Abstract
Molecular and virulence characteristics of CTX-M-producing and non-extended-spectrum-β-lactamase (non-ESBL)-producing Klebsiella pneumoniae isolates were compared. Lack of shared characteristics between the two groups suggested that most CTX-M-producing K. pneumoniae isolates in South Korea did not occur by transfer of blaCTX-M into susceptible strains. Conjugation assays confirmed that the plasmid with the blaCTX-M-15 gene confers virulence as well as antimicrobial resistance, suggesting that a CTX-M-15-producing clone such as ST11 may have a selective advantage even without antibiotic pressure.
TEXT
In parallel with the use of antibiotic drugs, the prevalence of Escherichia coli and Klebsiella pneumoniae strains producing CTX-M-type extended-spectrum β-lactamases (ESBLs) has increased worldwide (1). In addition to CTX-M-15-producing E. coli, CTX-M-15 has increased in prevalence in K. pneumoniae worldwide (2–4). In K. pneumoniae, blaCTX-M-15 is carried mainly by IncFII-type plasmids (5). Production of the SHV-type ESBLs in K. pneumoniae is associated with an increased tendency to invade epithelial cells and expression of fimbrial adhesions (6). Although highly virulent clones expressing CTX-M-type β-lactamase, such as E. coli ST131, have been reported (7), the association of CTX-M enzyme production with virulence and fitness in K. pneumoniae is not clear. In this study, we compared genotypic and phenotypic characteristics between CTX-M-producing and non-ESBL-producing K. pneumoniae isolates from South Korea. In addition, the fitness cost of carrying the blaCTX-M-15 gene-bearing plasmid was investigated.
In this study, 98 K. pneumoniae isolates, which were collected from patients with bacteremia from nine South Korean hospitals as a part of multicenter surveillance study from September to December 2008, were included (8). Thirty-three isolates were found to express blaCTX-M genes (18 blaCTX-M-14 and 15 blaCTX-M-15), while 65 isolates did not produce any ESBL. ESBL activity was confirmed using the double-disc method. In vitro antimicrobial susceptibility testing was performed by a broth microdilution method, according to the CLSI guidelines (9). Multilocus sequence typing (MLST) was performed as described previously (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html) with some modifications. Pulsed-field gel electrophoresis (PFGE) was performed for all ST11 K. pneumoniae isolates (10). PCR assays were performed to monitor for the presence of genes previously found to be associated with virulence in K. pneumoniae (11, 12). The string test was used to determine the hypermucoviscosity phenotype (13). α-Hemolysin production was detected using a 5% sheep blood agar plate (14).
The transfer of the plasmid carrying blaCTX-M-15 was accomplished using an E. coli DH5α strain as described previously (15). The plasmid carrying the blaCTX-M-15 gene from an ST11 K. pneumoniae isolate, K01-12226, was used (5). The plasmid carrying blaCTX-M-15 was transferred into five non-ESBL-producing ST11 K. pneumoniae isolates, K01-1054, K01-6053, K01-7096, K01-8102, and K01-8139, from E. coli DH5α as a donor (8). The presence of blaCTX-M-15 in transconjugants was confirmed by PCR. A serum sensitivity assay was performed on clinical isolates and transconjugants, as previously described (16). Fisher's exact test was used to determine the significance of differences in serum resistance between strains using SPSS version 11.5 (SPSS, Chicago, IL, USA).
The CTX-M-producing K. pneumoniae isolates showed significantly greater resistance to most antibiotics except ampicillin, amikacin, and imipenem than did non-ESBL-producing isolates (P < 0.05) (Table 1). While cf29a, a gene in E. coli encoding adhesin CS31A and associated with diarrhea in humans, was found in only one CTX-M-14-producing, ST11 isolate, it was present in 14 non-ESBL-producing isolates (P = 0.018) (Table 1). Eight (72.7%) of 11 non-ESBL-producing ST11 isolates possessed the cf29a gene, and four ST163 isolates tested positive for it. In addition to cf29a, allS, which encodes an activator of the allantoin regulon, was found more frequently in non-ESBL-producing isolates (P = 0.05).
TABLE 1.
Antimicrobial resistance and virulence factors of CTX-M-producing and non-ESBL-producing K. pneumoniae isolates
| Antimicrobial resistance or virulence factor | No. (%) of isolates |
Pa | ||
|---|---|---|---|---|
| Total (n = 98) | CTX-M-producing (n = 33) | Non-ESBL-producing (n = 65) | ||
| Resistance to antimicrobial agent | ||||
| Ampicillin | 97 (99.0) | 33 (100) | 64 (98.5) | 0.474 |
| Ceftazidime | 50 (51.0) | 31 (93.9) | 19 (29.2) | <0.001 |
| Cefotaxime | 47 (48.0) | 28 (84.8) | 19 (29.2) | <0.001 |
| Aztreonam | 42 (42.9) | 27 (81.8) | 15 (23.1) | <0.001 |
| Amikacin | 11 (11.2) | 2 (6.1) | 9 (13.8) | 0.327 |
| Gentamicin | 50 (51.0) | 33 (100) | 17 (26.2) | <0.001 |
| Ciprofloxacin | 43 (43.9) | 20 (60.6) | 23 (35.4) | 0.017 |
| Imipenem | 1 (1.0) | 1 (3) | 0 | 0.287 |
| Trimethoprim-sulfamethoxazole | 45 (45.9) | 30 (90.9) | 15 (23.8) | <0.001 |
| Piperacillin-tazobactam | 52 (53.1) | 22 (66.7) | 30 (46.2) | 0.022 |
| Virulence factor | ||||
| fimH | 98 (100) | 33 (100) | 65 (100) | |
| oxyR | 98 (100) | 33 (100) | 65 (100) | |
| ureA | 98 (100) | 33 (100) | 65 (100) | |
| kfu | 97 (99.0) | 32 (96.9) | 65 (100) | 0.330 |
| wabG | 97 (99.0) | 33 (100) | 64 (94.1) | 0.549 |
| uge | 92 (93.8) | 33 (100) | 59 (90.7) | 0.174 |
| ramA | 91 (92.8) | 31 (93.9) | 60 (92.3) | 0.579 |
| allS | 31 (31.6) | 4 (12.1) | 27 (41.5) | 0.005 |
| iutA | 29 (29.5) | 8 (24.2) | 21 (32.3) | 0.494 |
| rmpA | 24 (24.4) | 4 (12.1) | 20 (30.7) | 0.080 |
| wca | 22 (22.4) | 5 (15.1) | 17 (26.1) | 0.310 |
| cf29a | 15 (15.3) | 1 (3.0) | 14 (21.5) | 0.018 |
| magA | 13 (13.2) | 2 (6.0) | 11 (16.9) | 0.134 |
| ybtQ | 2 (2.0) | 0 | 2 (3.0) | 0.549 |
P values between CTX-M-producing and non-ESBL-producing K. pneumoniae isolates.
In MLST analysis, a total of 52 different STs were identified among the 98 K. pneumoniae isolates: 19 STs among the 33 CTX-M-producing isolates and 36 among the 65 non-ESBL-producing isolates (Table 2). Only three STs (ST11, ST15, and ST48) were detected in both CTX-M-producing and non-ESBL-producing isolates. This suggests that most CTX-M-producing K. pneumoniae isolates in South Korea did not occur by transfer of the blaCTX-M gene into susceptible strains. In PFGE analysis, 16 ST11 isolates did not show exactly the same restriction pattern, but similarities in pattern could be identified. In particular, an approximately 388-kb fragment was found only in CTX-M-15-producing K. pneumoniae isolates. It was revealed to be a plasmid carrying blaCTX-M-15, by PCR. In addition, PFGE analysis of transconjugants receiving a plasmid showed an additional 388-kb band (not shown). It may be premature to conclude that CTX-M-15-producing ST11 isolates arise by transfer of a blaCTX-M-15-bearing plasmid into susceptible strains unique to South Korea, since ST11 is distributed among CTX-M-15-producing clones worldwide (17).
TABLE 2.
MLST analysis of CTX-M-producing and non-ESBL-producing K. pneumoniae isolates
| Clonal complex (CC) | ST | Allelic profilea | No. of isolates |
||
|---|---|---|---|---|---|
| Total (n = 98) | CTX-M-producing (n = 33) | Non-ESBL-producing (n = 65) | |||
| CC11 | 11 | 3-3-1-1-1-1-4 | 16 | 5 | 11 |
| 258 | 3-3-1-1-1-1-79 | 1 | 1 | ||
| 340 | 3-3-1-1-1-1-18 | 1 | 1 | ||
| CC631 | 163 | 2-1-1-1-9-1-12 | 10 | 10 | |
| 23 | 2-1-1-1-9-4-12 | 3 | 3 | ||
| 17 | 2-1-1-1-4-4-4 | 1 | 1 | ||
| 18 | 2-1-1-1-4-1-4 | 1 | 1 | ||
| 631 | 2-1-1-1-9-4-4 | 1 | 1 | ||
| 1059 | 2-1-1-1-12-1-4 | 1 | 1 | ||
| CC298 | 36 | 2-1-2-1-7-1-7 | 3 | 3 | |
| 298 | 2-1-2-1-1-1-7 | 1 | 1 | ||
| 966 | 2-1-2-1-1-1-68 | 1 | 1 | ||
| CC375 | 375 | 43-1-2-1-10-4-3 | 2 | 2 | |
| 65 | 2-1-2-1-10-4-13 | 1 | 1 | ||
| 1053 | 16-1-2-1-10-4-13 | 1 | 1 | ||
| CC469 | 469 | 2-1-2-1-10-1-4 | 2 | 2 | |
| 35 | 2-1-2-1-10-1-19 | 1 | 1 | ||
| CC1063 | 1063 | 2-3-1-1-9-4-193 | 1 | 1 | |
| 1064 | 2-3-1-1-1-1-193 | 1 | 1 | ||
| Singleton | 15 | 1-1-1-1-1-1-1 | 8 | 7 | 1 |
| 86 | 9-4-2-1-1-1-27 | 5 | 5 | ||
| 48 | 2-5-2-2-7-1-10 | 4 | 2 | 2 | |
| 34 | 2-3-6-1-9-7-4 | 2 | 2 | ||
| 101 | 2-6-1-5-4-1-6 | 2 | 2 | ||
| 12 | 6-3-1-1-12-1-4 | 1 | 1 | ||
| 76 | 4-1-1-1-21-1-35 | 1 | 1 | ||
| 105 | 2-3-2-1-1-4-18 | 1 | 1 | ||
| 110 | 2-6-1-3-8-1-44 | 1 | 1 | ||
| 147 | 2-6-1-3-8-1-44 | 1 | 1 | ||
| 165 | 2-1-13-2-23-1-19 | 1 | 1 | ||
| 222 | 2-1-2-2-7-4-4 | 1 | 1 | ||
| 300 | 2-1-19-1-9-4-34 | 1 | 1 | ||
| 317 | 10-1-2-1-9-27-18 | 1 | 1 | ||
| 372 | 2-1-2-1-1-15-4 | 1 | 1 | ||
| 380 | 2-1-1-1-1-4-19 | 1 | 1 | ||
| 412 | 2-1-2-1-9-1-112 | 1 | 1 | ||
| 502 | 2-53-3-10-4-18 | 1 | 1 | ||
| 518 | 2-3-1-1-7-4-87 | 1 | 1 | ||
| 537 | 6-3-1-4-12-4-4 | 1 | 1 | ||
| 538 | 2-1-2-20-9-1-14 | 1 | 1 | ||
| 1026 | 2-1-2-35-10-24-19 | 1 | 1 | ||
| 1050 | 16-8-21-31-92-17-67 | 1 | 1 | ||
| 1051 | 2-1-11-1-9-10-9 | 1 | 1 | ||
| 1052 | 2-1-1-1-17-1-42 | 1 | 1 | ||
| 1054 | 2-3-1-1-12-4-12 | 1 | 1 | ||
| 1055 | 16-24-21-1-1-17-1 | 1 | 1 | ||
| 1056 | 16-62-21-40-153-40-67 | 1 | 1 | ||
| 1057 | 16-24-21-27-47-17-134 | 1 | 1 | ||
| 1058 | 43-1-2-1-10-1-12 | 1 | 1 | ||
| 1060 | 16-18-21-21-52-30-75 | 1 | 1 | ||
| 1061 | 2-3-1-20-61-4-181 | 1 | 1 | ||
| 1062 | 2-3-2-2-162-1-4 | 1 | 1 | ||
Allelic profile, gapA-infB-mdh-pgi-phoE-rpoB-tonB.
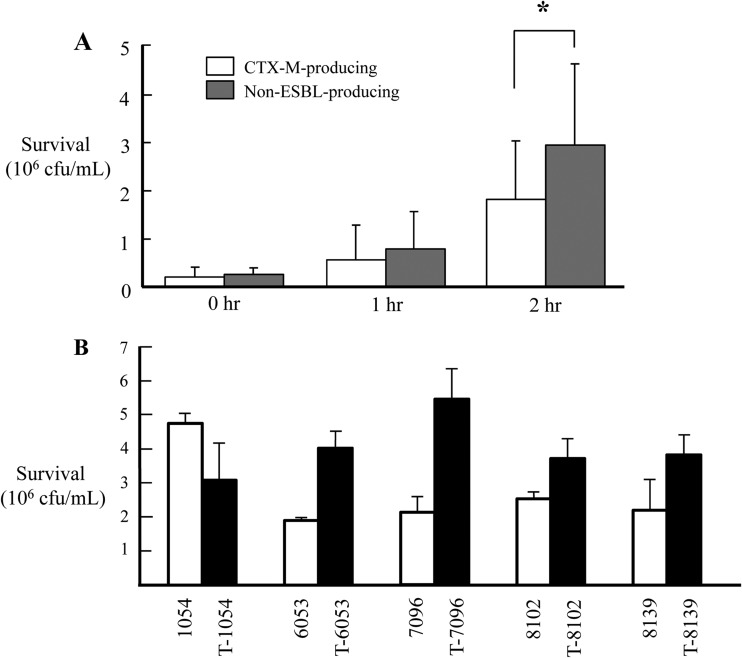
Non-ESBL-producing K. pneumoniae isolates showed higher serum resistance than did CTX-M-producing isolates (P = 0.004) (Fig. 1A). In addition, the hypermucoviscosity phenotype was more frequently identified in non-ESBL-producing isolates; only three CTX-M-producing isolates (9.1%) expressed the hypermucoviscosity phenotype compared to 19 non-ESBL-producing isolates (29.2%) (P = 0.038). None of the K. pneumoniae isolates produced α-hemolysin.
FIG 1.
Results of serum resistance assay are shown here as CFU viability. Error bars indicate standard deviations. (A) Serum resistance assay for all CTX-M-producing and non-ESBL-producing K. pneumoniae isolates. Significance is shown for the difference between CTX-M-producing and non-ESBL-producing K. pneumoniae isolates (*, P < 0.05). (B) Survival rate of each non-ESBL-producing K. pneumoniae isolate and its transconjugant after incubation for 2 h.
To understand the effects of the plasmid on fitness measures such as growth rate and serum resistance, plasmids carrying the blaCTX-M-15 gene of an ST11 isolate were transferred to non-ESBL-producing ST11 isolates by conjugation. All of these transconjugants showed substantial increases in MICs for cephalosporins, with one exception (cefotaxime for T-6053). The CTX-M-producing, non-ESBL-producing, and transconjugant strains showed no significant differences in growth rates. Although serum resistance did not differ between CTX-M-producing and non-ESBL-producing ST11 K. pneumoniae isolates, transconjugants showed higher survival rates against serum than did their host isolates and donors (Fig. 1B). Comparing the survival rates against serum in pairs of transconjugant and host isolates, transconjugants showed significantly higher serum resistance than did their host isolates, except for the pair that included K01-1054 and T-1054. The traT gene in the plasmid may contribute to serum resistance, which explains the increased survival rates against serum in the transconjugants.
As a whole, non-ESBL-producing K. pneumoniae isolates were assumed to be more virulent than CTX-M-producing isolates. First of all, non-ESBL-producing isolates showed a higher rate of resistance against human serum than did CTX-M-producing K. pneumoniae isolates. Second, the hypermucoviscosity phenotype was more frequently found in non-ESBL-producing isolates. In addition, while CTX-M-producing isolates contained 7.64 virulence factors on average, 8.54 virulence factors, on average, were identified in non-ESBL-producing isolates (P = 0.005).
However, serum resistance did not differ between CTX-M-15-producing and non-ESBL-producing isolates of ST11. More importantly, four out of five transconjugants showed higher serum resistance than did their hosts (Fig. 1B). This suggests that plasmids with the blaCTX-M-15 gene may confer virulence as well as antimicrobial resistance, although only one kind of plasmid (IncFII) was tested in this study. Thus, CTX-M-15-producing K. pneumoniae isolates may carry more than one positively selective trait, implying that antimicrobial-resistant strains could increase in prevalence even without antimicrobial pressure. Although increased serum resistance is generally associated with decreased virulence and fitness, credible evidence to the contrary has emerged (18). Global dissemination of highly pathogenic and resistant clones would be cause for great concern (7).
ACKNOWLEDGMENTS
The K. pneumoniae strains used in this study were obtained from the Asian Bacterial Bank (ABB) of the Asia Pacific Foundation for Infectious Diseases (APFID; Seoul, South Korea).
This research was supported by the Basic Science Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT and Future Planning (2010-0004848). J. Shin was supported partly by the Basic Science Research Program through the NRF funded by the Ministry of Education (2013R1A1A2062884).
Footnotes
Published ahead of print 10 February 2014
REFERENCES
- 1.Canton R, Coque TM. 2006. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9:466–475. 10.1016/j.mib.2006.08.011 [DOI] [PubMed] [Google Scholar]
- 2.Coelho A, González-López JJ, Miró E, Alonso-Tarrés C, Mirelis B, Larrosa MN, Bartolomé RM, Andreu A, Navarro F, Johnson JR, Prats G. 2010. Characterization of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. Int. J. Antimicrob. Agents 36:73–78. 10.1016/j.ijantimicag.2010.03.005 [DOI] [PubMed] [Google Scholar]
- 3.Ensor VM, Jamal W, Rotimi VO, Evans JT, Hawkey PM. 2009. Predominance of CTXM-15 extended-spectrum β-lactamases in diverse Escherichia coli and Klebsiella pneumoniae from hospital and community patients in Kuwait. Int. J. Antimicrob. Agents 33:487–489. 10.1016/j.ijantimicag.2008.10.011 [DOI] [PubMed] [Google Scholar]
- 4.Ko KS, Lee JY, Baek JY, Suh JY, Lee MY, Choi JY, Yeom JS, Kim YS, Jung SI, Shin SY, Heo ST, Kwon KT, Son JS, Kim SW, Chang HH, Ki HK, Chung DR, Peck KR, Song JH. 2010. Predominance of an ST11 extended-spectrum β-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J. Med. Microbiol. 59:822–828. 10.1099/jmm.0.018119-0 [DOI] [PubMed] [Google Scholar]
- 5.Shin J, Choi MJ, Ko KS. 2012. Replicon sequence typing of IncF plasmids and the genetic environments of blaCTX-M-15 indicate multiple acquisitions of blaCTX-M-15 in Escherichia coli and Klebsiella pneumoniae isolates from South Korea. J. Antimicrob. Chemother. 67:1853–1857. 10.1093/jac/dks143 [DOI] [PubMed] [Google Scholar]
- 6.Sahly H, Navon-Venezia S, Roesler L, Hay A, Carmeli Y, Podschun R, Hennequin C, Forestier C, Ofek I. 2008. Extended-spectrum β-lactamase production is associated with an increase in cell invasion and expression of fimbrial adhesions in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52:3029–3034. 10.1128/AAC.00010-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol. Lett. 35:736–755. 10.1111/j.1574-6976.2011.00268.x [DOI] [PubMed] [Google Scholar]
- 8.Shin J, Kim DH, Ko KS. 2011. Comparison of CTX-M-14- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae isolates from patients with bacteremia. J. Infect. 63:39–47. 10.1016/j.jinf.2011.05.003 [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2011. Performance standards for antimicrobial susceptibility testing. 21st informational supplement. Document M100-S21 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10.Samuelsen O, Naseer U, Tofteland S, Skutlaberg DH, Onken A, Hjetland R, Sundsfjord A, Giske CG. 2009. Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J. Antimicrob. Chemother. 63:654–658. 10.1093/jac/dkp018 [DOI] [PubMed] [Google Scholar]
- 11.Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. 10.1371/journal.pone.0004982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hennequin C, Forestier C. 2009. oxyR, a LysR-type regulator involved in Klebsiella pneumoniae mucosal and abiotic colonization. Infect. Immun. 77:5449–5457. 10.1128/IAI.00837-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiskur BJ, Hunt JJ, Callegan MC. 2008. Hypermucoviscosity as a virulence factor in experimental Klebsiella pneumoniae endophthalmitis. Invest. Ophthalmol. Vis. Sci. 49:4931–4938. 10.1167/iovs.08-2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koczura R, Kaznowski A. 2003. Occurrence of the Yersinia high-pathogenicity island and iron uptake systems in clinical isolates of Klebsiella pneumoniae. Microb. Pathog. 35:197–202. 10.1016/S0882-4010(03)00125-6 [DOI] [PubMed] [Google Scholar]
- 15.Wollheim C, Guerra IMF, Conte VD, Hoffman SP, Schreiner FJ, Delamare AP, Barth AL, Echeverrigaray S, Costa SO. 2011. Nosocomial and community infections due to class A extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella spp. in southern Brazil. Braz. J. Infect. Dis. 15:138–143. 10.1590/S1413-86702011000200008 [DOI] [PubMed] [Google Scholar]
- 16.Dozois CM, Fairbrother JM, Harel J, Bosse M. 1992. pap- and pil-related DNA sequences and other virulence determinants associated with Escherichia coli isolated from septicemic chickens and turkeys. Infect. Immun. 60:2648–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damjanova I, Tóth A, Pászti J, Hajbel-Vékony G, Jakab M, Berta J, Milch H, Füzi M. 2008. Expansion and countrywide dissemination of ST11, ST15, and ST147 ciprofloxacin-resistant CTX-M-15-type β-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005—the new ‘MRSAs'? J. Antimicrob. Chemother. 62:978–985. 10.1093/jac/dkn287 [DOI] [PubMed] [Google Scholar]
- 18.Beceiro A, Tamás M, Bou G. 2013. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 26:185–230. 10.1128/CMR.00059-12 [DOI] [PMC free article] [PubMed] [Google Scholar]