Abstract
Pterostilbene (PTE) is a stilbene-derived phytoalexin that originates from several natural plant sources. In this study, we evaluated the activity of PTE against Candida albicans biofilms and explored the underlying mechanisms. In 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) reduction assays, biofilm biomass measurement, confocal laser scanning microscopy, and scanning electron microscopy, we found that ≤16 μg/ml PTE had a significant effect against C. albicans biofilms in vitro, while it had no fungicidal effect on planktonic C. albicans cells, which suggested a unique antibiofilm effect of PTE. Then we found that PTE could inhibit biofilm formation and destroy the maintenance of mature biofilms. At 4 μg/ml, PTE decreased cellular surface hydrophobicity (CSH) and suppressed hyphal formation. Gene expression microarrays and real-time reverse transcription-PCR showed that exposure of C. albicans to 16 μg/ml PTE altered the expression of genes that function in morphological transition, ergosterol biosynthesis, oxidoreductase activity, and cell surface and protein unfolding processes (heat shock proteins). Filamentation-related genes, especially those regulated by the Ras/cyclic AMP (cAMP) pathway, including ECE1, ALS3, HWP1, HGC1, and RAS1 itself, were downregulated upon PTE treatment, indicating that the antibiofilm effect of PTE was related to the Ras/cAMP pathway. Then, we found that the addition of exogenous cAMP reverted the PTE-induced filamentous growth defect. Finally, with a rat central venous catheter infection model, we confirmed the in vivo activity of PTE against C. albicans biofilms. Collectively, PTE had strong activities against C. albicans biofilms both in vitro and in vivo, and these activities were associated with the Ras/cAMP pathway.
INTRODUCTION
Candida albicans is the most common fungal pathogen, and it causes superficial to life-threatening infections (1). Infections caused by C. albicans remain the predominant nosocomial fungal infections, due to the increasing population of patients whose immune systems are compromised by AIDS or immunosuppressant or anticancer therapy (2, 3). The number of available antifungal drugs is limited, and repeated exposure to these limited antifungal agents has led to the rapid development of drug resistance (4).
Biofilms are microbial communities composed of cells adhering to abiotic or biotic surfaces, and these adherent cells are frequently embedded in a polymeric extracellular matrix (5). Unlike planktonic cells, C. albicans biofilms display resistance to a wide variety of clinical antifungal agents, including amphotericin B and fluconazole (6, 7), and they play a contributing role in the process of C. albicans infections (8, 9). Therefore, there is an urgent need to develop new antifungal agents against C. albicans biofilms.
Pterostilbene (PTE) (Fig. 1) is a stilbene-derived phytoalexin that originates from several natural plant sources, such as Pterocarpus marsupium (the Indian kino tree), Pterocarpus santalinus (red sandalwood), Vitis vinifera (common grape vine), and Vaccinium ashei (rabbiteye blueberry) (10–13). It has been reported that PTE has potential health benefits as an anticancer, anti-inflammatory, antioxidant, and analgesic agent (14–17). Meanwhile, it has strong activity against some plant fungal pathogens, such as Phomopsis viticola, Phomopsis obscurans, and Botrytis cinerea (18–20). Nevertheless, its activity against C. albicans has not yet been investigated.
FIG 1.
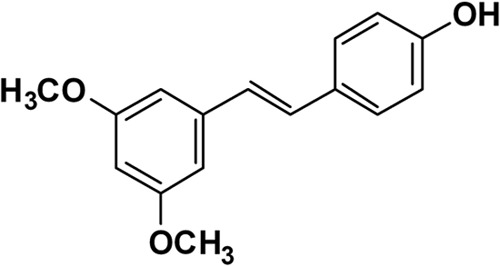
Chemical structure of PTE.
In this study, we evaluated the activities of PTE against C. albicans biofilms in vitro and in vivo, and we found that the antibiofilm activities of PTE are associated with the Ras/cAMP pathway.
MATERIALS AND METHODS
Strains, culture, and agents.
C. albicans strains SC5314, Y0109, 0304103, and 01010 were used in this study. Strains were routinely grown in YPD (1% yeast extract, 2% peptone, and 2% dextrose) liquid medium at 30°C in a shaking incubator. PTE was synthesized by the Guangzhou Institute of Chemistry of the Chinese Academy of Sciences (Guangzhou, China). The level of purity of the synthesized PTE was ≥99% based on high-performance liquid chromatography (21). For in vitro experiments, 6.4 mg/ml PTE in dimethyl sulfoxide (DMSO) was used as a stock and added to the culture suspensions to obtain the required PTE concentrations. For in vivo experiments, PTE was dissolved in 0.3 M 2-hydroxypropyl-β-cyclodextrin (HP-β-CyD) solution prepared with isotonic saline (22). Other media used included YPD plus 10% (vol/vol) fetal calf serum (FCS) (23), Spider medium (23), Lee's medium (pH 6.8) (24), synthetic low-ammonium dextrose (SLAD) medium (25), and RPMI 1640 medium (26).
Time-kill curve assay.
For the time-kill assay, exponentially growing C. albicans cells were washed with phosphate-buffered saline (PBS), resuspended with RPMI 1640 medium to 1 × 106 cells/ml, and divided into 4 cultures. Different concentrations of PTE were added to the C. albicans suspensions. The samples were cultured at 30°C under constant shaking (200 rpm). Portions of cell suspensions were withdrawn at predetermined time points (0, 2, 4, and 6 h). Serial dilutions were plated on YPD agar to determine the CFU/ml of the cell suspensions. CFU were determined after incubation for 48 h at 30°C. Three independent experiments were performed (27).
Antifungal susceptibility testing.
Antifungal susceptibility testing was carried out in 96-well microtiter plates (Greiner, Germany) using the broth microdilution protocol of the Clinical and Laboratory Standards Institute methods (M27-A3) with a few modifications (26, 28, 29). Briefly, the initial concentration of fungal suspension in RPMI 1640 medium was 103 CFU/ml, and the final concentrations ranged from 0.125 to 64 μg/ml for fluconazole and 0.5 to 32 μg/ml for PTE. Plates were incubated at 35°C for 24 h. The growth inhibition was determined by using a spectrophotometer. The optical density at 630 nm (OD630) was measured, and background optical densities were subtracted from that of each well. Each strain was tested in triplicate. The MIC80s were determined, i.e., the lowest concentrations of the drugs that inhibited growth by 80%.
In vitro biofilm formation assay.
The in vitro biofilm formation assay was carried out as described by Ramage et al. (30) with slight modifications. Briefly, the assay was performed in a 96-well tissue culture plate by seeding with 100-μl aliquots of cell suspensions (1.0 × 106 cells/ml) in RPMI 1640 medium and incubating them statically at 37°C. After 90 min for adhesion, the medium was aspirated, nonadherent cells were removed, and fresh RPMI 1640 was added. The plate was further incubated at 37°C for 24 h until formation of mature biofilms. A semiquantitative measure of the formed biofilms was calculated with a 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) reduction assay (31). To detect the effect of PTE on the formation of biofilms, different concentrations of PTE were added to fresh RPMI 1640 after 90 min of adhesion. The plates were further incubated at 37°C for 24 h. To detect the effect of PTE on mature biofilms, C. albicans biofilms were formed at 37°C for 24 h as described above. The biofilm supernatant was then discarded, and fresh RPMI 1640 medium containing different concentrations of PTE was added. The plates were incubated at 37°C for an additional 24 h to observe the antibiofilm effect of PTE.
Measurement of biofilm biomass.
Biofilm biomass was measured as described by Nobile et al. (32) with slight modifications. Sterile 1.5- by 1.5-cm silicone squares were weighed, pretreated with bovine serum overnight, and washed with PBS before inoculation. Exponentially growing C. albicans cells were diluted to an OD600 of 0.2 with Spider medium, and the suspension was added to a sterile 12-well plate with one prepared silicone square in each well. The inoculated plate was incubated with gentle agitation (150 rpm) for 90 min at 37°C until adhesion occurred. To remove nonadherent cells, the squares were washed with 2 ml PBS and then moved to a fresh 12-well plate containing 2 ml fresh Spider medium. For PTE treatment groups, PTE was added to the fresh Spider medium. The plate was incubated at 37°C for an additional 60 h at 75 rpm agitation to allow biofilm formation. The silicone squares with attached biofilms were removed from the wells, dried overnight, and weighed the following day. The total biomass of each biofilm was calculated by subtracting the weight of the silicone prior to biofilm growth from the weight of the silicone after the drying period, with adjustment for the weight of control silicone squares exposed to no cells. The mean dry biomass was calculated from five independent samples. Statistical significance was determined by an analysis of variance (ANOVA). A P value of less than 0.05 was considered statistically significant.
CLSM.
Confocal laser scanning microscopy (CLSM) was performed as described previously (33) to determine the inhibitory effect of PTE on biofilm formation. In brief, 35-mm clear-wall glass-bottom dishes (catalog number 70670–02; Ted Pella, Inc.) were inoculated with C. albicans statically at 37°C for 90 min to allow adhesion. After removing nonadherent cells, the dishes were incubated with fresh RPMI 1640 medium at 37°C for 24 h to allow biofilm formation. For PTE treatment groups, PTE was added to the fresh RPMI 1640 medium after 90 min of adhesion. The dishes were transferred to a new 6-well plates and incubated at 37°C for 45 min in 4 ml PBS containing the fluorescent stain FUN-1 (10 μM; Molecular Probes, Eugene, OR) and concanavalin A (ConA; 25 μg/ml; Molecular Probes)-Alexa Fluor 488 conjugate. FUN-1 (excitation wavelength of 543 nm and emission wavelength of 560 nm; long-pass filter) is converted to an orange-red cylindrical intravascular structure by metabolically active cells, while the ConA-Alexa Fluor 488 conjugate (excitation wavelength of 488 nm and emission wavelength of 505 nm; long-pass filter) binds to glucose and mannose residues of cell wall polysaccharides and emits green fluorescence. After incubation with the dyes, the dishes were flipped and C. albicans cells were observed under a Leica TCS SP2 CLSM.
SEM.
For scanning electron microscopy (SEM), glass disks coated with poly-l-lysine hydrobromide (catalog number P6282; Sigma) were used to develop biofilms. The disks were inoculated with C. albicans and incubated statically at 37°C for 90 min to allow adhesion. After removing non-dherent cells, the disks were incubated with fresh RPMI 1640 medium at 37°C for 24 h. For PTE treatment groups, PTE was added with the fresh RPMI 1640 medium after 90 min of adhesion. Biofilms were washed and placed in a fixative consisting of 2% (vol/vol) glutaraldehyde in 0.15 M sodium cacodylate buffer (pH 7.2) for 2 h. The samples were rinsed twice in cacodylate buffer, garnished with 1% osmic acid for 2 h, dehydrated in an ascending ethanol series, treated with hexamethyl-disilazane (Polyscience Europe GmbH, Eppelheim, Germany), and dried overnight. The specimens were coated with gold and observed through a Philips XL-30 SEM (Philips, The Netherlands) in high-vacuum mode (34).
CSH assay.
C. albicans cellular surface hydrophobicity (CSH) was measured in a water-hydrocarbon two-phase assay as described previously (31). Briefly, the formed C. albicans biofilms were removed from the flask surface to obtain a cell suspension (OD600, 1.0 in YPD medium). Then, 1.2 ml of suspension was pipetted into a clean glass tube and overlaid with 0.3 ml of octane. The mixture was vortexed for 3 min and then allowed to stand at room temperature for another 3 min for phase separation. Then, the OD600 of the aqueous phase was determined. The OD600 for the group without the octane overlay was used as the control. Three repeats were performed for each group. Relative hydrophobicity was calculated as follows: [(OD600 of control − OD600 after octane overlay)/OD600 of control] × 100%.
Gene expression microarrays.
Exponentially growing C. albicans cells were washed with PBS and resuspended with RPMI 1640 medium to 1 × 106 cells/ml. Eighty milliliters of the C. albicans suspension was added to 150-mm by 25-mm polystyrene cell culture dishes (Corning). The dishes were incubated statically at 37°C to allow initial adhesion and the production of germ tubes. After incubation for 90 min, the medium was removed and replaced with 80 ml fresh RPMI 1640 medium containing 16 μg/ml PTE, or DMSO as the control. The dishes were further incubated statically at 37°C for 1 h. C. albicans cells were collected, washed once with PBS, and stored in liquid nitrogen rapidly. Triplicate independent experiments were conducted on each sample for the following microarray experiments.
For biological replicates, triplicate independent experiments were carried out for the microarray assay. Total RNA was isolated by the CapitalBio Corporation by using the hot phenol method (35) and purified with a NucleoSpin Extract II kit (Macherey-Nagel Corp., Germany) following the manufacturer's protocol. cDNA labeled with a fluorescent dye (Cy5 or Cy3-dCTP) was produced by Eberwine's linear RNA amplification method and subsequent enzymatic reaction. This procedure has been previously described (36–37), and the procedure has been improved by using CapitalBio cRNA amplification and labeling kit to produce higher yields of labeled cDNA. In detail, double-stranded cDNAs (containing the T7 RNA polymerase promoter sequence) were synthesized from 1 μg total RNA using the CbcScript reverse transcriptase with the cDNA synthesis system according to the manufacturer's protocol (Capitalbio) with T7 oligo(dT). The T7 oligo(dT) primer was provided in the kit, and has the sequence 5′-AAACGACGGCCAGTGAATTGTAATACGACTCACTATAGGCGCTTTTTTTTTTTTTTTTT- 3′. After completion of double-stranded cDNA (dsDNA) synthesis using DNA polymerase and RNase H, the dsDNA products were purified using a PCR NucleoSpin Extract II kit and eluted with 30 μl elution buffer. The eluted double-stranded cDNA products were vacuum evaporated to 16 μl and subjected to 40 μl in vitro transcription reactions at 37°C for 4 to 14 h using T7 enzyme mix. The amplified cRNA was purified using the RNA cleanup kit. The Klenow enzyme labeling strategy was adopted after reverse transcription using CbcScript II reverse transcriptase. Briefly, 2 μg amplified RNA was mixed with 4 μg random nanomer, denatured at 65°C for 5 min, and cooled on ice. Then, 5 μl of 4× first-strand buffer, 2 μl of 0.1 M dithiothreitol, and 1.5 μl CbcScript II reverse transcriptase were added. The mixtures were incubated at 25°C for 10 min and then at 37°C for 90 min. The cDNA products were purified using a PCR NucleoSpin Extract II kit and vacuum evaporated to 14 μl. The cDNA was mixed with 4 μg random nanomer, heated to 95°C for 3 min, and snap-cooled on ice for 5 min. Then, 5 μl Klenow buffer, deoxynucleoside triphosphate, and Cy5-dCTP or Cy3-dCTP (GE Healthcare) were added to final concentrations of 240 μM dATP, 240 μM dGTP, 240 μM dTTP, 120 μM dCTP, and 40 μM Cy-dCTP. A 1.2-μl volume of Klenow enzyme was then added, and the reaction was performed at 37°C for 90 min.
Labeled cDNA was purified with a PCR NucleoSpin Extract II kit and resuspended in elution buffer. Labeled controls and test samples were quantitatively adjusted based on the efficiency of Cy5-dCTP or Cy3-dCTP incorporation and dissolved in 80 μl hybridization solution containing 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.2% SDS, 5× Denhardt's solution, and 25% formamide. DNA in hybridization solution was denatured at 95°C for 3 min prior to loading onto the microarray. Arrays were hybridized in a CapitalBio BioMixer II hybridization station overnight at a rotation speed of 8 rpm and a temperature of 42°C, and then washed in a Capitalbio SlideWasher (CapitalBio) with two consecutive solutions (0.2% SDS, 2× SSC at 42°C for 5 min, and then 0.2× SSC for 5 min at room temperature). Arrays were scanned with a confocal LuxScan scanner, and the images obtained were then analyzed using the LuxScan 3.0 software (both from CapitalBio). For individual channel data extraction, faint spots were removed if their intensities were below 400 units after the background was subtracted in both channels (Cy3 and Cy5). A space- and intensity-dependent normalization based on a LOWESS program was employed (38). To determine the significant differentially expressed genes, a significance analysis of microarrays (SAM, version 3.02) were performed (see Table S1 in the supplemental material). Genes with q value of <5%, a false-discovery rate (FDR) of <5%, and a change of ≥2-fold in C. albicans strains following PTE exposure were identified as significantly differentially expressed genes (see Table S2 in the supplemental material). A Gene Ontology (GO) enrichment search was conducted by using the Candida Genome Database GO term finder (http://www.candidagenome.org/cgi-bin/GO/goTermFinder) with default parameters (39), and a corrected P value of less than 0.05 was considered significant (see Tables S3 and S4 in the supplemental material).
Real-time RT-PCR.
Real-time reverse transcription-PCR (RT-PCR) was performed as described previously (40). Aliquots of the RNA preparations used in the microarray experiments (triplicate samples) were saved for the real-time RT-PCR study. cDNA was obtained through a reverse transcription reaction performed with a reverse transcription kit (TaKaRa Biotechnology, Dalian, China). Real-time PCR was performed with SYBR green I (TaKaRa) and the LightCycler real-time PCR system (Roche Diagnostics, GmbH, Mannheim, Germany). Primers used in the real-time RT-PCR are shown in Table S5 of the supplemental material. The thermal cycling conditions comprised an initial step at 95°C for 2 min, followed by 40 cycles at 95°C for 10 s, 60°C for 20 s, and 72°C for 30 s. The change in fluorescence of SYBR green I in every cycle was monitored, and the threshold cycle (CT) above background was determined. The CT value for 18S rRNA was subtracted from that of the gene of interest to obtain a ΔCT value. The ΔCT value of the untreated sample was subtracted from the ΔCT value of the PTE-treated sample to obtain a ΔΔCT value. The relative change of the gene expression level was calculated using the formula 2−ΔΔCT.
In vivo biofilm model.
For the in vivo study, an overnight routine culture of C. albicans SC5314 was washed and resuspended with normal saline. A rat central venous catheter infection model (41) was used to demonstrate the antibiofilm effect of PTE in vivo. Briefly, 12 female Sprague-Dawley rats (Shanghai SLAC Laboratory Animal Center, China) weighing about 300 g were used in this study. The rats were anesthetized, placed in a supine position, and prepared in a sterile fashion. A vertical incision was made on the skin of the anterior neck right to the midline. The external jugular vein was identified, and a longitudinal incision was made in the vein wall. For each rat, a sterile, heparinized polyethylene catheter (PE 90; inner diameter, 0.76 mm, and outer diameter, 1.27 mm; Smiths Medical) was placed in the opening and advanced to a site above the right atrium (about 2 cm) 24 h prior to C. albicans infection. The infection was established by intraluminal instillation of 500 μl C. albicans cells (1 × 106 cells/ml). After 4 h of indwelling, the catheter content was withdrawn and the catheter was flushed with 500 μl liquid containing different concentrations of PTE (0, 16, 32, and 64 μg/ml; 3 rats per group). After 72 h of incubation, animals were sacrificed by CO2 asphyxiation, and the catheters were removed. The distal 2 cm of each catheter was cut from the entire catheter length and imaged using SEM as described above.
The animal experiments were approved by the Animal Ethics Committee of the Second Military Medical University (Shanghai, China). All surgical procedures were performed under sodium pentobarbital anesthesia. To minimize suffering or substantial distress and to enhance animal welfare during the study, rats were recorded and clinically inspected during the whole study (at least twice daily, depending on trauma severity). Daily monitoring and examinations were carried out to ensure that there were no cases of dehydration, to control the body temperature, and in general, to prevent any severe discomfort of the animals.
Microarray data accession number.
All of the transcription data were deposited in the NCBI Gene Expression Omnibus (GEO) database and assigned accession number GSE43690 and platform identification number GPL9728.
RESULTS
Fungicidal effect of PTE.
It has been reported that PTE has strong activity against some plant fungal pathogens (18–20). Nevertheless, its activity against C. albicans has not yet been investigated. In this study, we first evaluated the fungicidal effect of PTE on C. albicans. However, very weak fungicidal activity of PTE was found. As shown in Fig. 2, time-kill curves indicated that ≤16 μg/ml PTE had no significant fungicidal effect on C. albicans, as the SC5314 strain grew to more than 5 × 106 CFU/ml after 6 h of treatment, similar to that with the control group without PTE treatment (Fig. 2). In contrast, 32 μg/ml PTE exhibited strong antifungal activity (P < 0.001), as only about 1 × 104 CFU/ml survived after 6 h of treatment (Fig. 2). In addition, the results of antifungal susceptibility tests indicated that PTE had antifungal activity, with a MIC80 of PTE for both fluconazole-susceptible strains and fluconazole-resistant strains of 32 μg/ml (Table 1).
FIG 2.
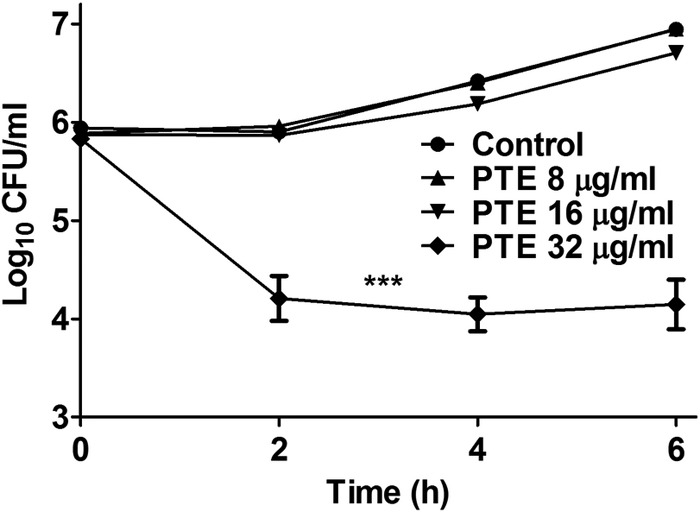
Time-kill curves for different concentrations of PTE against C. albicans strain SC5314. A 6.4-mg/ml PTE concentration in DMSO was used as a stock and added to 1 × 106 cells/ml of C. albicans suspensions in RPMI 1640 medium to obtain the indicated PTE concentrations. A C. albicans suspension in RPMI 1640 medium with 0.5% DMSO was used as the control. The data shown are means ± standard deviations from three independent experiments. Statistical significance among the groups was determined by an ANOVA. ***, P < 0.001 compared to the control group.
TABLE 1.
MIC80 of PTE and fluconazole against different C. albicans strains
| Fluconazole resistance profile and strain | MIC80 (μg/ml) |
|
|---|---|---|
| PTE | Fluconazole | |
| Fluconazole-susceptible strains | ||
| SC5314 | 32 | 0.5 |
| Y0109 | 32 | 0.5 |
| Fluconazole-resistant strains | ||
| 0304103 | 32 | >64 |
| 01010 | 32 | >64 |
PTE inhibits the formation of C. albicans biofilms in vitro.
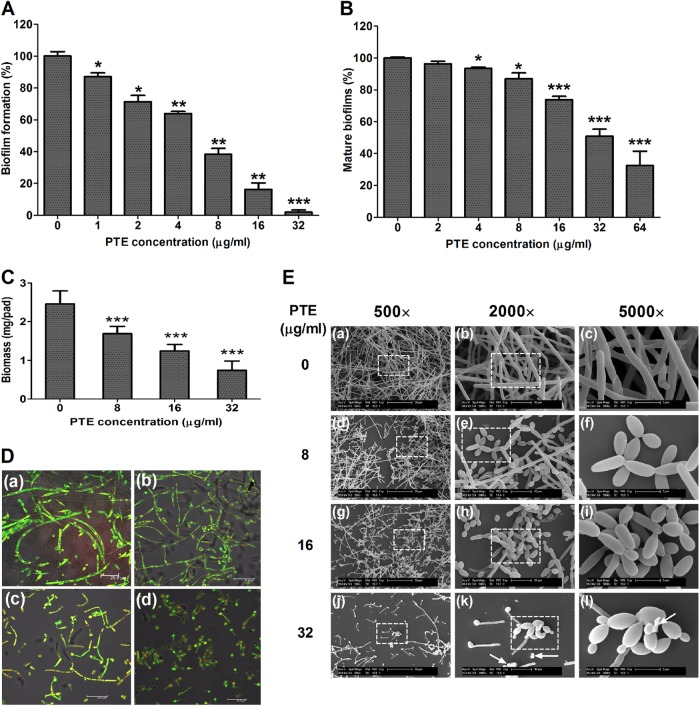
We then evaluated the effect of PTE on C. albicans biofilm formation in an XTT reduction assay and determined the antibiofilm effect of PTE. Addition of PTE to C. albicans cells after 90 min of adhesion inhibited biofilm formation in a dose-dependent manner (Fig. 3A). More specifically, 1 μg/ml PTE inhibited biofilm formation by about 17% (P < 0.05), and this antibiofilm effect increased as the PTE concentration increased. In the 16-μg/ml PTE group, the biofilm formation rate was about 18% relative to the control group (without PTE treatment). Notably, under the condition that the biofilms were mature after 24 h of incubation at 37°C, PTE also inhibited biofilms in a dose-dependent manner (Fig. 3B). At 4 μg/ml, PTE inhibited mature biofilms by about 7% (P < 0.05). The antibiofilm effect increased with the PTE concentration. In the 64-μg/ml PTE group, the maintenance rate of biofilm was about 30% compared with the control group without PTE treatment. Collectively, PTE exhibited an antibiofilm effect.
FIG 3.
PTE inhibits C. albicans biofilm formation in vitro. (A) Effects of different concentrations of PTE on biofilm formation. (B) Effects of different concentrations of PTE on the maintenance of mature biofilms. Biofilm formation was evaluated in an XTT reduction assay, and the results are presented as the percent biofilm dry weight relative to control biofilms formed without PTE treatment. Biofilm formation results represent means ± standard deviations for five independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001, compared to the control biofilms. (C) Effects of different concentrations of PTE on biofilms formed on silicone pads. Standard deviations are depicted and based on 5 silicone pad measurements. ***, P < 0.001. (D) Effects of different concentrations of PTE on biofilm formation, shown in CLSM images. (a) Control; (b) 8 μg/ml PTE; (c) 16 μg/ml PTE; (d) 32 μg/ml PTE. (E) Effects of different concentrations of PTE on biofilm formation, shown in SEM images. White arrows indicate cell fragments. Images in the dashed boxes were enlarged (images in right-most column).
The results of biofilm biomass determination also confirmed the results of the XTT reduction assay, indicating that PTE inhibited biofilm formation in a dose-dependent manner (Fig. 3C). Biofilm formation was seriously affected in the 8-μg/ml PTE group compared with the control group without PTE treatment (P < 0.001). With increasing PTE, the effect on biofilms was more obvious.
The antibiofilm effect of PTE was further confirmed by CSLM and SEM. Compared with the normal C. albicans biofilm, with true hyphae criss-crossing (Fig. 3D, panel a, and E, panels a to c), C. albicans biofilm formation was disrupted by PTE in a dose-dependent manner (Fig. 3D and E). The 8-μg/ml dose of PTE led to a reduction in cell density and a defect in filamentation (Fig. 3D, panel b, and E panels d to f). When the PTE concentration was increased to 16 μg/ml and 32 μg/ml, cell density was further reduced and the defect in filamentation became more obvious (Fig. 3D, panels c and d, and E, panels g to l).
PTE decreases CSH of C. albicans.
Knowing that adhesion and morphological transition are two important stages for C. albicans biofilm formation (42) and that there is a positive correlation between CSH and adhesion of C. albicans (43–45), we first examined the effect of PTE on CSH of C. albicans. The normal CSH of C. albicans was found to be 0.67 (Fig. 4). The 1-μg/ml PTE dose significantly decreased CSH to 0.47 (P < 0.001) (Fig. 4). In addition, PTE decreased the CSH of C. albicans in a dose-dependent manner (Fig. 4). The CSH of C. albicans decreased to 0.03 in the 16-μg/ml PTE group (Fig. 4). Collectively, PTE decreased the CSH of C. albicans.
FIG 4.
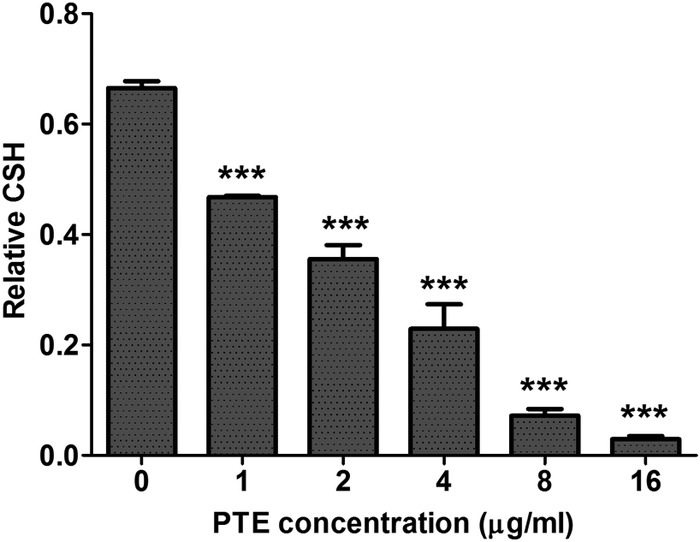
Effects of different concentrations of PTE on CSH of C. albicans SC5314. CSH was evaluated in a water-hydrocarbon two-phase assay. Standard deviations are depicted and are based on three independent experiments. ***, P < 0.001.
PTE inhibits hyphal formation of C. albicans.
The activity of PTE on the yeast-to-hypha morphological transition of C. albicans was further evaluated by growing C. albicans in several media known to induce morphological transition, including YPD plus FCS (liquid and solid media), Spider (liquid and solid media), Lee (liquid and solid media), and SLAD (liquid) media. Without PTE treatment, C. albicans formed hyphae in all the media tested. At 4 μg/ml, PTE inhibited the yeast-to-hypha morphological transition to some extent and inhibited the morphological transition in a dose-dependent manner (Fig. 5). The addition of 16 μg/ml PTE totally disrupted the formation of true hyphae in all the media tested: yeast-form cells and pseudohyphae were observed in YPD plus FCS liquid medium (Fig. 5); only yeast-form cells were observed in Spider, Lee, and SLAD liquid media (Fig. 5); only smooth-edged colonies were observed on YPD plus FCS, Spider, and Lee solid media (see Fig. S1 in the supplemental material).
FIG 5.
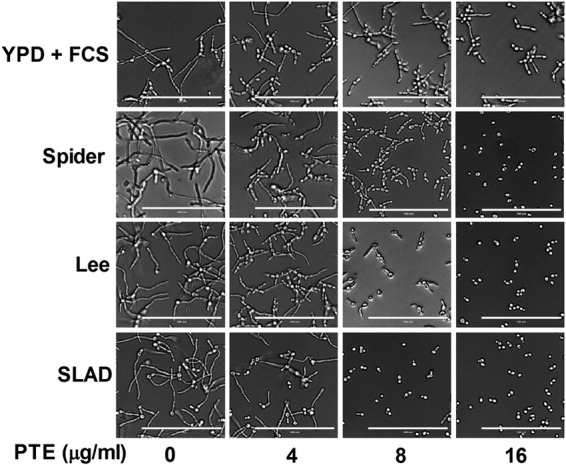
Effects of different concentrations of PTE on hyphal formation. Exponentially growing C. albicans SC5314 cells were transferred to hypha-inducing liquid media. The cellular morphology was photographed after incubation at 37°C for 3.5 h. Bar, 100 μm.
Exposure to PTE alters C. albicans gene expression.
In the microarray experiments, changes in C. albicans gene expression after PTE treatment were observed. The 16-μg/ml PTE dose was used in the assay because it had a significant antibiofilm effect against C. albicans but had little fungicidal effect on planktonic C. albicans cells. It was found that 307 genes were differentially expressed after exposure to PTE for 1 h, including downregulation of 193 genes and upregulation of 114 genes (cutoff value of 2) (see Table S2 in the supplemental material). Through GO term analysis, we found that the differentially expressed genes could be assigned to several categories. Of note, the downregulated categories included the ergosterol biosynthesis process, oxidoreductase activity function, and the cell surface component (see Table S3 in the supplemental material), and the upregulated categories included the protein unfolding process (heat shock proteins) (see Table S4 in the supplemental material). More specifically, 15 ergosterol biosynthesis-related genes, 19 oxidoreductase genes, and 18 cell surface-related genes were downregulated (Table 2), while 3 protein unfolding process-related genes (heat shock protein genes) were upregulated (Table 3). Since PTE inhibits biofilm/filamentation of C. albicans, we further focused on some important biofilm/filamentation-related genes. Notably, RAS1 (the RAS signal transduction GTPase gene [46]), ECE1 (cell elongation protein gene [47]), SAP5 (secreted aspartyl proteinase gene [48]), and SAM2 (S-adenosylmethionine synthetase gene [49]) were downregulated (Table 2), while ESC4 (a gene playing a role in inhibiting biofilm formation [50]) and YWP1 (a gene that encodes yeast cell wall protein [51]) were upregulated (Table 3), which was consistent with the antibiofilm/antifilamentation effects of PTE.
TABLE 2.
Important genes downregulated in PTE-treated C. albicans compared to C. albicans without treatment
| Functional group and gene namea | Descriptiona | Extent of regulation (fold change relative to control)b |
|---|---|---|
| Filamentation | ||
| orf19.4749 | Predicted ORF from assembly 19; hypha-induced expression, regulated by Cyr1p, Ras1p, Efg1p | 0.17 ± 0.08 |
| ALS10 | Agglutinin-like sequence (ALS) family protein | 0.17 ± 0.13 |
| ECE1 | Protein comprising eight 34-residue repeats; expression specific to hyphae, increases with extent of elongation of the cell | 0.19 ± 0.12 |
| MET14 | Predicted role in sulfur metabolism; induced upon biofilm formation; possibly adherence induced | 0.29 ± 0.09 |
| orf19.6920 | Predicted ORF from assembly 19; induced upon biofilm formation | 0.31 ± 0.18 |
| SAP5 | Secreted aspartyl proteinase | 0.32 ± 0.03 |
| SAP4 | Secreted aspartyl proteinase | 0.32 ± 0.14 |
| MET15 | O-Acetylhomoserine O-acetylserine sulfhydrylase; involved in sulfur amino acid biosynthesis; biofilms, possibly adherence induced | 0.34 ± 0.17 |
| RAS1 | RAS signal transduction GTPase; regulates cAMP and MAP kinase pathways; involved in hyphal induction, virulence, and heat shock sensitivity | 0.42 ± 0.08 |
| orf19.5905 | Predicted ORF from assembly 19; induced upon biofilm formation | 0.43 ± 0.21 |
| SAM2 | S-Adenosylmethionine synthetase; localizes to surface of hyphae, not yeast cells | 0.48 ± 0.17 |
| Ergosterol biosynthetic process | ||
| ERG2 | C-8 sterol isomerase; enzyme of ergosterol biosynthesis pathway; converts fecosterol to episterol | 0.23 ± 0.05 |
| ERG1 | Squalene epoxidase, catalyzes epoxidation of squalene to 2,3(S)-oxidosqualene in the ergosterol biosynthetic pathway | 0.23 ± 0.11 |
| ERG6 | Delta(24)-sterol C-methyltransferase, converts zymosterol to fecosterol in ergosterol biosynthesis | 0.24 ± 0.06 |
| UPC2 | Transcription factor involved in regulation of ergosterol biosynthesis genes and sterol uptake; binds ERG2 promoter | 0.24 ± 0.17 |
| ERG9 | Putative farnesyl-diphosphate farnesyl transferase (squalene synthase) involved in the sterol biosynthesis pathway | 0.26 ± 0.06 |
| ERG5 | Putative C-22 sterol desaturase (fungal C-22 sterol desaturases are cytochrome P450 enzymes of ergosterol biosynthesis) | 0.26 ± 0.09 |
| ERG26 | C-3 sterol dehydrogenase, catalyzes the second of three steps in ergosterol biosynthesis | 0.28 ± 0.08 |
| ERG3 | C-5 sterol desaturase; introduces C-5(6) double bond into episterol in ergosterol biosynthesis | 0.32 ± 0.07 |
| ERG25 | Putative C-4 methyl sterol oxidase with role in C-4 demethylation of ergosterol biosynthesis intermediates | 0.32 ± 0.09 |
| ERG24 | C-14 sterol reductase, has a role in ergosterol biosynthesis | 0.35 ± 0.07 |
| ERG4 | Protein described as similar to sterol C-24 reductase | 0.36 ± 0.06 |
| ERG11 | Lanosterol 14-α-demethylase, member of cytochrome P450 family that functions in ergosterol biosynthesis | 0.38 ± 0.09 |
| ERG27 | 3-Keto sterol reductase of ergosterol biosynthesis; acts in C-4 sterol demethylation with Erg25p and Erg26p | 0.41 ± 0.15 |
| ERG13 | Protein similar to S. cerevisiae Erg13p, which is involved in ergosterol biosynthesis | 0.42 ± 0.01 |
| ERG10 | Protein described as similar to acetyl-CoA acetyltransferase; protein of ergosterol biosynthesis | 0.42 ± 0.12 |
| Oxidoreductase activity | ||
| CFL11 | Protein similar to ferric reductase Fre10p | 0.06 ± 0.05 |
| ILV5 | Ketol-acid reductoisomerase; regulated by Gcn4; macrophage-repressed protein | 0.17 ± 0.06 |
| orf19.2286 | Putative deoxyhypusine hydroxylase; protein level decreases in stationary-phase cultures; required for biofilm formation | 0.23 ± 0.04 |
| MAE1 | Putative malate permease; induced during macrophage infection; putative peroxisome targeting signal | 0.28 ± 0.13 |
| orf19.6899 | Putative oxidoreductase; mutation confers hypersensitivity to toxic ergosterol analogue | 0.35 ± 0.20 |
| orf19.5517 | Similar to alcohol dehydrogenases; induced by benomyl treatment, nitric oxide; oxidative stress induced via Cap1 | 0.36 ± 0.07 |
| IDP1 | Putative isocitrate dehydrogenase; transcriptionally induced by interaction with macrophage | 0.36 ± 0.12 |
| GCV2 | Glycine decarboxylase P-subunit; protein of glycine catabolism; repressed by Efg1 | 0.38 ± 0.07 |
| MCS7 | Uncharacterized | 0.40 ± 0.13 |
| GPD1 | Glycerol-3-phosphate dehydrogenase; glycerol biosynthesis; regulated by Tsa1, Tsa1B under H2O2 stress conditions | 0.40 ± 0.16 |
| CBP1 | Corticosteroid binding protein; contains possible NAD/FAD binding region; regulated by Nrg1, Tup1 | 0.41 ± 0.05 |
| orf19.3810 | Orthologue(s) has methylenetetrahydrofolate dehydrogenase (NAD+) activity | 0.42 ± 0.08 |
| SLD1 | Sphingolipid delta-8 desaturase; catalyzes desaturation at C-8 in the long-chain base moiety of ceramides in glucosylceramide synthesis | 0.43 ± 0.20 |
| orf19.5136 | Putative pyridoxamine 5′-phosphate oxidase; planktonic growth and early-stage flow model biofilm induced | 0.44 ± 0.15 |
| orf19.225 | Predicted 2-hydroxyacid dehydrogenase; Hap43-repressed gene | 0.46 ± 0.11 |
| HOM6 | Putative homoserine dehydrogenase; Gcn4 regulated; macrophage-induced protein | 0.48 ± 0.07 |
| SOU1 | Enzyme involved in utilization of l-sorbose; has sorbitol dehydrogenase, fructose reductase, and sorbose reductase activities | 0.49 ± 0.09 |
| orf19.1306 | Uncharacterized | 0.49 ± 0.16 |
| orf19.3515 | Putative 3-hydroxyanthranilic acid dioxygenase, involved in NAD biosynthesis; Hap43p-repressed gene | 0.50 ± 0.19 |
| Cell surface | ||
| SOD5 | Cu- and Zn-containing superoxide dismutase; protects against oxidative stress | 0.08 ± 0.06 |
| SCW11 | Cell wall protein; rat catheter and Spider biofilm repressed | 0.16 ± 0.05 |
| CHT3 | Major chitinase; secreted; farnesol upregulated in biofilms; regulated by Efg1p, Cyr1p, Ras1p | 0.16 ± 0.10 |
| FGR41 | Putative GPI-anchored adhesion-like protein; transposon mutation affects filamentous growth; Spider biofilm repressed | 0.17 ± 0.08 |
| PGA44 | Putative GPI-anchored protein | 0.18 ± 0.05 |
| PGA10 | GPI-anchored membrane protein; required for RPMI biofilm formation, Bcr1 induced in biofilm | 0.19 ± 0.07 |
| PGA38 | Putative adhesion-like GPI-anchored protein | 0.20 ± 0.02 |
| PGA34 | Putative GPI-anchored protein; induced in oral pharyngeal candidiasis; flow model biofilm induced; Spider biofilm induced | 0.20 ± 0.05 |
| GPD2 | Surface protein; induced by cell wall regeneration, macrophage/pseudohypha growth; Spider biofilm induced | 0.24 ± 0.06 |
| PGA26 | GPI-anchored adhesion-like protein of the cell wall; role in cell wall integrity | 0.26 ± 0.07 |
| EXG2 | GPI-anchored cell wall protein; induced during cell wall regeneration | 0.27 ± 0.03 |
| ENG1 | Endo-1,3-beta-glucanase; flow model biofilm induced; rat catheter biofilm repressed | 0.28 ± 0.12 |
| RHD3 | GPI-anchored yeast-associated cell wall protein; induced by high iron levels | 0.30 ± 0.11 |
| PGA45 | Putative GPI-anchored cell wall protein; Mob2-dependent hyphal regulation; flow model biofilm induced | 0.30 ± 0.14 |
| CRH11 | GPI-anchored cell wall transglycosylase; predicted glycosyl hydrolase domain | 0.34 ± 0.09 |
| SIT1 | Transporter of ferrichrome siderophores; rat catheter and Spider biofilm induced | 0.35 ± 0.15 |
| PLB4.5 | Putative GPI anchor; repressed during cell wall regeneration | 0.42 ± 0.19 |
| PGA41 | Putative GPI-anchored protein; adhesion-like protein | 0.45 ± 0.03 |
As reported in the CGD database (http://www.candidagenome.org/). Abbreviations: ORF, open reading frame; CoA, coenzyme A.
Data are means ± standard deviations from three experiments.
TABLE 3.
Selected important genes upregulated in PTE-treated C. albicans compared to C. albicans without treatment
| Functional group and gene namea | Descriptiona | Extent of regulation (fold change relative to control)b |
|---|---|---|
| Filamentation | ||
| ESC4 | Protein similar to S. cerevisiae Esc4p, which represses transposition; transposon mutation affects filamentous growth; rat catheter biofilm repressed | 4.07 ± 2.76 |
| CCC1 | Protein described as predicted manganese transporter; shows colony morphology-related gene regulation by Ssn6p | 3.63 ± 0.62 |
| orf19.1584 | Predicted ORF from assembly 19; shows colony morphology-related gene regulation by Ssn6p | 3.20 ± 1.61 |
| YWP1 | Secreted yeast cell wall protein; possible role in dispersal in host; mutation causes increased adhesion and biofilm formation | 2.83 ± 0.38 |
| AAF1 | Possible regulatory protein with glutamine-rich domain; mRNA detected in yeast-form and hyphal cells | 2.76 ± 0.86 |
| DDR48 | Stress-associated protein; expression regulated by multiple filamentous growth pathways | 2.63 ± 0.90 |
| FGR46 | Protein lacking an orthologue in S. cerevisiae; transposon mutation affects filamentous growth | 2.48 ± 0.19 |
| CHT2 | GPI-linked chitinase required for normal filamentous growth; mRNA is localized to yeast-form buds and hyphal tips | 2.39 ± 0.35 |
| SFL1 | Transcription factor involved in negative regulation of flocculation and filamentous growth | 2.23 ± 0.09 |
| RIM8 | Protein required for alkaline pH-induced hyphal growth; shows colony morphology-related gene regulation by Ssn6p | 2.17 ± 0.19 |
| CAR1 | Transcription is regulated by Nrg1p, Mig1p, and Tup1p; shows colony morphology-related gene regulation by Ssn6p | 2.04 ± 0.47 |
| CEK1 | ERK family protein kinase, required for wild-type yeast-hypha switching, mating efficiency, and virulence in a mouse model; invasive hyphal growth under some conditions | 2.02 ± 0.45 |
| Protein unfolding (heat shock proteins) | ||
| HSP104 | Described as a heat shock protein; downregulated in biofilms upon treatment with farnesol | 5.66 ± 3.01 |
| HSP78 | Protein described as a heat shock protein; transcriptionally regulated by macrophage response | 3.12 ± 1.17 |
| SSC1 | Heat shock protein; localizes to surface of yeast-form cells, but not hyphae | 2.07 ± 0.31 |
As reported in the CGD database (http://www.candidagenome.org/). Abbreviations: ORF, open reading frame; ERK, extracellular signal-regulated kinase.
Data are means ± standard deviations from three experiments.
To validate the changes in gene expression identified by microarray analysis, we selected 8 genes (RAS1, ECE1, CEK1, YWP1, RIM8, ERG11, PGA10, and HSP78) for real-time RT-PCR analysis. These certain genes were selected because they are involved in filamentation (RAS1, ECE1, CEK1, YWP1, and RIM8), ergosterol biosynthesis (ERG11), cell surface (PGA10) and protein unfolding (HSP78), respectively, based on information in the CGD database (http://www.candidagenome.org/) and GO term analysis. The real-time RT-PCR results indicated that the expression levels of RAS1, ECE1, ERG11, and PGA10 were reduced, with relative fold changes of 0.18, 0.06, 0.20, and 0.13, respectively, while CEK1, YWP1, RIM8, and HSP78 were overexpressed after exposure to 16 μg/ml PTE, with relative fold changes of 1.22, 3.24, 1.22, and 2.68, respectively (Fig. 6A). Plotting the fold change obtained by real-time RT-PCR versus the value observed with the microarray analysis yielded an R2 value of 0.95 (see Fig. S2 in the supplemental material), indicating a good correlation between the two data sets.
FIG 6.
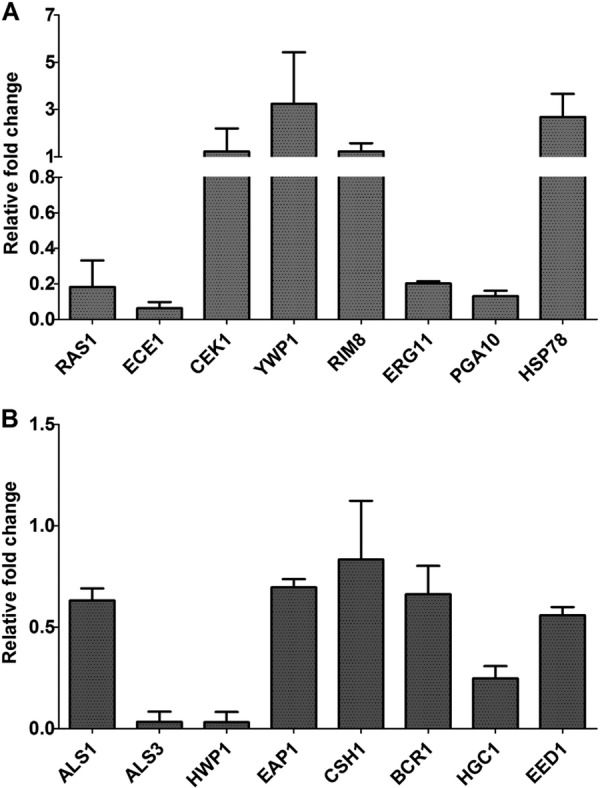
Real-time RT-PCR analysis results for some important biofilm/filamentation-related genes. (A) Analysis of genes identified as differentially expressed in microarray experiments. (B) Analysis of some other biofilm/filamentation-related genes. Gene expression fold changes are relative to results with the control group (C. albicans SC5314 without PTE treatment). 18S rRNA was used to normalize the expression data. Data are means ± standard deviations from three experiments.
As was detailed in the previous section, stringent criteria were used to identify differentially expressed genes in our microarray assay (intensities below 400 units were regarded as faint spots in microarray; q value < 5%, FDR < 5%, and fold change ≥2; if looser criteria had been used, many more genes would have been detected by the microarray). To avoid missing some important yeast-to-hypha transition genes, including ALS1, ALS3, HWP1, EAP1, CSH1, BCR1, HGC1, and EED1 (52, 53; http://www.candidagenome.org/), we further detected the change in expression levels of these genes by using real-time RT-PCR. No obvious change was found in the expression of ALS1, EAP1, CSH1, BCR1, or EED1 (relative fold change >0.5 or <2) (Fig. 6B). Nevertheless, the expression levels of ALS3, HWP1, and HGC1 were downregulated after PTE treatment, with the relative change being 0.03-, 0.03-, and 0.25-fold, respectively (Fig. 6B). Of note, HWP1 and ALS3 were not identified to be differentially expressed, because their intensities were below 400 units in the microarrays. HGC1 was downregulated, but the average ratio was not less than 0.5 in the microarray assay.
Exogenous cAMP reverts the morphogenesis defect caused by PTE.
Since a series of important genes in the Ras/cAMP pathway, including ECE1, ALS3, HWP1, HGC1, and RAS1 itself (54, 55), were downregulated after PTE treatment, we hypothesized that the antibiofilm effect of PTE was related to the downregulation of the Ras/cAMP pathway. The effect of exogenous cAMP on the PTE-induced morphological transition defect was investigated. Since YPD plus FCS and Spider media are the two most common media used to induce the morphological transition in C. albicans (23, 52) and PTE exhibited a stronger effect in Spider medium (Fig. 5), we chose Spider medium for the cAMP experiment. The results showed that exogenous cAMP restored the yeast-to-hypha morphological transition after PTE treatment (Fig. 7). Specifically, addition of 5 mM cAMP to solid or liquid Spider medium restored the hyphal formation in the 8-μg/ml PTE group (Fig. 7).
FIG 7.
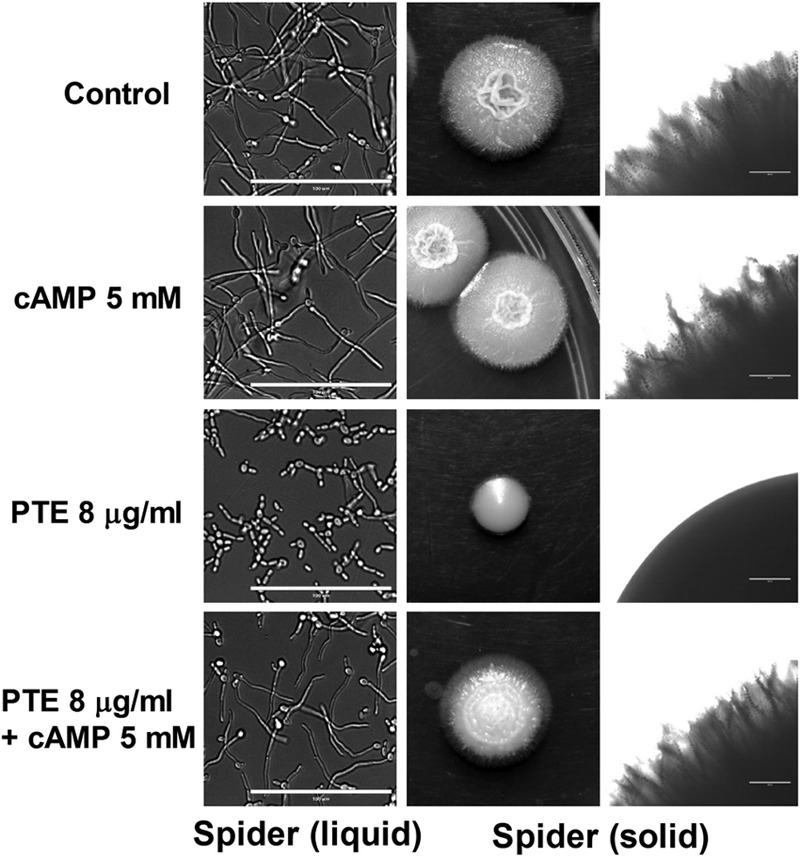
Exogenous cAMP reverts the morphological transition defect of C. albicans SC5314 caused by PTE. Exponentially growing C. albicans SC5314 cells were transferred to hypha-inducing media. In liquid Spider medium, the cellular morphology was photographed after incubation at 37°C for 3.5 h. On solid Spider medium, approximately 10 cells were plated, and the culture was incubated at 37°C for 5 days. Bars, 100 μm (left) and 400 μm (right).
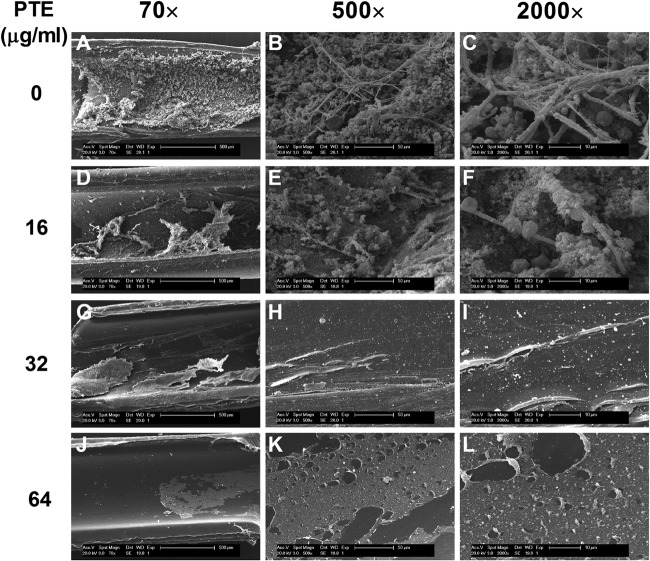
PTE inhibits C. albicans biofilm formation in vivo.
In the rat central venous catheter biofilm model, the antibiofilm effect of PTE in vivo was validated by inoculating animals via a central venous catheter with C. albicans and treating via the catheter with PTE intraluminally. The catheter was removed 72 h later, the luminal surface of which was observed by SEM. It was found that PTE inhibited C. albicans biofilm formation in the central venous catheter (Fig. 8), while thick biofilms were formed on the intraluminal surface of the catheter without PTE treatment, and these biofilms consisted of hyphae, yeast cells, and heterogeneous matrix (Fig. 8A to C). In the 16-μg/ml PTE group, C. albicans biofilm was defective, and only sparse cells were observed (Fig. 8D to F). In the 32-μg/ml or 64-μg/ml PTE group, no biofilm formation was observed, nor were fungal cells detected on the intraluminal surface of the catheter (Fig. 8G to I), indicating that PTE has antibiofilm activity in vivo.
FIG 8.
PTE inhibits C. albicans biofilm formation in vivo. SEM images show Candida albicans biofilms attached to the intraluminal surface of catheters. PTE inhibited C. albicans biofilm formation in a dose-dependent manner.
DISCUSSION
C. albicans biofilms constitute a threat to successful antifungal treatment (30). In this study, we revealed a significant effect of PTE against C. albicans biofilms, including biofilm formation and maintenance of mature biofilms in vitro. We also found that PTE could decrease CSH and suppress the yeast-to-hypha morphological transition. The results of microarrays and real-time RT-PCR indicated that some important filamentation genes, ergosterol biosynthesis genes, cell surface-related genes, and heat shock protein genes were differentially expressed after exposure to 16 μg/ml PTE. We further revealed that the effect of PTE was related to the Ras/cAMP pathway, and exogenous cAMP could restore the morphological transition of C. albicans under the condition of PTE exposure. Finally, we confirmed the antibiofilm effect of PTE in vivo.
To the best of our knowledge, this is the first report indicating that PTE has a strong antibiofilm effect against C. albicans in vitro and in vivo. It has been reported that PTE has a strong activity against some plant fungal pathogens, such as P. viticola, P. obscurans, and B. cinerea (18–20). For example, Sobolev et al. (18) found that 30 μM (about 8 μg/ml) PTE inhibited the growth of P. viticola by 50%, the growth of P. obscurans by 100%, and the growth of B. cinerea by 80% within 144 h. In this study, we found that PTE had a strong antibiofilm effect against C. albicans. More importantly, PTE could not only inhibit the formation of biofilms but also destroy the maintenance of mature biofilms. At 16 μg/ml (½× MIC), PTE reduced biofilm formation by about 82%, and 64 μg/ml (2× MIC) PTE destroyed the maintenance of mature biofilms by about 70%. The effect of PTE was outstanding compared with various other antifungal agents. Vila et al. (56) found that at 16× MIC, fluconazole could not inhibit the development of biofilms and that amphotericin B at 1× MIC inhibited the development of biofilms but could not affect the maintenance of mature biofilms. In contrast, our data indicate that PTE possesses good antibiofilm activity.
Of note, PTE possesses similar antifungal activity against both fluconazole-susceptible strains (SC5314 and Y0109) and fluconazole-resistant strains (strains 0304103 and 01010), with MIC80s of 32 μg/ml for all four strains tested. C. albicans 0304103 and 01010 are strains that overexpress multidrug resistance genes (CDR1 and MDR1 [unpublished data]) and exhibit strong fluconazole resistance (MIC80, >64 μg/ml). It can be inferred that PTE is quite different from fluconazole, at least with regard to C. albicans susceptibility and resistance.
The effect of resveratrol (3,5,4-trihydroxystilbene), a close analogue of PTE, on C. albicans has been investigated previously (57–59). However, its antifungal effect was much weaker than PTE. More specifically, Okamoto et al. revealed that the MIC50 of resveratrol against C. albicans SC5314 was about 200 μg/ml, and resveratrol (≥40 μg/ml) could inhibit the yeast-to-hypha morphological transition of C. albicans (57), while Collado et al. (58) and Weber et al. (59) reported that resveratrol had no antifungal activity against C. albicans. In this study, we found that PTE has a much stronger antifungal effect against C. albicans than resveratrol, with a MIC80 of 32 μg/ml and a minimal concentration inhibiting morphological transition of 4 μg/ml. We are performing further studies to reveal the structure-activity relationship of PTE.
Our data indicate that PTE may inhibit biofilm formation by preventing adhesion (with CSH as the indicator [45]) and morphological transition, rather than killing C. albicans cells. There are three known stages for biofilm formation: adhesion to biomaterial surfaces, growth to form an anchoring layer, and morphological transition to form a complex three-dimensional structure (40, 42, 60, 61). Our data indicate that 16 μg/ml (½× MIC), PTE significantly inhibited biofilm formation, obviously decreased the CSH, and apparently inhibited the yeast-to-hypha morphological transition, while PTE at 16 μg/ml did not kill C. albicans. Thus, the antibiofilm formation effect of PTE seems attributable to its antiadhesion and anti-morphological transition activities.
To clarify the antibiofilm mechanism of PTE, microarray and real-time RT-PCR analyses were performed in this study. Of note, first of all, some adhesion- and hypha-specific genes, including RAS1, ECE1, ALS3, HWP1, and HGC1, were downregulated after the 16-μg/ml PTE treatment. Ras1 is a signal transduction GTPase that can induce hyphal formation by activating both the Ras/cAMP pathway and the mitogen-activated protein (MAP) kinase pathway in C. albicans (52). ECE1, ALS3, HWP1, and HGC1 are highly expressed in hyphae and play essential roles in the yeast-to-hypha morphological transition of C. albicans (47, 53, 62–64). The downregulation of these genes may contribute to the antibiofilm effect of PTE. Second, 15 ergosterol biosynthesis-related genes were downregulated after PTE treatment. It has been reported that ergosterol biosynthesis-related genes are upregulated during biofilm formation (65), and the inhibition of ergosterol biosynthesis can significantly inhibit the morphological transition and biofilm development for C. albicans (66). The downregulation of ergosterol biosynthesis may also contribute to the antibiofilm effect of PTE. Third, the expression of heat shock proteins was upregulated upon PTE treatment. Heat shock proteins are a group of molecular chaperones induced by stresses (67). It can be inferred that C. albicans cells are under stress during PTE treatment. Fourth, PTE exposure may affect cell surface components of C. albicans, which is in accordance with the findings with Saccharomyces cerevisiae reported by Pan et al. (68). Notably, in this study, glycosylphosphatidylinositol (GPI)-anchored protein-coding genes were downregulated in C. albicans after PTE treatment. GPI-anchored proteins are important components of the cell wall and play important roles in biofilm formation by C. albicans (53). The downregulation of GPI-anchored proteins and alteration of the cell surface may also be associated with the antibiofilm effect of PTE. Finally, ESC4 and YWP1 were found to be upregulated after PTE treatment. ESC4 encodes a protein that represses transposition in C. albicans, and its mutant was observed to have increased filamentous growth (50). YWP1 (yeast wall protein 1) encodes a yeast-specific cell wall protein, and its mutation was revealed to cause increased adhesion and biofilm formation (51). The upregulation of YWP1 and ESC4 may contribute to the antibiofilm effect of PTE, too. Collectively, the antibiofilm effect of PTE may be associated with the downregulation of morphological transition genes, downregulation of ergosterol biosynthesis, stress, alteration of the cell surface, and upregulation of YWP1 and ESC4.
Some downregulated genes after PTE treatment, including ECE1, ALS3, HWP1, HGC1, and ergosterol biosynthesis-related genes, are genes regulated by the Ras/cAMP pathway (54, 55, 69). Notably, RAS1 itself was downregulated upon PTE exposure. Thus, we speculate that the antibiofilm effect of PTE involves the downregulation of the Ras/cAMP pathway. Exogenous cAMP reverted the morphogenesis defect caused by PTE in this study, which further supports our speculation. Nevertheless, further studies are required to explore whether PTE has a direct effect on Ras1 or whether it affects some other factor that regulates the expression of Ras1.
PTE exhibited an antibiofilm effect in vivo. The pharmacokinetics, oral bioavailability, and metabolism of PTE have been studied in rats, and the results indicate that when administered orally, PTE has a high bioavailability (17, 22, 70). Kapetanovic et al. (70) revealed that approximately 80% of PTE was bioavailable following oral administration, while Lin et al. (22) reported that the bioavailability should be 95%. In addition, the half-life of PTE is up to 105 min, which is almost seven times longer than that of its analogue, resveratrol (14 min) (17). In addition, it has been reported that dietary administration of a high dose of PTE produced no toxicity in mice. More specifically, 3,000 mg/kg of body weight/day of PTE for 28 days did not affect biochemical or clinical parameters or the organ weights of the tested mice (71). These findings demonstrated that PTE possesses key preclinical characteristics of a potent antibiofilm agent.
Collectively, PTE has a strong antibiofilm effect against C. albicans in vitro and in vivo, which may be related to the Ras/cAMP pathway. PTE has high bioavailability and no toxicity for mice, which makes it ideally suited as a potent drug for the life-threatening biofilm-related infections caused by C. albicans. Further translational research is required to determine whether the antibiofilm effect of PTE is applicable in the clinical setting.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81273558, 81072678, and 90913008); the National Key Basic Research Program of China (2013CB531602); Shanghai Educational Development Foundation (2007CG51); Core Technology Program for Strategy Emerging Industries of Guangdong Province (2011A081401002); and the Science and Technology Program of Guangzhou City (2012J4300097).
We thank Dominique Sanglard (Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland) for kindly providing C. albicans SC5314 and Jun Gu (Changhai Hospital, Shanghai, China) for kindly providing the C. albicans strains Y0109, 0304103, and 01010.
Footnotes
Published ahead of print 10 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01583-13.
REFERENCES
- 1.Karkowska-Kuleta J, Rapala-Kozik M, Kozik A. 2009. Fungi pathogenic to humans: molecular bases of virulence of Candida albicans, Cryptococcus neoformans and Aspergillus fumigatus. Acta Biochim. Pol. 56:211–224 [PubMed] [Google Scholar]
- 2.Alangaden GJ. 2011. Nosocomial fungal infections: epidemiology, infection control, and prevention. Infect. Dis. Clin. North Am. 25:201–225. 10.1016/j.idc.2010.11.003 [DOI] [PubMed] [Google Scholar]
- 3.Jarvis WR. 1995. Epidemiology of nosocomial fungal infections, with emphasis on Candida species. Clin. Infect. Dis. 20:1526–1530 [DOI] [PubMed] [Google Scholar]
- 4.Miceli MH, Lee SA. 2011. Emerging moulds: epidemiological trends and antifungal resistance. Mycoses 54:e666–e678. 10.1111/j.1439-0507.2011.02032.x [DOI] [PubMed] [Google Scholar]
- 5.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167–193. 10.1128/CMR.15.2.167-193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandra J, Mukherjee PK, Leidich SD, Faddoul FF, Hoyer LL, Douglas LJ, Ghannoum MA. 2001. Antifungal resistance of Candidal biofilms formed on denture acrylic in vitro. J. Dent. Res. 80:903–908. 10.1177/00220345010800031101 [DOI] [PubMed] [Google Scholar]
- 7.Ramage G, Vandewalle K, Wickes BL, Lopez-Ribot JL. 2001. Characteristics of biofilm formation by Candida albicans. Rev. Iberoam. Micol. 18:163–170 [PubMed] [Google Scholar]
- 8.Berman J, Sudbery PE. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918–930. 10.1038/nrg948 [DOI] [PubMed] [Google Scholar]
- 9.Pukkila-Worley R, Peleg AY, Tampakakis E, Mylonakis E. 2009. Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot. Cell 8:1750–1758. 10.1128/EC.00163-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobolev VS, Cole RJ, Dorner JW, Yagen B. 1995. Isolation, purification, and liquid chromatographic determination of stilbene phytoalexins in peanuts. J. AOAC Int. 78:1177–1182 [Google Scholar]
- 11.Langcake P, Cornford CA, Pryce RJ. 1979. Identification of pterostilbene as aphytoalexin from Vitis vinifera leaves. Phytochemistry 18:1025–1027 [Google Scholar]
- 12.Seshadri TR. 1972. Polyphenols of Pterocarpus and Dalbergia woods. Phytochemistry 11:881–898 [Google Scholar]
- 13.Rimando AM, Kalt W, Magee JB, Dewey J, Ballington JR. 2004. Resveratrol, pterostilbene, and piceatannol in vaccinium berries. J. Agric. Food Chem. 52:4713–4719. 10.1021/jf040095e [DOI] [PubMed] [Google Scholar]
- 14.Cichocki M, Paluszczak J, Szaefer H, Piechowiak A, Rimando AM, Baer-Dubowska W. 2008. Pterostilbene is equally potent as resveratrol in inhibiting 12-O-tetradecanoylphorbol-13-acetate activated NFκB, AP-1, COX-2, and iNOS in mouse epidermis. Mol. Nutr. Food Res. 52(Suppl 1):S62–S70. 10.1002/mnfr.200700466 [DOI] [PubMed] [Google Scholar]
- 15.McCormack D, Schneider J, McDonald D, McFadden D. 2011. The antiproliferative effects of pterostilbene on breast cancer in vitro are via inhibition of constitutive and leptin-induced Janus kinase/signal transducer and activator of transcription activation. Am. J. Surg. 202:541–544. 10.1016/j.amjsurg.2011.06.020 [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Ding L, Wang X, Zhang J, Han W, Feng L, Sun J, Jin H, Wang XJ. 2012. Pterostilbene simultaneously induces apoptosis, cell cycle arrest and cyto-protective autophagy in breast cancer cells. Am. J. Transl. Res. 4:44–51 [PMC free article] [PubMed] [Google Scholar]
- 17.Remsberg CM, Yanez JA, Ohgami Y, Vega-Villa KR, Rimando AM, Davies NM. 2008. Pharmacometrics of pterostilbene: preclinical pharmacokinetics and metabolism, anticancer, antiinflammatory, antioxidant and analgesic activity. Phytother. Res. 22:169–179. 10.1002/ptr.2277 [DOI] [PubMed] [Google Scholar]
- 18.Sobolev VS, Khan SI, Tabanca N, Wedge DE, Manly SP, Cutler SJ, Coy MR, Becnel JJ, Neff SA, Gloer JB. 2011. Biological activity of peanut (Arachis hypogaea) phytoalexins and selected natural and synthetic stilbenoids. J. Agric. Food Chem. 59:1673–1682. 10.1021/jf104742n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pont V, Pezet R. 1990. Relation between the chemical structure and the biological activity of hydroxystilbenes against Botrytis cinerea. J. Phytopathol. 130:1–8 [Google Scholar]
- 20.Adrian M, Jeandet P, Veneau J, Weston LA, Bessis R. 1997. Biological activity of resveratrol, a stilbenic compound from grapevines, against Botrytis cinerea, the causal agent for gray mold. J. Chem. Ecol. 23:1689–1702 [Google Scholar]
- 21.Zou Y, Huang Q, Huang TK, Ni QC, Zhang ES, Xu TL, Yuan M, Li J. 2013. CuI/1,10-phen/PEG promoted decarboxylation of 2,3-diarylacrylic acids: synthesis of stilbenes under neutral and microwave conditions with an in situ generated recyclable catalyst. Org. Biomol. Chem. 11:6967–6974. 10.1039/c3ob41588k [DOI] [PubMed] [Google Scholar]
- 22.Lin HS, Yue BD, Ho PC. 2009. Determination of pterostilbene in rat plasma by a simple HPLC-UV method and its application in pre-clinical pharmacokinetic study. Biomed. Chromatogr. 23:1308–1315. 10.1002/bmc.1254 [DOI] [PubMed] [Google Scholar]
- 23.Maidan MM, De Rop L, Serneels J, Exler S, Rupp S, Tournu H, Thevelein JM, Van Dijck P. 2005. The G protein-coupled receptor Gpr1 and the Gα protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol. Biol. Cell 16:1971–1986. 10.1091/mbc.E04-09-0780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KL, Buckley HR, Campbell CC. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148–153 [DOI] [PubMed] [Google Scholar]
- 25.Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077–1090 [DOI] [PubMed] [Google Scholar]
- 26.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts, vol 28, no. 14. Approved standard, 3rd ed. CLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 27.An M, Shen H, Cao Y, Zhang J, Cai Y, Wang R, Jiang Y. 2009. Allicin enhances the oxidative damage effect of amphotericin B against Candida albicans. Int. J. Antimicrob. Agents 33:258–263. 10.1016/j.ijantimicag.2008.09.014 [DOI] [PubMed] [Google Scholar]
- 28.Quan H, Cao YY, Xu Z, Zhao JX, Gao PH, Qin XF, Jiang YY. 2006. Potent in vitro synergism of fluconazole and berberine chloride against clinical isolates of Candida albicans resistant to fluconazole. Antimicrob. Agents Chemother. 50:1096–1099. 10.1128/AAC.50.3.1096-1099.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y, Wang Y, Yan L, Liang RM, Dai BD, Tang RJ, Gao PH, Jiang YY. 2009. Proteomic analysis reveals a synergistic mechanism of fluconazole and berberine against fluconazole-resistant Candida albicans: endogenous ROS augmentation. J. Proteome Res. 8:5296–5304. 10.1021/pr9005074 [DOI] [PubMed] [Google Scholar]
- 30.Ramage G, Saville SP, Wickes BL, Lopez-Ribot JL. 2002. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 68:5459–5463. 10.1128/AEM.68.11.5459-5463.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klotz SA, Drutz DJ, Zajic JE. 1985. Factors governing adherence of Candida species to plastic surfaces. Infect. Immun. 50:97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2(7):e63. 10.1371/journal.ppat.0020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao Y, Dai B, Wang Y, Huang S, Xu Y, Gao P, Zhu Z, Jiang Y. 2008. In vitro activity of baicalein against Candida albicans biofilms. Int. J. Antimicrob. Agents 32:73–77. 10.1016/j.ijantimicag.2008.01.026 [DOI] [PubMed] [Google Scholar]
- 34.Braga PC, Culici M, Alfieri M, Dal Sasso M. 2008. Thymol inhibits Candida albicans biofilm formation and mature biofilm. Int. J. Antimicrob. Agents 31:472–477. 10.1016/j.ijantimicag.2007.12.013 [DOI] [PubMed] [Google Scholar]
- 35.Köhrer K, Domdey H. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398–405 [DOI] [PubMed] [Google Scholar]
- 36.Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao W, Fang H, Kawasaki ES, Hager J, Tikhonova IR. 2006. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) Project. Nat. Biotechnol. 24:1140–1150. 10.1038/nbt1242 [DOI] [PubMed] [Google Scholar]
- 37.Guo Y, Guo H, Zhang L, Xie H, Zhao X, Wang F, Li Z, Wang Y, Ma S, Tao J, Wang W, Zhou Y, Yang W, Cheng J. 2005. Genomic analysis of anti-hepatitis B virus (HBV) activity by small interfering RNA and lamivudine in stable HBV-producing cells. J. Virol. 79:14392–14403. 10.1128/JVI.79.22.14392-14403.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. 10.1093/nar/30.4.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. 2004. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20:3710–3715. 10.1093/bioinformatics/bth456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li DD, Wang Y, Dai BD, Li XX, Zhao LX, Cao YB, Yan L, Jiang YY. 2013. ECM17-dependent methionine/cysteine biosynthesis contributes to biofilm formation in Candida albicans. Fungal Genet. Biol. 51:50–59. 10.1016/j.fgb.2012.11.010 [DOI] [PubMed] [Google Scholar]
- 41.Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. 2004. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect. Immun. 72:6023–6031. 10.1128/IAI.72.10.6023-6031.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seneviratne CJ, Jin L, Samaranayake LP. 2008. Biofilm lifestyle of Candida: a mini review. Oral Dis. 14:582–590. 10.1111/j.1601-0825.2007.01424.x [DOI] [PubMed] [Google Scholar]
- 43.Luo G, Samaranayake LP. 2002. Candida glabrata, an emerging fungal pathogen, exhibits superior relative cell surface hydrophobicity and adhesion to denture acrylic surfaces compared with Candida albicans. APMIS 110:601–610. 10.1034/j.1600-0463.2002.1100902.x [DOI] [PubMed] [Google Scholar]
- 44.Pompilio A, Piccolomini R, Picciani C, D'Antonio D, Savini V, Di Bonaventura G. 2008. Factors associated with adherence to and biofilm formation on polystyrene by Stenotrophomonas maltophilia: the role of cell surface hydrophobicity and motility. FEMS Microbiol. Lett. 287:41–47. 10.1111/j.1574-6968.2008.01292.x [DOI] [PubMed] [Google Scholar]
- 45.Samaranayake YH, Wu PC, Samaranayake LP, So M. 1995. Relationship between the cell surface hydrophobicity and adherence of Candida krusei and Candida albicans to epithelial and denture acrylic surfaces. APMIS 103:707–713 [PubMed] [Google Scholar]
- 46.Zhu Y, Fang HM, Wang YM, Zeng GS, Zheng XD, Wang Y. 2009. Ras1 and Ras2 play antagonistic roles in regulating cellular cAMP level, stationary-phase entry and stress response in Candida albicans. Mol. Microbiol. 74:862–875. 10.1111/j.1365-2958.2009.06898.x [DOI] [PubMed] [Google Scholar]
- 47.Birse CE, Irwin MY, Fonzi WA, Sypherd PS. 1993. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect. Immun. 61:3648–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beggah S, Lechenne B, Reichard U, Foundling S, Monod M. 2000. Intra- and intermolecular events direct the propeptide-mediated maturation of the Candida albicans secreted aspartic proteinase Sap1p. Microbiology 146:2765–2773 [DOI] [PubMed] [Google Scholar]
- 49.Urban C, Sohn K, Lottspeich F, Brunner H, Rupp S. 2003. Identification of cell surface determinants in Candida albicans reveals Tsa1p, a protein differentially localized in the cell. FEBS Lett. 544:228–235 http://dx.doi.org/10.1016/S0014-5793(03)00455-1 [DOI] [PubMed] [Google Scholar]
- 50.Uhl MA, Biery M, Craig N, Johnson AD. 2003. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 22:2668–2678. 10.1093/emboj/cdg256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Granger BL, Flenniken ML, Davis DA, Mitchell AP, Cutler JE. 2005. Yeast wall protein 1 of Candida albicans. Microbiology 151:1631–1644. 10.1099/mic.0.27663-0 [DOI] [PubMed] [Google Scholar]
- 52.Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9:737–748. 10.1038/nrmicro2636 [DOI] [PubMed] [Google Scholar]
- 53.Chaffin WL. 2008. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 72:495–544. 10.1128/MMBR.00032-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Braun BR, Johnson AD. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hogan DA, Sundstrom P. 2009. The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future Microbiol. 4:1263–1270. 10.2217/fmb.09.106 [DOI] [PubMed] [Google Scholar]
- 56.Vila TV, Ishida K, de Souza W, Prousis K, Calogeropoulou T, Rozental S. 2013. Effect of alkylphospholipids on Candida albicans biofilm formation and maturation. J. Antimicrob. Chemother. 68:113–125. 10.1093/jac/dks353 [DOI] [PubMed] [Google Scholar]
- 57.Okamoto-Shibayama K, Sato Y, Azuma T. 2010. Resveratrol impaired the morphological transition of Candida albicans under various hyphae-inducing conditions. J. Microbiol. Biotechnol. 20:942–945 [DOI] [PubMed] [Google Scholar]
- 58.Collado-Gonzalez M, Guirao-Abad JP, Sanchez-Fresneda R, Belchi-Navarro S, Arguelles JC. 2012. Resveratrol lacks antifungal activity against Candida albicans. World J. Microbiol. Biotechnol. 28:2441–2446. 10.1007/s11274-012-1042-1 [DOI] [PubMed] [Google Scholar]
- 59.Weber K, Schulz B, Ruhnke M. 2011. Resveratrol and its antifungal activity against Candida species. Mycoses 54:30–33. 10.1111/j.1439-0507.2009.01763.x [DOI] [PubMed] [Google Scholar]
- 60.Nobile CJ, Mitchell AP. 2006. Genetics and genomics of Candida albicans biofilm formation. Cell. Microbiol. 8:1382–1391. 10.1111/j.1462-5822.2006.00761.x [DOI] [PubMed] [Google Scholar]
- 61.Ramage G, Saville SP, Thomas DP, Lopez-Ribot JL. 2005. Candida biofilms: an update. Eukaryot. Cell 4:633–638. 10.1128/EC.4.4.633-638.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sundstrom P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4:461–469. 10.1046/j.1462-5822.2002.00206.x [DOI] [PubMed] [Google Scholar]
- 63.Tronchin G, Pihet M, Lopes-Bezerra LM, Bouchara JP. 2008. Adherence mechanisms in human pathogenic fungi. Med. Mycol. 46:749–772. 10.1080/136937802206435 [DOI] [PubMed] [Google Scholar]
- 64.Zheng X, Wang Y. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23:1845–1856. 10.1038/sj.emboj.7600195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nett JE, Lepak AJ, Marchillo K, Andes DR. 2009. Time course global gene expression analysis of an in vivo Candida biofilm. J. Infect. Dis. 200:307–313. 10.1086/599838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rajput SB, Karuppayil SM. 2013. β-Asarone, an active principle of Acorus calamus rhizome, inhibits morphogenesis, biofilm formation and ergosterol biosynthesis in Candida albicans. Phytomedicine 20:139–142. 10.1016/j.phymed.2012.09.029 [DOI] [PubMed] [Google Scholar]
- 67.Burnie JP, Carter TL, Hodgetts SJ, Matthews RC. 2006. Fungal heat-shock proteins in human disease. FEMS Microbiol. Rev. 30:53–88. 10.1111/j.1574-6976.2005.00001.x [DOI] [PubMed] [Google Scholar]
- 68.Pan Z, Agarwal AK, Xu T, Feng Q, Baerson SR, Duke SO, Rimando AM. 2008. Identification of molecular pathways affected by pterostilbene, a natural dimethylether analog of resveratrol. BMC Med. Genomics 1:7. 10.1186/1755-8794-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bahn YS, Molenda M, Staab JF, Lyman CA, Gordon LJ, Sundstrom P. 2007. Genome-wide transcriptional profiling of the cyclic AMP-dependent signaling pathway during morphogenic transitions of Candida albicans. Eukaryot. Cell 6:2376–2390. 10.1128/EC.00318-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kapetanovic IM, Muzzio M, Huang Z, Thompson TN, McCormick DL. 2011. Pharmacokinetics, oral bioavailability, and metabolic profile of resveratrol and its dimethylether analog, pterostilbene, in rats. Cancer Chemother. Pharmacol. 68:593–601. 10.1007/s00280-010-1525-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruiz MJ, Fernandez M, Pico Y, Manes J, Asensi M, Carda C, Asensio G, Estrela JM. 2009. Dietary administration of high doses of pterostilbene and quercetin to mice is not toxic. J. Agric. Food Chem. 57:3180–3186. 10.1021/jf803579e [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.