4°C and 6,000 × g for 5 min and washed twice with 10 ml of ice-cold PBS. The cells were resuspended in 0.5 ml of PBS and then disrupted by ultrasonication. The supernatant was taken after centrifugation at 15,000 × g for 5 min and filtered with a 0.45-μm filter. High-performance liquid chromatography (HPLC) analysis was performed on a Beckman Coulter 126 HPLC, equipped with a photodiode array detector (model 168) and a model 508 autosampler (Beckman Coulter, Inc., Brea, CA). The mobile phase was monopotassium phosphate (0.05 M)-acetic acid (10%)-methanol (90:7:3, vol/vol/vol) at a flow rate of 1.0 ml/min. A universal reversed-phase Supelcosil ABZ+Plus column (150 mm by 4.6 mm by 5 μm; Supelco, Bellefonte, PA) was used at room temperature. A standard curve was prepared by measuring the peak area from PBS containing serially diluted roxarsone and used to determine the concentration of roxarsone in the samples.
RESULTS
Arsenic resistance varies in Campylobacter strains of different origins.
To determine if arsenic resistance in Campylobacter is associated with the origin of isolation, we examined the MICs of various arsenic compounds in C. jejuni isolates from different animal species. In total, 131 C. jejuni isolates (35 from chickens, 27 from humans, 35 from sheep, and 34 from turkeys) were evaluated by the agar dilution method (Tables 3, 4, and 5). In general, MICs of roxarsone, arsenite, and arsenate ranged from 4 to 256 μg/ml, 1 to 256 μg/ml, and 16 to >1,024 μg/ml, respectively, and the MIC50 and MIC90 of the sheep isolates were lower than those of the chicken, human, and turkey isolates (Tables 3, 4, and 5). Statistical analysis with the Kruskal-Wallis test (nonparametric analysis of variance [ANOVA]) indicated that the MICs of C. jejuni from different animal species are significantly different in terms of resistance to roxarsone (P < 0.005), arsenite (P < 0.002), and arsenate (P < 0.0001). Furthermore, Dunn's multiple-comparison test indicated that the MICs of roxarsone for the sheep isolates are significantly lower than those for the human (P < 0.01), chicken (P < 0.05), and turkey (P < 0.05) isolates, the MICs of arsenite for the sheep isolates were significantly lower than those for the chicken isolates (P < 0.001), the MICs of arsenate for the sheep isolates were significantly lower than those for the chicken (P < 0.001) and turkey (P < 0.001) isolates, and the MICs of roxarsone, arsenite, and arsenate for the human, chicken, and turkey isolates were not significantly different (P > 0.05).
TABLE 3.
Roxarsone MIC distributions in C. jejuni isolates of different origins
| Isolate source | Total no. | No. of isolates inhibited by ROX concn (μg/ml) |
MIC50/MIC90 (μg/ml) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | |||
| Chicken | 35 | 2 | 11 | 7 | 4 | 11 | 0 | 0 | 0 | 16/64 |
| Human | 27 | 5 | 4 | 4 | 5 | 5 | 3 | 1 | 0 | 32/128 |
| Sheep | 35 | 1 | 23 | 6 | 3 | 2 | 0 | 0 | 0 | 8/32 |
| Turkey | 34 | 0 | 7 | 18 | 7 | 0 | 1 | 1 | 0 | 16/32 |
TABLE 4.
Arsenite MIC distributions in C. jejuni isolates of different origins
| Isolate source | Total no. | No. of isolates inhibited by As(III) concn (μg/ml) |
MIC50/MIC90 (μg/ml) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | |||
| Chicken | 35 | 0 | 2 | 1 | 8 | 9 | 7 | 8 | 0 | 0 | 16/64 |
| Human | 27 | 2 | 0 | 3 | 9 | 6 | 0 | 5 | 2 | 0 | 8/64 |
| Sheep | 35 | 0 | 0 | 8 | 23 | 0 | 4 | 0 | 0 | 0 | 8/32 |
| Turkey | 34 | 0 | 1 | 10 | 9 | 5 | 2 | 6 | 0 | 1 | 8/64 |
TABLE 5.
Arsenate MIC distributions in C. jejuni isolates of different origins
| Isolates | Total no. | No. of isolates inhibited by As(V) concn (μg/ml) |
MIC50/MIC90 (μg/ml) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 32 | 64 | 128 | 256 | 512 | 1,024 | >1,024 | |||
| Chicken | 35 | 0 | 5 | 10 | 1 | 9 | 6 | 4 | 0 | 256/1,024 |
| Human | 27 | 6 | 6 | 4 | 1 | 4 | 4 | 0 | 2 | 64/512 |
| Sheep | 35 | 6 | 19 | 2 | 2 | 6 | 0 | 0 | 0 | 32/256 |
| Turkey | 34 | 1 | 4 | 4 | 13 | 3 | 3 | 3 | 3 | 128/1,024 |
Genetic features of arsP.
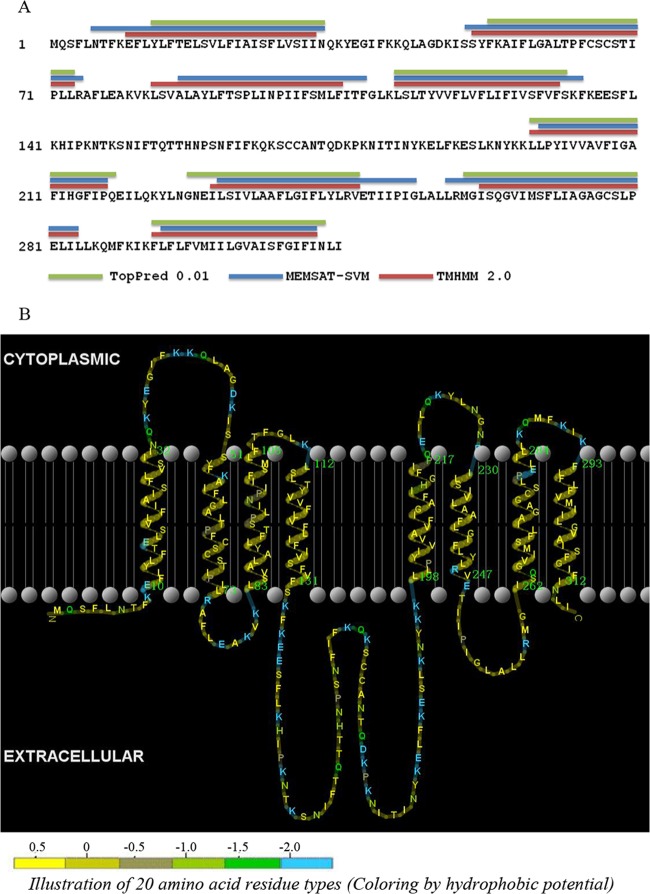
arsP (cje1730) is the first gene in the 4-gene ars operon and encodes a putative transmembrane permease (315 amino acids [aa]) (Fig. 1A). The transmembrane helices (TMH) of ArsP were predicted by using different algorithms, including TopPred 0.01, MEMSAT-SVM, and TMHMM 2.0. Generally, the three methods predicted similar topologies for ArsP. Results from MEMSAT-SVM and TMHMM 2.0 indicated that ArsP has eight putative TMH, while TopPred 0.01 failed to predict the third TMH (Fig. 2). All three methods predicted a large hydrophilic loop (∼60 aa) in the central portion (between the fourth and fifth helices) of the protein (Fig. 2). The large central hydrophilic loop is predicted to be outside the inner membrane, facing the periplasmic space. Thus, the predicted topology of ArsP is quite different from those of ArsB and Acr3. Additionally, ArsP shares little sequence homology with ArsB or Acr3, which are transporters for As(III). According to the transporter classification database (TCDB; http://www.tcdb.org), ArsP (TCID: 9.B.28.1.1) belongs to Duf318 family of putative permeases. The database indicates that none of the members of the Duf318 family has been functionally characterized. These observations suggest that ArsP may have a distinct substrate specificity. Based on BLAST search of the GenBank database, ArsP homologues were found in 27 bacterial and archaeal genera (see Fig. S1 in the supplemental material for phylogenetic tree), including at least 68 species. The amino acid lengths of the ArsP homologues range from 294 to 365 aa, and TMHMM 2.0 predicted that all of them contain similar TMH with large hydrophilic loops between the fourth and fifth TMH, ranging from 37 to 109 amino acids in length. Multiple-sequence alignment indicated that the ArsP homologues are highly conserved in the transmembrane regions but quite diverse in the central loop region (see Fig. S2 in the supplemental material). Using WebLogo, a highly conserved signature motif, TPFCSCSTIP, located in the second transmembrane domain was identified among the ArsP homologues (Fig. 3). Since arsP is in the ars operon, we hypothesized that ArsP plays a role in arsenic resistance in Campylobacter.
FIG 1.
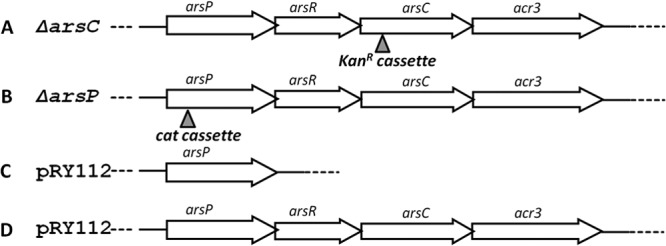
Diagrams showing the genomic organization of arsP and its flanking genes as well as cloning of the ars genes into C. jejuni NCTC 11168. (A) The ars operon in C. jejuni CB5-28 and inactivation of arsC by insertion of a kanamycin resistance cassette. (B) The ars operon in C. jejuni CB5-28 and inactivation of arsP by insertion of a choramphenicol resistance cassette. (C) Cloning of the single arsP gene from CB5-28 to shuttle plasmid pRY112, which was then transferred into NCTC 11168 by conjugation. (D) Cloning of the entire ars operon from CB5-28 to shuttle plasmid pRY112, which was then transferred into NCTC 11168 by conjugation.
FIG 2.
Predicted membrane topologies of ArsP. (A) The prediction of TMH was based on the amino acid sequence of ArsP in C. jejuni CB5-28 and was done by using three algorithms, including TopPred 0.01(green), MEMSAT-SVM (blue), and TMHMM 2.0 (red). (B) Visual representation of ArsP transmembrane topology. This is based on the TMHMM prediction and is shown by TMRPres2D. The hydrophobic potential of the 20 amino acid residue types is illustrated by different colors. The model predicts that a large hydrophilic loop (∼60 aa) is in the central portion (between the fourth and fifth helices) of ArsP.
FIG 3.
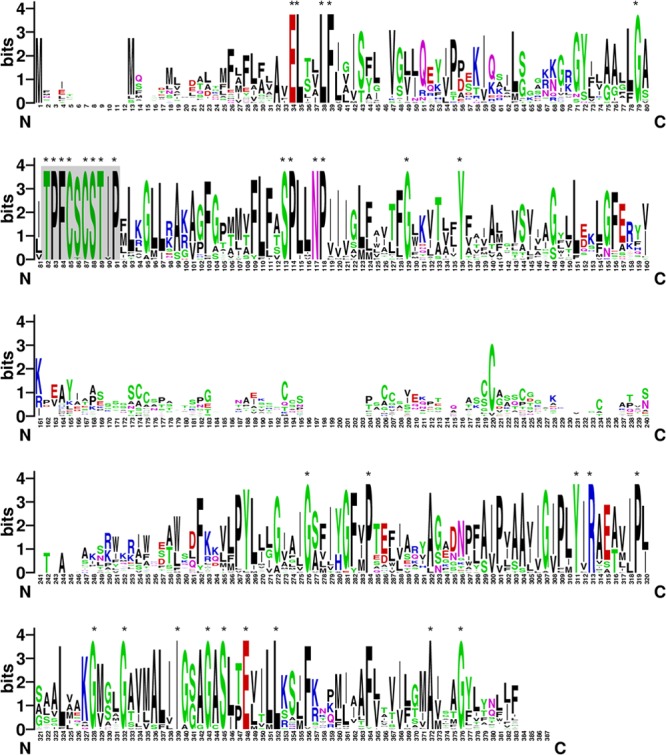
Identification of a signature motif among ArsP homologues by WebLogo. The sequence logo was constructed from the alignment of 27 ArsP homologues. The letter size is proportional to the degree of amino acid conservation. The position of signature motif TPFCSCSTIP is shaded gray.
Correlation of arsP with increased resistance to roxarsone in different C. jejuni strains.
To examine the association between arsP and the enhanced resistance to roxarsone, the presence of arsP in different C. jejuni isolates was determined using PCR. In total, 54 C. jejuni isolates (19 from humans, 21 from chickens, and 14 from turkeys) were used. Statistical analysis of the PCR and MIC data with Wilcoxon rank sum test with continuity correction indicated that the presence of an intact arsP gene is significantly (P < 0.001) associated with elevated resistance to roxarsone (Table 6). The PCR results also showed that an arsP gene is not always linked with the acr3 gene (member of 4-gene operon) (Table 6). Six of the strains (CT4-17, F1587, 11168, W28752, W56246, and X39768) are negative for both arsP and acr3 as determined by PCR amplification, while only 3 of the strains (CB5-28, RM1221, and S13530) are positive for both arsP and acr3. Notably, most of the strains contain either arsP or acr3 (Table 6). The association of arsP with the elevated MIC of roxarsone suggested its possible contribution to roxarsone resistance in C. jejuni.
TABLE 6.
Presence of arsP detected by PCR and its association with elevated resistance to roxarsone
| Strain | MIC (μg/ml) | Presence of intact arsPa | Source |
|---|---|---|---|
| CB3-15 | 8 | − | Chicken |
| CB3-18 | 8 | − | Chicken |
| CB3-23 | 8 | − | Chicken |
| CB3-6 | 8 | − | Chicken |
| CB3-9 | 8 | − | Chicken |
| CB7-22 | 8 | − | Chicken |
| CT3-19 | 16 | − | Turkey |
| CT3-7 | 8 | − | Turkey |
| CT4-10 | 16 | − | Turkey |
| CT4-15 | 16 | − | Turkey |
| CT4-17 | 32 | − | Turkey |
| CT4-4 | 16 | − | Turkey |
| CT4-6 | 16 | − | Turkey |
| CT5-1 | 16 | − | Turkey |
| CT5-10 | 16 | − | Turkey |
| CT5-12 | 16 | − | Turkey |
| CT5-2 | 8 | − | Turkey |
| CT5-9 | 8 | − | Turkey |
| E46972 | 128 | − | Human |
| F1587 | 4 | − | Human |
| M33323 | 8 | −b | Human |
| M36292 | 32 | − | Human |
| M402 | 16 | − | Human |
| M76297 | 16 | − | Human |
| NCTC 11168 | 8 | −b | Human |
| T37957A | 32 | − | Human |
| W28752 | 8 | − | Human |
| W56246 | 4 | − | Human |
| X39768 | 128 | − | Human |
| CB1-10 | 64 | + | Chicken |
| CB1-15 | 64 | + | Chicken |
| CB1-17 | 32 | + | Chicken |
| CB1-19 | 64 | + | Chicken |
| CB1-9 | 64 | + | Chicken |
| CB2-12 | 16 | + | Chicken |
| CB2-13 | 16 | + | Chicken |
| CB2-5 | 32 | + | Chicken |
| CB2-8 | 16 | + | Chicken |
| CB2-9 | 32 | + | Chicken |
| CB3-3 | 64 | + | Chicken |
| CB4-1 | 64 | + | Chicken |
| CB4-4 | 128 | + | Chicken |
| CB5-28 | 32 | + | Chicken |
| RM1221 | 64 | + | Chicken |
| CT3-2 | 32 | + | Turkey |
| CT4-20 | 128 | + | Turkey |
| H307369 | 32 | + | Human |
| H49024 | 64 | + | Human |
| M37523 | 32 | + | Human |
| S13530 | 64 | + | Human |
| W64861 | 128 | + | Human |
| X77136 | 16 | + | Human |
| cjs47645 | 64 | + | Human |
| Clev9100 | 64 | + | Human |
−, absence of genes by PCR detection; +, presence of genes by PCR detection.
Presence of frameshift mutations in arsP.
Role of arsP in the arsenic resistance.
To determine the role of arsP in arsenic resistance in C. jejuni, insertional mutagenesis was used to construct an isogenic mutant. The arsP mutant was compared with the parent strain CB5-28 and CB5-28 ΔarsC for susceptibility to arsenic compounds using the agar dilution method. According to MIC results from the agar dilution method, inactivation of arsP resulted in a 4-fold reduction in the MICs of roxarsone, arsenite, and arsenate, respectively, while inactivation of the downstream arsC gene resulted in 4-fold and 32-fold reductions in the MICs of arsenite and arsenate, respectively, but did not affect the MIC of roxarsone (Table 7). This finding suggests that arsP is associated with roxarsone resistance in Campylobacter. Since the arsP mutation might cause a polar effect on the expression of the downstream genes in the ars operon, we further analyzed the function of arsP by cloning this gene into NCTC 11168, which lacks arsP and does not contain a functional ars operon (Fig. 1). According to the MIC results, acquisition of the single arsP gene in NCTC 11168 resulted in an 16-fold increase in the MICs of roxarsone but had no effects on the MICs of arsenite or arsenate (Table 7), indicating that arsP specifically contributes to the resistance to roxarsone but not to arsenite or arsenate. When the entire ars operon was cloned into NCTC 11168 (Fig. 1), it resulted in 8-fold, 8-fold, and 2-fold increases in the MICs of roxarsone, arsenite, and arsenate, respectively. In addition to roxarsone, we also examined the MICs of three other organoarsenic compounds: arsanilic acid, carbarsone, and nitarsone. The results indicated that inactivation of arsP in CB5-28 resulted in 2-fold and 8-fold reductions in the MICs of arsanilic acid and nitarsone, respectively, but did not affect the MIC of carbarsone (Table 7). In addition, acquisition of the single arsP gene in NCTC 11168 resulted in 2-fold and 64-fold increases in the MICs of arsanilic acid and nitarsone, respectively, but did not affect the MIC of carbarsone (Table 7). These results demonstrate the specific and varied role of ArsP in the resistance to organoarsenic compounds.
TABLE 7.
MICs of roxarsone, arsenite, arsenate, arsanilic acid, carbarsone, and nitarsone in various C. jejuni constructs
| Strain | MIC (μg/ml)a |
|||||
|---|---|---|---|---|---|---|
| Roxarsone | Arsenite | Arsenate | Arsanilic acid | Carbarsone | Nitarsone | |
| NCTC 11168 | 8 | 16 | 1,024 | 1,250 | 5,000 | 4 |
| 11168+arsP | 128 (↑16) | 16 | 1,024 | 2,500 (↑2) | 5,000 | 256 (↑64) |
| 11168+4ars | 64 (↑8) | 128 (↑8) | 2,048 (↑2) | 2,500 (↑2) | 5,000 | 128 (↑32) |
| CB5-28 | 32 | 64 | 2,048 | 2,500 | 5,000 | 32 |
| CB5-28 ΔarsP | 8 (↓4) | 16 (↓4) | 512 (↓4) | 1,250 (↓2) | 5,000 | 4 (↓8) |
| CB5-28 ΔarsC | 32 | 16 (↓4) | 64 (↓32) | 2,500 | 5,000 | 32 |
The numbers in parentheses indicate fold changes over the wild-type control, either increased (↑) or decreased (↓).
ArsP did not confer resistance to the other antibiotics.
To examine if ArsP contributes to the resistance of other antimicrobials, we compared the susceptibilities of the wild-type and genetically modified Campylobacter strains to azithromycin, ciprofloxacin, erythromycin, gentamicin, tetracycline, florfenicol, nalidixic acid, telithromycin, and clindamycin using commercially prepared Sensititre broth microdilution plates. The results showed no differences between the wild type and the genetically modified strains in the susceptibilities to these compounds (data not shown), indicating that arsP does not confer resistance to these tested antibiotics.
The arsP gene is not inducible by roxarsone.
To determine if the transcriptional level of arsP gene is inducible by roxarsone, C. jejuni CB5-28 was cultured in MH broth with different concentrations of roxarsone. The transcriptional level of arsP in cultures supplemented with or without roxarsone did not differ as measured by quantitative reverse transcription-PCR (qRT-PCR) (data not shown). This result suggests that arsP is not inducible by roxarsone.
ArsP reduces intracellular accumulation of roxarsone.
An accumulation assay using HPLC was performed to assess whether ArsP functions as an efflux transporter for roxarsone. The wavelength was 337 nm and the retention time was 5.2 min for roxarsone (Fig. 4B). According to the standard curve, a linear regression equation was obtained: y = (x − 287.54)/11,176, with an R2 of 0.9999, where y is the concentration of roxarsone and x is the peak area of the sample. Using this equation, we were able to measure the concentrations of roxarsone in different samples. The NCTC 11168 wild type, which does not have a functional arsP gene, accumulated 44.8% more roxarsone than 11168+arsP (containing an intact arsP gene) (Fig. 4A). This result suggests that ArsP functions as an efflux transporter, extruding roxarsone out of Campylobacter cells.
FIG 4.
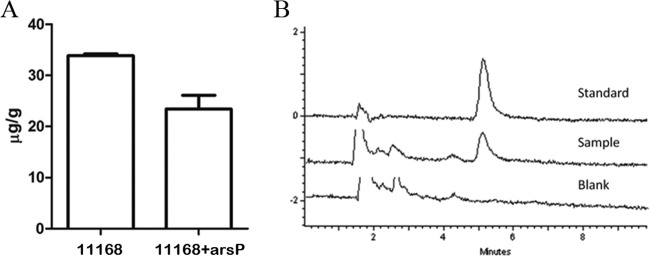
Intracellular accumulation of roxarsone as measured by HPLC. (A) Accumulation of roxarsone in wild-type NCTC 11168 and 11168+arsP. Each bar represents the mean and standard deviation of triplicate samples in one representative experiment (P < 0.02). (B) Representative chromatogram of the HPLC assay to determine roxarsone accumulation in Campylobacter. The peak at 5.2 min represents roxarsone. The standard is PBS buffer with added roxarsone. The sample is a PBS cell lysate of NCTC 11168 treated with roxarsone. The blank control is a PBS cell lysate of NCTC 11168.
DISCUSSION
The results from this study identify ArsP as a novel membrane transporter mediating resistance to organic arsenic in C. jejuni. This conclusion is supported by multiple pieces of evidence. The presence of ArsP is associated with elevated MICs of roxarsone (Table 6). Inactivation of arsP resulted in 2-, 8-, and 4-fold reductions in the MICs of arsanilic acid, nitarsone, and roxarsone, respectively, while cloning of arsP into a C. jejuni strain lacking a functional arsP showed 2-fold, 64-fold, and 16-fold increases in the MICs of arsanilic acid, nitarsone, and roxarsone, respectively, but had no effect on the MICs of inorganic arsenic compounds and antibiotics. Additionally, ArsP reduced intracellular accumulation of roxarsone in C. jejuni, and the presence of the arsP gene is associated with elevated resistance to roxarsone in various strains. These results indicate that ArsP functions as a resistance mechanism specific for organic arsenic. This finding enriches our knowledge of various mechanisms for arsenic resistance in Campylobacter. Based on these results and previously known information regarding arsenic resistance in Campylobacter, a model showing the various arsenical detoxification mechanisms is presented in Fig. 5.
FIG 5.
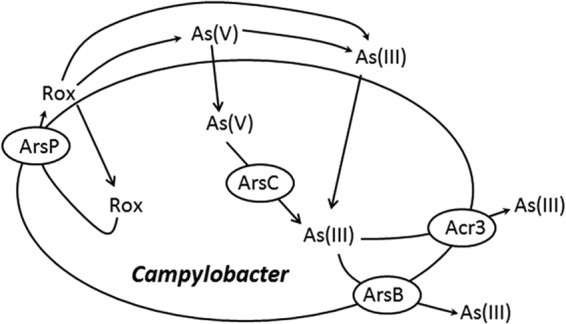
Model illustrating the currently known arsenical detoxification mechanisms in Campylobacter. Campylobacter has an arsenic reductase (ArsC), which converts arsenate to arsenite, two types of arsenite transporters (ArsB and Acr3), which extrude arsenite, and the ArsP transporter, which specifically extrudes roxarsone (Rox) out of the cells.
Due to the use of organoarsenics in poultry production, Campylobacter spp. in poultry must be able to deal with the toxicity and selective pressure from arsenic compounds. A previous study indicated that the Campylobacter isolates from conventional poultry products had significantly higher roxarsone MICs than those isolated from antimicrobial-free poultry products, suggesting the association of the roxarsone use in conventional poultry facilities with the development of roxarsone resistance in Campylobacter spp. in poultry (38). In this study, we demonstrated that the presence of an intact arsP gene is associated with increased resistance to roxarsone in various C. jejuni isolates (Table 6). This observation plus the findings from the mutagenesis and cloning experiments established ArsP as a key resistance mechanism for roxarsone, providing a molecular explanation for the prevalence of roxarsone-resistant Campylobacter in poultry. Although arsP is found to be in the same operon as arsC and acr3 in certain C. jejuni strains, it is functionally distinct from the other two genes, which confer resistance to arsenite and arsenate (9). Additionally, it was found that arsP does not have to be associated with arsC and acr3, which was also shown in a recent study by Noormohamed and Fakhr on the prevalence of ars genes in C. jejuni isolates from retail meats (39). These findings suggest that arsP evolved differently from other ars genes. Interestingly, the study by Noormohamedemail and Fakhr (39) reported a much higher detection rate (94.7%) of arsP in their C. jejuni isolates than that reported in this study (∼50%). The difference in the arsP detection rates is probably due to the different origins of the isolates.
In this study, we tested the MICs of arsenic compounds for the C. jejuni isolates from chickens, turkeys, humans, and sheep. An interesting finding is that the MICs of roxarsone for the sheep isolates were significantly lower than those for isolates from humans, chickens, and turkeys. This is probably due to the fact that the sheep industry does not use roxarsone as a feed additive, while roxarsone was extensively used on chicken and turkey farms in the United States. Although the use of roxarsone was banned in the European Union in 1999 and the manufacturer voluntarily suspended sale of roxarsone in the United States in 2011, roxarsone is still approved for use in poultry and livestock industry in many other countries (http://www.fda.gov/AnimalVeterinary/SafetyHealth/ProductSafetyInformation/ucm257540.htm). Interestingly, human C. jejuni isolates showed a level of roxarsone resistance similar to that of the turkey and chicken isolates. This could be explained by the fact that poultry is the main reservoir for human C. jejuni infections.
Notably, ArsP shows substrate specificity, as mutation of the gene or acquisition of the gene only affected the MICs of organic arsenic and did not have any effect on the MICs of inorganic arsenic and other antibiotics (Table 7). Even among the organoarsenic compounds examined in this study, ArsP was more effective against roxarsone and nitarsone than arsanilic acid and had no effect on carbarsone (Table 7). The difference in the effects is likely due to the structural differences among the organoarsenic compounds. Specifically, the structural differences among the tested compounds lie in the modifications of the benzene ring by a nitro group (roxarsone and nitarsone), an amino group (arsanilic acid), or a carbamide group (carbarsone). Thus, it appears that ArsP functions more efficiently with phenylarsenic compounds modified by nitro groups than with those modified by other groups. It should be pointed out that we tested only a limited number of organoarsenic compounds and did not evaluate the ability of ArsP to transport 4-hydroxy-3-nitrobenzene or related nitroaromatic compounds. Considering the wide distribution of ArsP homologues in various bacterial organisms, it is possible that ArsP has a function beyond organoarsenic resistance. This possibility remains to be determined in future studies.
Compared to other known arsenic efflux transporters (such as ArsB and Acr3), ArsP is structurally unique in having 8 transmembrane domains and a large hydrophilic and centrally located loop (Fig. 2). These structural features may have determined the substrate specificity of ArsP. Interestingly, the ArsP in C. jejuni contains 5 cysteines, which are in positions 65, 67, 169, 170, and 277 (Fig. 2A). 169C and 170C are located in the nonconserved central loop region, while 65C and 67C are located in the highly conserved signature motif of the second transmembrane domain (Fig. 2; see also Fig. S2 in the supplemental material). It is unknown if they are redox reactive and if they are involved in the interaction with the substrates. Further structural and chemical analyses are required to answer these questions.
Although ArsP contributes to organoarsenic resistance, its expression is not inducible by roxarsone. Previously it was revealed that arsP is regulated by ArsR, which is encoded in the ars operon that includes arsP, arsC, and acr3 (9). The induction of the ars operon is through the inhibition of ArsR binding to the promoter DNA, which is mediated by conformational change in ArsR triggered by arsenite binding (40). Thus, arsenite is the true inducer of ArsR, and the arsenate-mediated induction is via bioconversion of arsenate to arsenite by ArsC reductase in Campylobacter. In this study, the presence of roxarsone in culture did not induce the expression of arsP. This result indicates that roxarsone does not interact with ArsR and does not directly induce the ars operon. However, roxarsone can be converted to inorganic arsenic in chicken litter through biological processes (5) and is a potential indirect inducer for the ars operon in the poultry production environment. Additionally, the MICs of roxarsone for the isolates containing the arsP genes range from 16 to 128 μg/ml (Table 6). The wide range of MICs suggests the differential expression of arsP or the existence of unknown regulatory mechanisms for arsP in those isolates. This possibility remains to be determined in future studies.
The results from the roxarsone accumulation assay (Fig. 4) suggest that AsrP functions as an efflux transporter. This finding is consistent with the predicted function of ArsP as a permease. However, the energy source required for ArsP to transport organic arsenic is unknown at this stage. According to the TCDB database (http://www.tcdb.org), ArsP is distantly related to both primary (ABC transporter ThiW; 3.A.1.26.2) and secondary (TrpP; 2.A.88.4.1) transporters. A motif search did not identify an ATP-binding site in ArsP, but this does not exclude the possibility that ArsP utilizes ATP by partnering with a ATPase. Additionally, we compared the MICs of roxarsone and nitarsone in C. jejuni CB5-28 in the presence and absence of carbonyl cyanide m-chlorophenylhydrazine (CCCP), an inhibitor of the membrane proton gradient, and found that the presence of CCCP made no difference in the MICs (data not shown). This result suggests that ArsP may not utilize the proton gradient for transport of organic arsenic. The energy sources for ArsP function remain to be determined in future studies.
In summary, our study identified a novel transporter that contributes to organic arsenic resistance in Campylobacter. To the best of our knowledge, ArsP is the first characterized mechanism for bacterial detoxification of organic arsenic and appears to be quite different in structure and function from previously known transporters for inorganic arsenic, such as ArsB and Acr3. Additionally, ArsP represents the first functionally characterized member in the Duf318 family of transporters. Given that ArsP homologues are present in a number of bacterial and archaeal species and that they are highly conserved in their transmembrane topologies, it is possible that they may play a similar role in the resistance to organic arsenic or similar compounds. Thus, findings in this study may be useful for elucidating functions of ArsP homologues in other organisms. Furthermore, the identified role of ArsP in organic arsenic resistance suggests that it likely contributes to the ecological fitness of Campylobacter in poultry and livestock species, in which organic arsenic compounds are commonly used for production. Finally, results from this study and our previous work (9) indicate that the arsenic resistance mechanisms in Campylobacter do not confer cross-resistance to clinically used antibiotics. These findings significantly expand our knowledge of antimicrobial resistance in a major food-borne pathogen, reveal a new mechanism for organic arsenic resistance in bacterial organisms, and provide new insights into the adaptive mechanisms of Campylobacter in animal reservoirs.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by grant number R01DK063008 from the National Institute of Diabetes and Digestive and Kidney Diseases. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the funding agencies.
We thank Bing Wang (Iowa State University) for help in statistical analysis.
Footnotes
Published ahead of print 13 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02137-13.
REFERENCES
- 1.Ruiz-Palacios GM. 2007. The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 44:701–703. 10.1086/509936 [DOI] [PubMed] [Google Scholar]
- 2.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15. 10.3201/eid1701.09-1101p1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humphrey T, O'Brien S, Madsen M. 2007. Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 117:237–257. 10.1016/j.ijfoodmicro.2007.01.006 [DOI] [PubMed] [Google Scholar]
- 4.Chapman HD, Johnson ZB. 2002. Use of antibiotics and roxarsone in broiler chickens in the USA: analysis for the years 1995 to 2000. Poult. Sci. 81:356–364 [DOI] [PubMed] [Google Scholar]
- 5.Garbarino JR, Bednar AJ, Rutherford DW, Beyer RS, Wershaw RL. 2003. Environmental fate of roxarsone in poultry litter. I. Degradation of roxarsone during composting. Environ. Sci. Technol. 37:1509–1514. 10.1021/es026219q [DOI] [PubMed] [Google Scholar]
- 6.Stolz JF, Perera E, Kilonzo B, Kail B, Crable B, Fisher E, Ranganathan M, Wormer L, Basu P. 2007. Biotransformation of 3-nitro-4-hydroxybenzene arsonic acid (roxarsone) and release of inorganic arsenic by Clostridium species. Environ. Sci. Technol. 41:818–823. 10.1021/es061802i [DOI] [PubMed] [Google Scholar]
- 7.Jackson BP, Bertsch PM. 2001. Determination of arsenic speciation in poultry wastes by IC-ICP-MS. Environ. Sci. Technol. 35:4868–4873. 10.1021/es0107172 [DOI] [PubMed] [Google Scholar]
- 8.Rosen BP. 2002. Biochemistry of arsenic detoxification. FEBS Lett. 529:86–92. 10.1016/S0014-5793(02)03186-1 [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Jeon B, Sahin O, Zhang Q. 2009. Identification of an arsenic resistance and arsenic-sensing system in Campylobacter jejuni. Appl. Environ. Microbiol. 75:5064–5073. 10.1128/AEM.00149-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C. 2006. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 103:2075–2080. 10.1073/pnas.0506836103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butcher BG, Deane SM, Rawlings DE. 2000. The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli. Appl. Environ. Microbiol. 66:1826–1833. 10.1128/AEM.66.5.1826-1833.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butcher BG, Rawlings DE. 2002. The divergent chromosomal ars operon of Acidithiobacillus ferrooxidans is regulated by an atypical ArsR protein. Microbiology 148:3983–3992 [DOI] [PubMed] [Google Scholar]
- 13.Cai J, Salmon K, DuBow MS. 1998. A chromosomal ars operon homologue of Pseudomonas aeruginosa confers increased resistance to arsenic and antimony in Escherichia coli. Microbiology 144(Part 10):2705–2713 [DOI] [PubMed] [Google Scholar]
- 14.Diorio C, Cai J, Marmor J, Shinder R, DuBow MS. 1995. An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J. Bacteriol. 177:2050–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López-Maury L, Florencio FJ, Reyes JC. 2003. Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 185:5363–5371. 10.1128/JB.185.18.5363-5371.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato T, Kobayashi Y. 1998. The ars operon in the skin element of Bacillus subtilis confers resistance to arsenate and arsenite. J. Bacteriol. 180:1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki K, Wakao N, Kimura T, Sakka K, Ohmiya K. 1998. Expression and regulation of the arsenic resistance operon of Acidiphilium multivorum AIU 301 plasmid pKW301 in Escherichia coli. Appl. Environ. Microbiol. 64:411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dey S, Rosen BP. 1995. Dual mode of energy coupling by the oxyanion-translocating ArsB protein. J. Bacteriol. 177:385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saltikov CW, Olson BH. 2002. Homology of Escherichia coli R773 arsA, arsB, and arsC genes in arsenic-resistant bacteria isolated from raw sewage and arsenic-enriched creek waters. Appl. Environ. Microbiol. 68:280–288. 10.1128/AEM.68.1.280-288.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji G, Silver S. 1992. Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc. Natl. Acad. Sci. U. S. A. 89:9474–9478. 10.1073/pnas.89.20.9474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ajees AA, Yang J, Rosen BP. 2011. The ArsD As(III) metallochaperone. Biometals 24:391–399. 10.1007/s10534-010-9398-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Rawat S, Stemmler TL, Rosen BP. 2010. Arsenic binding and transfer by the ArsD As(III) metallochaperone. Biochemistry 49:3658–3666. 10.1021/bi100026a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin YF, Walmsley AR, Rosen BP. 2006. An arsenic metallochaperone for an arsenic detoxification pump. Proc. Natl. Acad. Sci. U. S. A. 103:15617–15622. 10.1073/pnas.0603974103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neyt C, Iriarte M, Thi VH, Cornelis GR. 1997. Virulence and arsenic resistance in yersiniae. J. Bacteriol. 179:612–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye J, Yang HC, Rosen BP, Bhattacharjee H. 2007. Crystal structure of the flavoprotein ArsH from Sinorhizobium meliloti. FEBS Lett. 581:3996–4000. 10.1016/j.febslet.2007.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenstein R, Nikoleit K, Gotz F. 1994. Binding of ArsR, the repressor of the Staphylococcus xylosus (pSX267) arsenic resistance operon to a sequence with dyad symmetry within the ars promoter. Mol. Gen. Genet. 242:566–572. 10.1007/BF00285280 [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Rosen BP. 1991. The ArsR protein is a trans-acting regulatory protein. Mol. Microbiol. 5:1331–1336. 10.1111/j.1365-2958.1991.tb00779.x [DOI] [PubMed] [Google Scholar]
- 28.Zhang YB, Monchy S, Greenberg B, Mergeay M, Gang O, Taghavi S, van der Lelie D. 2009. ArsR arsenic-resistance regulatory protein from Cupriavidus metallidurans CH34. Antonie Van Leeuwenhoek 96:161–170. 10.1007/s10482-009-9313-z [DOI] [PubMed] [Google Scholar]
- 29.Murphy JN, Saltikov CW. 2009. The ArsR repressor mediates arsenite-dependent regulation of arsenate respiration and detoxification operons of Shewanella sp. strain ANA-3. J. Bacteriol. 191:6722–6731. 10.1128/JB.00801-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papadopoulos JS, Agarwala R. 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23:1073–1079. 10.1093/bioinformatics/btm076 [DOI] [PubMed] [Google Scholar]
- 31.Claros MG, von Heijne G. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685–686 [DOI] [PubMed] [Google Scholar]
- 32.Nugent T, Jones DT. 2009. Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics 10:159. 10.1186/1471-2105-10-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 34.Spyropoulos IC, Liakopoulos TD, Bagos PG, Hamodrakas SJ. 2004. TMRPres2D: high quality visual representation of transmembrane protein models. Bioinformatics 20:3258–3260. 10.1093/bioinformatics/bth358 [DOI] [PubMed] [Google Scholar]
- 35.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. CLSI M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 37.Shen Z, Han J, Wang Y, Sahin O, Zhang Q. 2013. The contribution of ArsB to arsenic resistance in Campylobacter jejuni. PLoS One 8:e58894. 10.1371/journal.pone.0058894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sapkota AR, Price LB, Silbergeld EK, Schwab KJ. 2006. Arsenic resistance in Campylobacter spp. isolated from retail poultry products. Appl. Environ. Microbiol. 72:3069–3071. 10.1128/AEM.72.4.3069-3071.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noormohamed A, Fakhr MK. 2013. Arsenic resistance and prevalence of arsenic resistance genes in Campylobacter jejuni and Campylobacter coli isolated from retail meats. Int. J. Environ. Res. Public Health 10:3453–3464. 10.3390/ijerph10083453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi W, Dong J, Scott RA, Ksenzenko MY, Rosen BP. 1996. The role of arsenic-thiol interactions in metalloregulation of the ars operon. J. Biol. Chem. 271:9291–9297. 10.1074/jbc.271.16.9291 [DOI] [PubMed] [Google Scholar]
- 41.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. 10.1016/0378-1119(85)90120-9 [DOI] [PubMed] [Google Scholar]
- 42.Yao R, Alm RA, Trust TJ, Guerry P. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127–130. 10.1016/0378-1119(93)90355-7 [DOI] [PubMed] [Google Scholar]
- 43.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668. 10.1038/35001088 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.