Abstract
DNA gyrase is a type II topoisomerase that catalyzes the introduction of negative supercoils in the genomes of eubacteria. Fluoroquinolones (FQs), successful as drugs clinically, target the enzyme to trap the gyrase-DNA complex, leading to the accumulation of double-strand breaks in the genome. Mycobacteria are less susceptible to commonly used FQs. However, an 8-methoxy-substituted FQ, moxifloxacin (MFX), is a potent antimycobacterial, and a higher susceptibility of mycobacterial gyrase to MFX has been demonstrated. Although several models explain the mechanism of FQ action and gyrase-DNA-FQ interaction, the basis for the differential susceptibility of mycobacterial gyrase to various FQs is not understood. We have addressed the basis of the differential susceptibility of the gyrase and revisited the mode of action of FQs. We demonstrate that FQs bind both Escherichia coli and Mycobacterium tuberculosis gyrases in the absence of DNA and that the addition of DNA enhances the drug binding. The FQs bind primarily to the GyrA subunit of mycobacterial gyrase, while in E. coli holoenzyme is the target. The binding of MFX to GyrA of M. tuberculosis correlates with its effectiveness as a better inhibitor of the enzyme and its efficacy in cell killing.
INTRODUCTION
The DNA gyrase is an essential enzyme responsible for the maintenance of DNA topology in eubacteria. The enzyme catalyzes ATP-dependent negative supercoiling of relaxed circular DNA (1). It is composed of two GyrA and two GyrB subunits resulting in a heterotetrameric holoenzyme. Negative supercoiling by DNA gyrase involves a series of sequential events. The enzyme cleaves duplex DNA through which another segment of the same DNA molecule is transferred. The cleaved DNA is resealed followed by the release of substrate DNA, completing one round of DNA supercoiling (2–4). Owing to its indispensability for bacterial survival and its absence in mammals, gyrase has been a much sought-after drug target, culminating in the characterization of a number of inhibitors with diverse mechanisms of action. Among them, coumarins (5) and cyclothialidines (6) inhibit ATPase activity, and quinolones (7), CcdB (8), and microcin B17 (9) arrest the gyrase-DNA covalent complex. Several other compounds inhibit the enzyme activity (10–15). Among all of these molecules, fluorine substituted quinolones (FQs) act as bactericidal agents by poisoning the gyrase-DNA complex. Arrest of gyrase-DNA covalent complex leads to the accumulation of double-strand DNA breaks, triggering the activation of SOS pathways, depletion of reducing equivalents, and generation of reactive oxygen intermediates causing cell death (16, 17). Due to this potent bactericidal effect, FQs are used for the treatment of a wide range of bacterial infections (18).
Among all bacterial infections, tuberculosis (TB) continues to pose a major global challenge. The treatment of TB includes a combination of several drugs. FQs are now used to treat infection caused by rifampin-resistant tubercle bacilli (19). Although DNA gyrase from many pathogens is inhibited by FQs, Mycobacterium tuberculosis showed a lower susceptibility to the commonly used FQs, e.g., ciprofloxacin (CFX) and ofloxacin (OFX) (20). However, newer-generation FQs, moxifloxacin (MFX) and gatifloxacin (GFX), showed higher activity against M. tuberculosis and M. smegmatis. The studies with purified enzymes showed that MFX is a better inhibitor of both M. smegmatis and M. tuberculosis gyrases (20, 21; our unpublished results).
We sought here to understand the differential FQ susceptibility of gyrases from mycobacteria to explain why MFX inhibits the enzymes at a lower concentration than CFX. While investigating the basis for differential FQ susceptibility, we revisited the mechanism of action of the FQs. Although the FQs were initially suggested to be interacting with DNA (22), the interaction with both DNA and the enzyme was apparent from subsequent studies (23–27). Now, we provide a further insight into the mechanism of FQ action. We demonstrate the direct binding of the FQs to the enzyme, in addition to the gyrase-DNA complex. The differential susceptibility seems to be governed by the differences in the interaction pattern of the FQs with the enzyme. Surprisingly, we found that FQs bind primarily to GyrA of mycobacterial gyrases, a mode of interaction distinct from that of the Escherichia coli gyrase.
MATERIALS AND METHODS
Chemicals and enzymes.
The 72- and 143-bp oligonucleotides used in the present study were procured and prepared for the studies as described earlier (28). CFX, OFX, GFX, oxolinic acid (OXO), DNase I, proteinase K, IPTG (isopropyl-β-d-thiogalactopyranoside), and salmon sperm DNA were from Sigma-Aldrich (USA). MFX hydrochloride was a gift from H. Dornauer and B. N. Roy (Lupin, Ltd., India). E. coli DNA gyrase (29), M. smegmatis DNA gyrase (21), and E. coli Topo IV (30) subunits were purified as explained previously. The purification of GyrA subunit from the FQ-resistant strain of M. smegmatis mc2155 (31) was carried out as described previously (21). The purification was carried out using an automated Åkta purifier from GE Healthcare. MFX and CFX stock solutions and the subsequent dilutions were prepared in 25 mM Tris-HCl (pH 7.4) immediately before the fluorescence-based experiments.
Purification of M. tuberculosis DNA gyrase.
The open reading frame (ORF) encoding GyrA was cloned in pET20b, and the subunit was overexpressed in the E. coli BL21/pLysS strain (induction with 0.3 mM IPTG at an A595 of 0.8 for 3 h at 25°C). The cells were harvested and resuspended in buffer A (50 mM Tris-HCl [pH 7.4], 5% [vol/vol] glycerol, 0.1 mM EDTA, 50 mM NaCl, 2 mM 2-mercaptoethanol). The cells were lysed, and the S100 supernatant was subjected to 55% ammonium sulfate precipitation. The ammonium sulfate pellet was dissolved in buffer A and dialyzed against the same buffer. The dialyzed protein was loaded onto a Hi-Trap heparin column (GE Healthcare) and eluted with a linear gradient of 200 to 700 mM NaCl. The fractions containing GyrA were dialyzed against buffer A and loaded onto a Hi-Trap MonoQ column (GE Healthcare). Elution was carried out by using a linear gradient of 300 to 800 mM NaCl. The fractions containing GyrA were dialyzed against buffer A and loaded onto a Hi-Trap SP Sepharose column (GE Healthcare), followed by elution with 200 to 800 mM NaCl. The fractions were dialyzed against buffer A, followed by buffer B (100 mM KCl, Tris-HCl [pH 7.4], 2 mM 2-mercaptoethanol, 50% [vol/vol] glycerol), and then stored at −70°C. The ORF encoding GyrB was cloned in pET20b, and overexpression was carried out using similar parameters. It was purified by passing through Hi-Trap heparin and MonoQ columns using 100 to 300 mM NaCl for elution. The protein fractions were dialyzed against buffer A, followed by buffer B and stored at −70°C. The holoenzyme was reconstituted by mixing the GyrA and GyrB subunits in a 1:2 GyrA/GyrB molar ratio. The D94G GyrA of M. smegmatis was mixed with a twofold excess of wild-type M. smegmatis GyrB to reconstitute the mutant holoenzyme.
Enzyme assays.
DNA gyrase assays were carried out with E. coli, M. smegmatis, and M. tuberculosis enzymes. Plasmid and oligonucleotide cleavage experiments were carried out using the methods described earlier (21). A total of 200 ng of pUC18 plasmid and 100 nM M. tuberculosis DNA gyrase were incubated in the reaction buffer at 37°C for 30 min, followed by the addition of sodium dodecyl sulfate (SDS) to a final concentration of 0.1% and 10 μg of proteinase K/ml. The incubation was continued for another 30 min. For oligonucleotide cleavage experiments, 5′-γ-32P-labeled 143-bp DNA and DNA gyrase from M. smegmatis or E. coli were used. For FQ competition experiments, M. smegmatis DNA gyrase was incubated with 5′-γ-32P-labeled 143-bp DNA and the FQs (as described in the figure legends). To monitor the FQ mediated trapping of gyrase-DNA covalent complexes, 50 nM DNA gyrase from M. smegmatis was incubated with 5′-γ-32P-labeled 72-bp DNA and 25 nM CFX or MFX in supercoiling reaction buffer (21) for 15 min at 37°C. The trapped enzyme-DNA complexes were resolved by using 5% native polyacrylamide gel electrophoresis and visualized with a phosphorimager (FLA 5000; GE Healthcare). The fold increase in the formation of enzyme-DNA complex in the presence of FQs was calculated with respect to the complex formed in the absence of the drug.
Fluorescence measurements.
The fluorescence measurements were carried out with a Hitachi-7000 spectrofluorimeter fitted with a polarizer. All of the fluorescence measurements were taken using a slit width of 10 mm, a PMT voltage of 700 V, and an integration time of 0.1 s. The assays were carried out at specific excitation/emission wavelengths of 335/470 nm (for MFX) and 327/420 nm (for CFX), respectively. Fluorescence anisotropy titrations (32) were carried out at fixed concentrations of the FQs and with increasing concentrations of DNA gyrase from M. tuberculosis, M. smegmatis, E. coli, and Topo IV from E. coli or their subunits (as specified in the figure legends) in a binding buffer containing 10 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 100 mM KCl, and 2 mM dithiothreitol at 37°C. For DNase I-proteinase K treatment, the reactions without FQs were incubated with 10 μg of proteinase K/ml or 10 U of DNase I for 6 h, followed by the addition of the FQs. For FQ-DNA binding experiments, increasing concentrations of salmon sperm DNA were added to 10 nM CFX or MFX. To monitor the binding of FQs to preformed enzyme-DNA complex, 200 nM M. tuberculosis DNA gyrase was preincubated with salmon sperm DNA (200 bp/holoenzyme, 4°C, 10 min). Incubations were continued at 37°C for 10 min after the addition of 10 nM CFX or MFX. The binding of the FQs to the D94G mutant of M. smegmatis GyrA or the holoenzyme was carried out by incubating 50 nM GyrA or the holoenzyme with 10 nM CFX/MFX at 37°C for 10 min. The equations used to calculate anisotropy are given in Table S1 in the supplemental material. In the fluorescence-quenching experiments to monitor the drug-gyrase interaction, a 50 nM concentration of M. tuberculosis DNA gyrase was incubated with various concentrations (100 to 800 nM) of CFX or MFX. The ratio of change in the intrinsic fluorescence of the protein (excitation, 295 nm; emission, 340 nm) at a given drug concentration (ΔF) and the change at infinite concentrations of the drug (ΔFmax) were plotted against the drug concentration (33). The plot was fitted and analyzed by the Hill binding equation in GraphPad Prism 5. The drug-enzyme interaction was also monitored by the spin column gel filtration method (22).
RESULTS
DNA cleavage in the presence of FQs.
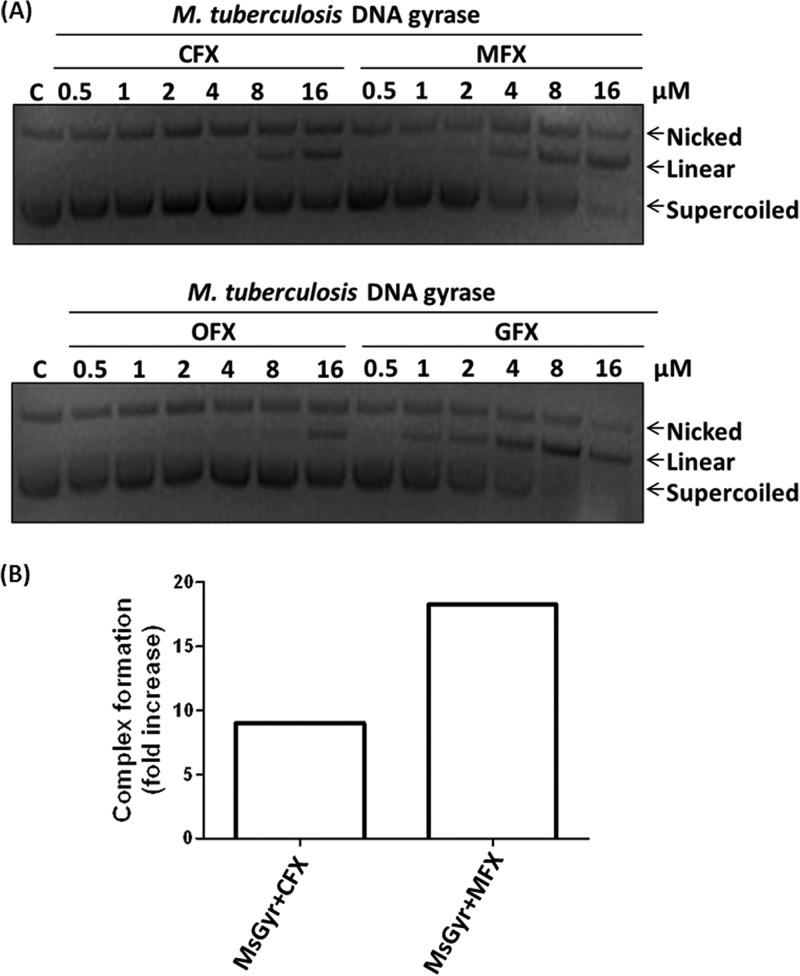
Although CFX and OFX are widely used FQs for the treatment of a variety of bacterial infections, several studies showed their lower efficacy against M. tuberculosis (34–37). However, the FQs with methoxy substitution at C-8 (MFX and GFX) exhibited potent antimycobacterial activity (38) and also inhibited mycobacterial DNA gyrase at low concentrations (20, 21; unpublished results). The DNA supercoiling reaction catalyzed by gyrase involves two sequential transesterification reactions and an intermediate strand passage step to pass the intact duplex DNA through the cleaved DNA (39). The FQs trap the covalent complex formed by gyrase with DNA during the first trans-esterification reaction, converting the enzyme into a double-strand cleaving endonuclease (34). To analyze the basis of the differential susceptibility of the mycobacterial gyrase to the FQs, DNA cleavage experiments were carried out with M. tuberculosis DNA gyrase. The cleavage induced by MFX or GFX was two to three times more than with CFX or OFX (Fig. 1A). A similar pattern was observed with M. smegmatis DNA gyrase (data not shown). These results are consistent with the results obtained in DNA supercoiling assays where 2- to 3-fold-higher inhibition was seen with MFX (20). The increased cleavage seen in the presence of MFX could be due to increased complex formation between the DNA and the enzyme in the presence of the compound. Electrophoretic mobility shift assays were carried out to assess the extent of complex formation in the presence of the two FQs by incubating 5′-end-labeled 72-bp DNA with the enzyme and 25 nM CFX or MFX. An increase in the enzyme-DNA complex was seen, indicating the formation of an FQ stabilized complex. Importantly, the extent of complex formation was 2-fold more with MFX than with CFX (Fig. 1B).
FIG 1.
Fluoroquinolone (FQ) sensitivity of mycobacterial gyrase. (A) FQ-induced plasmid DNA cleavage with M. tuberculosis DNA gyrase. A 100 nM concentration of the enzyme was incubated with 200 ng of pUC18 plasmid DNA and various concentrations (0.5 to 16 μM) of the different FQs (as indicated). The cleavage products were resolved on 1% agarose gel. Lane C, pUC18 DNA control. (B) Trapping of the gyrase-DNA complex by CFX and MFX. A total of 50 nM M. smegmatis DNA gyrase (MsGyr) was incubated with 5′-γ-32P-labeled 72-bp DNA in the presence of 25 nM CFX or MFX in supercoiling reaction buffer at 37°C for 15 min. The DNA-bound complex was resolved on a 5% native polyacrylamide gel and visualized using a phosphorimager. The fold increase in gyrase-DNA complex formation in the presence of CFX and MFX was calculated with respect to the complex formed in the absence of any drug.
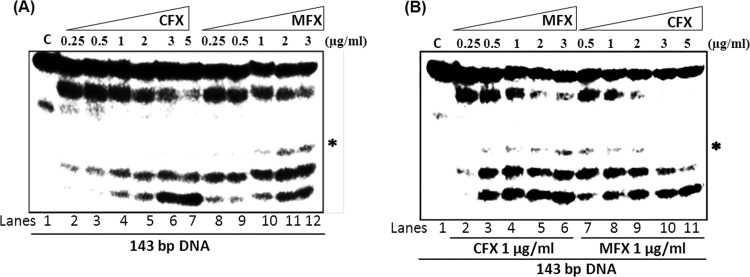
Earlier studies indicated that quinolones and FQs induce a different cleavage pattern by the E. coli DNA gyrase (40). However, the differential interaction of two FQs with a given gyrase has not been investigated. The FQs interact with the enzyme-DNA complex near the active site (10, 41, 42). Due to the differences in their chemical nature, the interaction with the FQs may involve different residues. The difference in the extent of complex formation and DNA cleavage seen in the presence of CFX and MFX may arise due to differences in their interaction. To evaluate whether this difference is also associated with a distinct cleavage pattern, assays were carried out with a linear DNA fragment labeled at the 5′ end. In the presence of CFX, three cleaved DNA fragments were seen, whereas MFX-induced cleavage resulted in an additional DNA fragment (Fig. 2A). In contrast, the cleavage pattern was similar in the presence of both CFX and MFX with E. coli enzyme (see Fig. S1 in the supplemental material). To investigate whether the interaction of CFX and MFX with the mycobacterial gyrase occurs at the same site, cleavage competition assays were carried out using a fixed concentration of one FQ and increasing the concentration of the other. Increasing the concentration of MFX led to the appearance of the MFX-specific cleavage product, whereas it gradually disappeared with an increase in CFX concentration (Fig. 2B). From these data, it is evident that these two FQs compete to the same binding site and, when bound, induce their respective characteristic cleavage patterns.
FIG 2.
Pattern of DNA cleavage by M. smegmatis DNA gyrase in the presence of different FQs. (A) DNA gyrase (100 nM) was incubated with 5′-γ-32P-labeled 143-bp DNA and various concentrations of CFX and MFX as described in Materials and Methods. Lane C, 143-bp DNA control (lane 1). The results obtained with enzyme and DNA with 0.25 to 5 μg of CFX/ml (lanes 2 to 7) and 0.25 to 3 μg of MFX/ml (lanes 8 to 12) are shown. (B) DNA cleavage reactions with a fixed concentration of one of the FQs and various concentrations of the other FQs and vice versa. Lane C, 143-bp DNA control (lane 1). The results obtained with enzyme and DNA with 1 μg of CFX/ml plus 0.25 to 3 μg of MFX/ml (lanes 2 to 6) or with enzyme and DNA with 1 μg of MFX/ml plus 0.25 to 5 μg of CFX/ml (lanes 7 to 11) are shown. The asterisk indicates the unique DNA cleavage product formed in the presence of MFX. The reactions were carried out in the supercoiling reaction buffer at 37°C for 30 min, and the reaction products were resolved on 8% denaturing polyacrylamide gels.
Interaction of the FQs with the M. tuberculosis DNA gyrase.
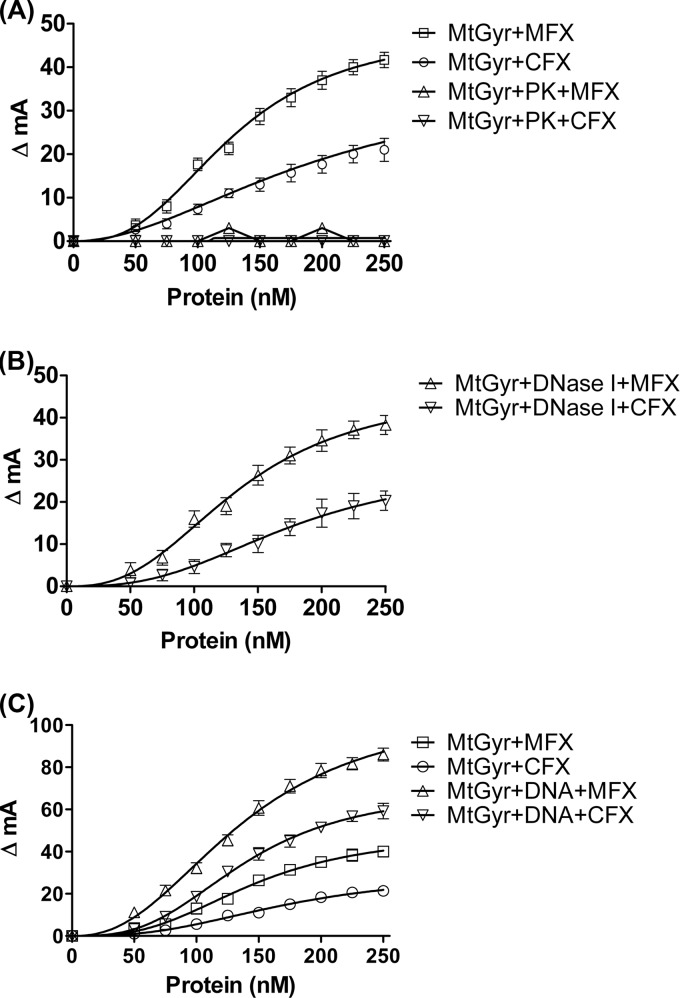
To understand the basis of the differential interaction of CFX and MFX with the M. tuberculosis gyrase and to evaluate the mechanism of action of the FQs, fluorescence anisotropy assays were carried out. The technique provides a sensitive and real-time measurement of the molecular interactions occurring in solution. The interaction of small fluorescent ligands with larger receptor protein molecules can be conveniently monitored by this technique (23). Many FQs are intrinsically fluorescent enabling the measurement of their interaction with the gyrase and/or DNA by a change in anisotropy. Initially, a set of control experiments were carried out before monitoring the interaction of the FQs with the enzyme-DNA complex. A 10 nM concentration of CFX or MFX was titrated with a range of enzyme concentrations. Surprisingly, a concentration-dependent increase in anisotropy was observed in these control experiments, which had only FQ and the enzyme. The binding of the FQs to the enzyme reached saturation at higher concentrations of the enzyme (Fig. 3A). Notably, MFX binding induced significantly higher changes in anisotropy compared to CFX (Fig. 3A). In experiments with 20 and 50 nM concentrations, the FQs also showed a similar pattern of interaction (data not shown).
FIG 3.
Interaction of FQs with M. tuberculosis DNA gyrase (MtGyr) measured by fluorescence anisotropy. ΔmA indicates the change in anisotropy (in milli-units). (A) 10 nM CFX and MFX were titrated using increasing concentrations of the enzyme. PK indicates titration carried out with the enzyme treated with proteinase K. The change in the anisotropy was plotted against the enzyme concentration (nM). (B) Binding of the FQs to the DNase I-treated enzyme. A 10 nM concentration of the drug was titrated with various concentrations of the DNase I-treated enzyme. (C) Binding of MFX and CFX with preincubated enzyme-DNA complex. Concentrations (10 nM) of the drugs were titrated with various concentrations of the enzyme-DNA complex. Titration of the drugs with enzyme but without DNA is shown as a control. Treatment of the enzyme with proteinase K (PK) and DNase I was carried out for 6 h before addition of the FQ, as described in Materials and Methods.
A number of previous studies had shown the requirement of DNA for gyrase-FQ interaction (24–26, 40). Moreover, the binding of FQs on their own to the enzyme was not apparent in these studies. Thus, to rule out the possibility that enzyme preparations contained contaminating DNA which could have contributed to the observed drug binding to the enzyme, the following experiments were carried out. First, the enzyme was treated with DNase I over an extended period of time (see Materials and Methods) before carrying out fluorescence measurements. The binding of both CFX and MFX to the enzyme remained unaltered (Fig. 3B). In another important control experiment, the enzyme was subjected to proteinase K treatment before the measurements. In the latter case, the change in anisotropy was negligible, indicating that in the experimental conditions used, the FQs bind to the enzyme directly (Fig. 3A). Next, to monitor the role of DNA in FQ-gyrase interactions, CFX or MFX were incubated with gyrase-DNA complex as described in Materials and Methods, which resulted in altered interactions, as seen by changes in anisotropy (Fig. 3C). However, similar to the pattern seen with free enzyme, the interaction of MFX with the enzyme-DNA complex was better than CFX. Fluorescence-quenching and spin column gel filtration experiments indicated that MFX bound M. tuberculosis gyrase better than CFX (see Fig. S2A and B in the supplemental material). When the binding of DNA alone to FQs was monitored in a separate experiment, surprisingly, a much lower change in anisotropy was observed (see Fig. S3 in the supplemental material). The implications of these findings are elaborated in the later part of the manuscript.
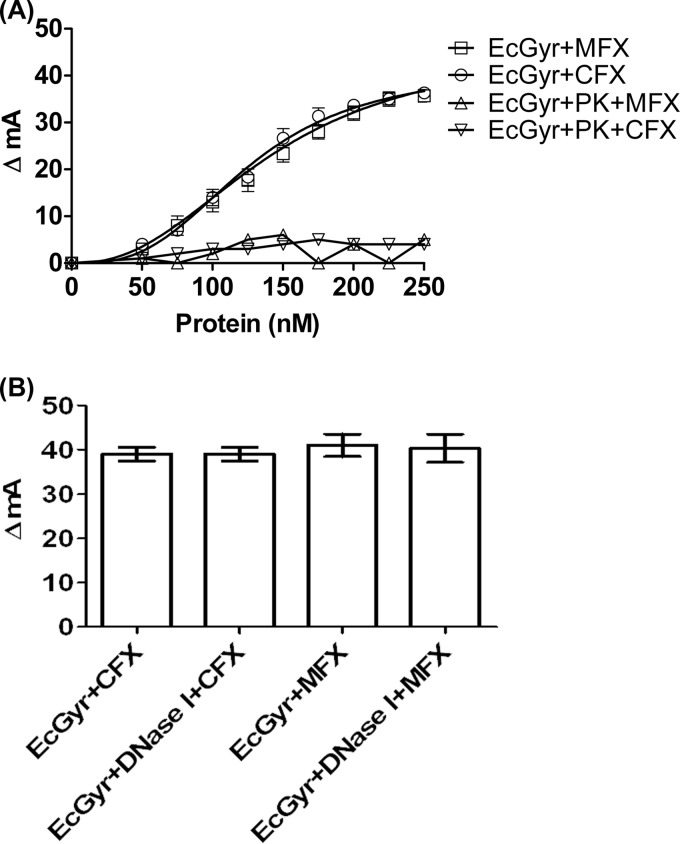
Although the experiments with the M. tuberculosis enzyme described above showed a direct interaction between FQs and the gyrase, in the case of E. coli gyrase FQ binding required the enzyme-DNA complex (23–27). To examine whether the DNA gyrases from two different bacteria do indeed exhibit different interaction patterns with the FQs, we carried out assays with the E. coli enzyme. Anisotropic changes were seen when the E. coli gyrase was incubated with either of the FQs (Fig. 4A). However, in contrast to the pattern seen in case of M. tuberculosis gyrase, the binding of both the FQs to E. coli enzyme seems to be comparable (Fig. 4A). The binding was remarkably poor in the case of OXO (data not shown), a weaker inhibitor of the E. coli enzyme (43). Most importantly, prior digestion of the enzyme samples with the DNase I, similar to the experiments with the M. tuberculosis enzyme, did not affect the binding of the FQs to the E. coli gyrase (Fig. 4B). However, with the proteinase K treatment to the enzyme, the binding of FQs was nearly abolished (Fig. 4A).
FIG 4.
Binding of FQs to E. coli DNA gyrase (EcGyr). (A) Titration of 10 nM MFX and CFX with various concentrations of the enzyme. The change in the anisotropy was plotted against the enzyme concentrations. (B) Effect of DNase I treatment on binding of MFX and CFX (10 nM) to EcGyr (250 nM). Proteinase K and DNase I treatments were carried out as described in the legend for Fig. 3.
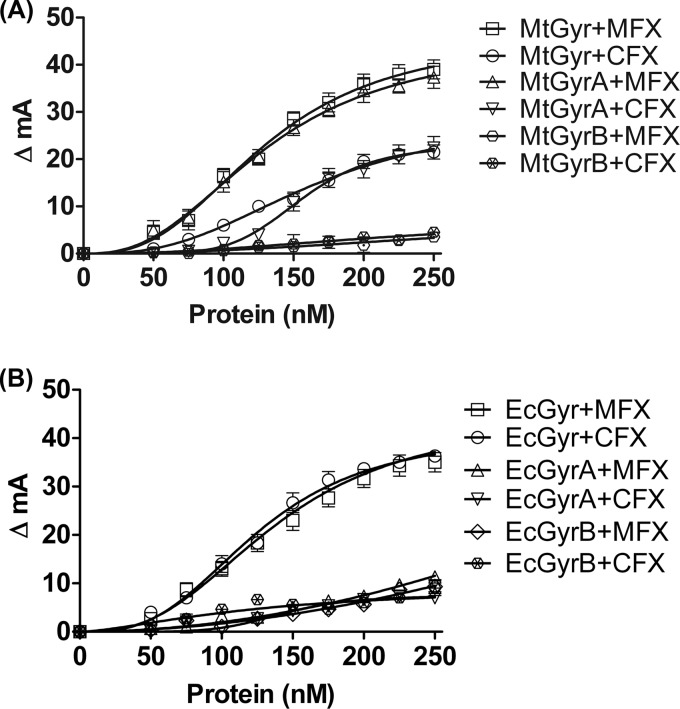
Although, from these studies, it is evident that FQs can bind directly to DNA gyrase holoenzymes from two different species, they do not reveal whether the drugs preferentially bind to only one of the subunits, or whether the assembled tetrameric holoenzyme is necessary for binding. To determine whether the FQs bind only to the holoenzyme or also to individual subunits, fluorescence anisotropy measurements were carried out with individual subunits of the M. tuberculosis gyrase. From the data shown in Fig 5A, it is evident that both FQs can bind to the GyrA subunit itself to an extent similar to that of the holoenzyme. Notably, the GyrA interaction with MFX seems to be markedly higher than to CFX (Fig. 5A); a low degree of binding of GyrB to FQs is seen (Fig. 5A). Together, these results imply that M. tuberculosis GyrA is primarily involved in interaction with the FQs, and the contribution from GyrB alone or GyrB in complex with GyrA, i.e., in the holoenzyme context, appears to be minimal. Interestingly, in the experiments with individual subunits of E. coli gyrase, we observed different results; the binding of either the GyrA or GyrB subunits of the E. coli enzyme was substantially lower, and only the assembled holoenzyme was competent for both CFX/MFX binding (Fig. 5B). These results are in contrast to those of an earlier study in which E. coli GyrA was shown to interact with CFX (58). FQ binding experiments were also carried out with Topo IV, another type II topoisomerase present in E. coli and its subunits ParC and ParE. Topo IV alone could bind to CFX and MFX in the absence of DNA; similar to E. coli gyrase subunits, the individual subunits of topo IV showed low degree of interaction with the FQs (see Fig. S4A and B in the supplemental material).
FIG 5.
FQ binding to DNA gyrase and its individual subunits. (A) Titration of CFX and MFX with increasing M. tuberculosis DNA gyrase (MtGyr) and its individual subunits (MtGyrA and MtGyrB). (B) Titration with E. coli DNA gyrase (EcGyr) and its individual subunits (EcGyrA and EcGyrB). The change in anisotropy was plotted against concentrations of the holoenzyme or individual subunits. CFX or MFX at 10 nM were used in these assays. MtGyrA, M. tuberculosis GyrA; MtGyrB, M. tuberculosis GyrB; EcGyrA, E. coli GyrA; EcGyrB, E. coli GyrB subunit.
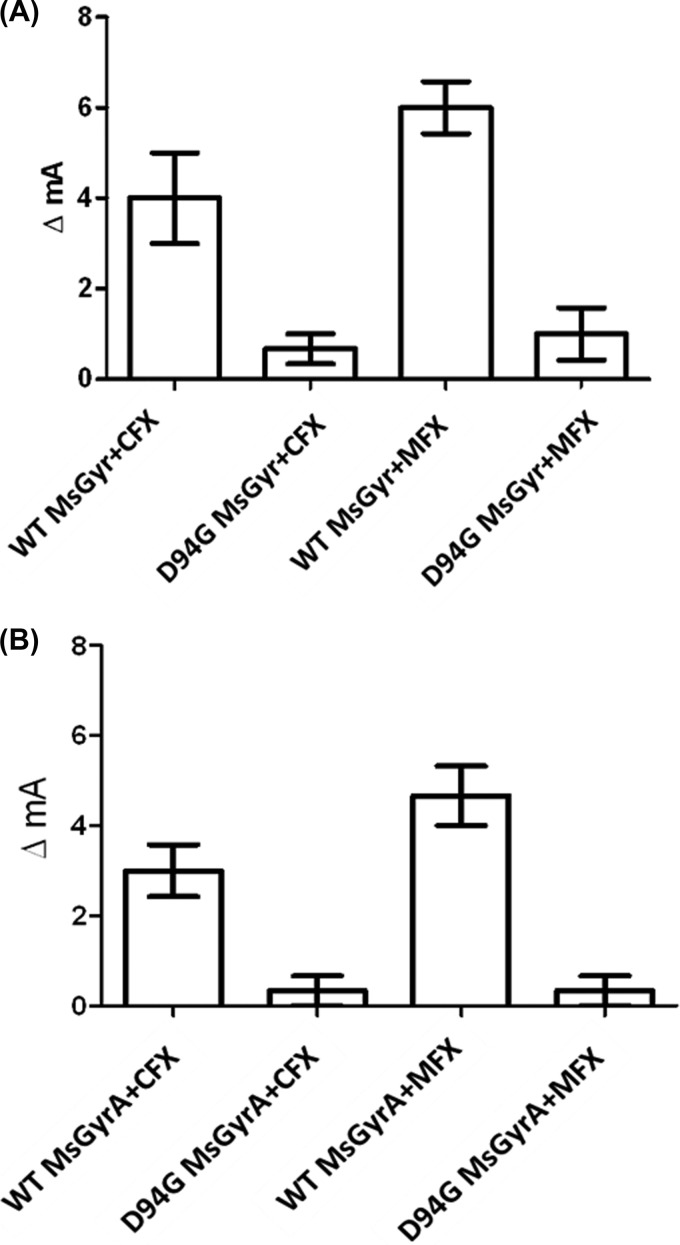
From the above-described experiments, it is apparent that CFX and MFX show a difference in the interaction with the M. tuberculosis gyrase, which explains the basis for their different inhibition potential. One of the most likely reasons for the FQ resistance acquired upon mutations in the quinolone resistance-determining region (QRDR) would be the weaker interaction of the FQs with the enzyme. To investigate whether a mutation in QRDR affects FQ-enzyme interaction, we chose one of the most common QRDR mutants. Among all QRDR mutations, serine 83 (S83) and aspartate 87 (D87) mutations in the GyrA subunit are most common in gyrases from various bacteria. Indeed, a mutation in D94 (corresponding to D87 in E. coli GyrA [see Fig. S5 in the supplemental material]) confers FQ resistance to both the M. tuberculosis and the M. smegmatis DNA gyrase (21, 44). Fluorescence anisotropy experiments showed a substantially weaker binding of FQs with either D94G GyrA alone or the M. smegmatis holoenzyme containing D94G GyrA (Fig. 6). However, the binding of CFX and MFX to the wild-type M. smegmatis GyrA or holoenzyme was similar to the results obtained with GyrA or the holoenzyme from M. tuberculosis.
FIG 6.
Effect of QRDR mutation in M. smegmatis GyrA on FQ binding. (A) Binding of CFX and MFX to the M. smegmatis DNA gyrase (MsGyr) reconstituted using wild-type (WT) or D94G GyrA. (B) Interaction of FQs with wild-type (WT) and D94G MsGyrA subunit. A 10 nM concentration of each of the FQs and a 50 nM concentration of the holoenzyme or the GyrA subunit were used.
DISCUSSION
By virtue of their ability to trap the gyrase-DNA covalent complex, leading to the generation of double-strand breaks in the genome, FQs have been subjected to intensive studies in order to find more effective derivatives (22–26, 45, 46). As a result, a number of new FQs with varied inhibitory potency against different bacteria have been characterized (18). In parallel, their mechanism of action has been also studied extensively to develop molecules with improved ability to induce double-strand cleavage. Initial efforts in understanding the mechanism involved ultrafiltration and equilibrium dialysis methods in order to monitor the enzyme and DNA interaction with [3H]norfloxacin. A strong binding of the drug to DNA, but an insignificant binding to the enzyme, was reported (22). In addition, the drug was shown to preferentially bind to the single-stranded DNA. Next, the DNA binding was shown to be dependent on the concentration of Mg2+ (46). Subsequent surface plasmon resonance studies on FQ-DNA interaction using single-stranded DNA showed a sequence-dependent binding with high affinity, whereas double-stranded DNA binding seemed to be sequence independent (40). In a separate study, the binding of norfloxacin to unique sites in the enzyme-DNA complex was observed, implicating the interaction of the FQs with both the gyrase and DNA (24). The interaction of the FQs with the enzyme-DNA complex was established by several other studies (23–27). However, in these studies, the direct binding of FQs to the enzyme itself was not apparent, and the basis for the varied FQ susceptibility of gyrases from different bacteria and the occurrence of a large number of quinolone-resistance mutations were unexplained to a large extent. In a recent study, however, the contribution of a water-metal ion bridge for Topo IV-quinolone interaction and its stabilization by two amino acid residues of the QRDR has been demonstrated (47, 48).
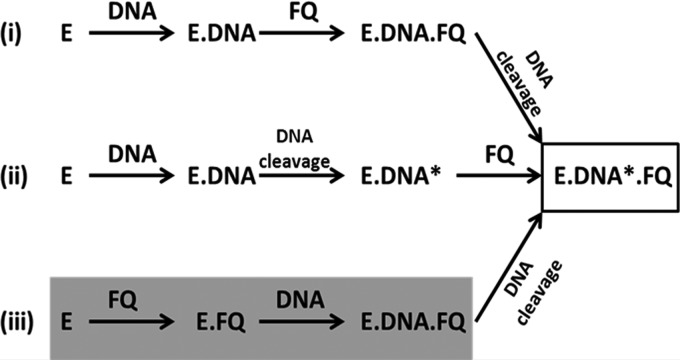
In order to address the problem and to account for the differential susceptibility of mycobacterial gyrases to FQs, we revisited the mechanism of FQ-gyrase interaction. We found two distinct ways in which the interaction of FQs with the enzyme is achieved. In the established mode known for more than a decade, FQs bind to the enzyme-DNA complex prior or after DNA cleavage (Fig. 7) (49). We described the binding of the enzyme to FQ before the binding to DNA, a mode of interaction indicated in an earlier work with E. coli gyrase (58). Moreover, from our studies, it appears that the binding of the drug to DNA is unlikely to be a major mechanism of drug action under physiological conditions. However, to investigate whether FQs bind DNA, we carried out experiments using the conditions described in earlier studies (22). When 25 mM KCl was used to monitor the interaction, the binding of CFX/MFX to DNA was seen as reported earlier (see Fig. S6 in the supplemental material) (22). However, binding of FQs to the enzyme alone was also observed, and binding of the drug to the enzyme-DNA complex was greater than the FQ binding to either the enzyme or DNA. Importantly, in assays where the salt concentration used was close to physiological levels, FQs exhibited a weak DNA-binding ability (see Fig. S3 in the supplemental material); the FQs bound the enzyme alone, as well the enzyme-DNA complex (Fig. 3A and C).
FIG 7.
FQ mechanisms of action. (i) Enzyme forms a noncovalent complex with the DNA to which the FQ binds. (ii) FQ binds to the enzyme-DNA covalent complex. (iii) FQ directly binds to the enzyme, followed by binding of this enzyme-FQ complex to DNA. All three modes converge at a common point where the enzyme-cleaved DNA covalent complex is trapped by the FQ. E, enzyme; FQ, fluoroquinolone; DNA*, cleaved DNA. Modes i and ii are based on data from Kampranis and Maxwell (49). The finding from the present study is highlighted in gray.
The binding of the FQs to the enzyme-DNA complex has been investigated extensively; the drugs may bind to the gyrase-DNA complex and induce the cleavage of the DNA. Alternatively, or additionally the drugs may bind to the gyrase-DNA covalent complex and stabilize the cleaved complex (49; summarized in Fig. 7). The net outcome of both the pathways is the accumulation of cleaved ternary complexes. The direct binding of FQ to the enzyme described here is likely to be an earlier step in the reaction (Fig. 7; see also Fig. S8 in the supplemental material). Normally, much of the DNA gyrase holoenzyme would be bound to the DNA to carry out the supercoiling function in vivo. However, some free subunits and holoenzymes generated due to dissociation from the DNA and continuous protein synthesis during the exponential phase of growth would be available for FQ binding. In one scenario, such FQ-gyrase complexes may occlude DNA binding, resulting in an inhibition of activity similar to the mechanism of inhibition observed with endogenous gyrase inhibitory proteins (50–52). A number of them directly bind to the gyrase and sequester the enzyme away from DNA, accounting for the phenotypic resistance against FQs (50–52). However, in contrast, we did see that the enzyme-FQ complex has also the ability to bind DNA (see Fig. S7 in the supplemental material), thus confining the inhibition mechanism to the established mode viz. double-stranded cleavage in the ternary complex (Fig. 7).
The direct binding of FQs to either the E. coli holoenzyme or the GyrA of the M. tuberculosis gyrase seems to account for the differential susceptibility seen with different drugs. The better binding of MFX to the GyrA of the latter enzyme seems to directly correlate to increased ternary complex formation and cleaved products. It also accounts for the higher inhibition of the supercoiling activity of the enzyme seen with MFX earlier (20, 21). The similar interaction of both CFX and MFX to the E. coli gyrase correlates well with the comparable DNA cleavage seen in the presence of the drugs (see Fig. S1 in the supplemental material). In support of this, the inhibition of the supercoiling activity of the E. coli enzyme was comparable to both CFX and MFX (21). Similarly, OXO, a weak inhibitor of the E. coli gyrase (43), demonstrated less binding (data not shown). Further, the findings presented here can account for the occurrence of a large number of FQ-resistant mutations in the QRDR of both GyrA and GyrB subunits, conferring various degrees of resistance. Given that the primary QRDR resides in the amino terminus of the GyrA subunit, mutations in the GyrA subunit result in high-level resistance, whereas those in GyrB tend to result in low-level resistance (53, 54). However, the frequency of the occurrence of mutations in the two genes seems to be highly varied in different organisms. For example, in a study analyzing the FQ resistance in M. tuberculosis, 711 of 806 FQ-resistant strains were found to contain a single-site mutation in the gyrase genes. Among these single site mutants, 97% (691/711) isolates contained mutations in gyrA and only 2.8% (20/711) in gyrB. In addition, among 109 isolates containing more than one mutation in the gyr locus, 103 isolates had multiple mutations in gyrA and only one isolate contained a double mutation in gyrB (55). In contrast, such a bias in mutation frequency is not seen in E. coli, where out of 25 spontaneous FQ-resistant strains selected in vitro, 13 had mutations in gyrA and the rest mapped to gyrB (54). Thus, the binding pattern of the FQs to the two gyrases seems to correlate well with the distribution of the resistance mutations. However, in the laboratory-conducted experiments, the mutation frequency in the particular allele (gyrA or gyrB) is dependent on the drug, its concentration chosen for the study, and a number of other variables (56).
Structural studies of the gyrase/Topo IV-DNA complex and FQs have been carried out with the objective of visualizing the molecular interactions in the ternary complex. In the structure of FQ-trapped Streptococcus pneumoniae Topo IV, two FQ molecules were shown to interact with the cleaved DNA, preventing the 3′-OH group from attacking the phosphotyrosine for religation (42), while the structure of the Acinetobacter baumannii Topo IV-DNA complex trapped with MFX showed Mg2+ bridged protein-FQ contact (41). The mutation of the residues involved in the Mg2+ bridged contact with FQs reduced the interaction (47), establishing the importance of the FQs binding to the enzyme-DNA complex for efficient inhibition. Given the high degree of similarity between the gyrase and Topo IV in the amino-terminal regions of GyrA and ParC, one would expect a similar interaction pattern between the gyrase and FQ. However, CFX bound to the Staphylococcus aureus gyrase-DNA complex showed a different orientation of the drug compared to the MFX-Topo IV complex (10). The docking of MFX on the M. tuberculosis DNA gyrase reaction core along with DNA indicated that the drug contacts both subunits in the enzyme-DNA complex (57). However, the overwhelming incidence of QRDR mutations mapping to the GyrA of M. tuberculosis (55) is an indication of a major role for GyrA in FQ binding. The GyrB binding shown in the docking model (57) may have a role in stabilizing the interaction in the ternary complex.
In conclusion, the binding of FQs to the enzyme itself and the enzyme-DNA complex may have physiological implications in the development of resistance and also account for the differential susceptibility of the enzyme to different FQs. The frequency of distribution of mutations in GyrA and GyrB seems to indicate whether a given FQ preferentially binds to one of the subunits or the holoenzyme from different bacteria. The high-affinity binding of the FQs to the gyrase itself, demonstrated here, may open up avenues for structural studies of the complex between the drug and the enzyme, paving the way for the generation of more potent lead molecules against a variety of bacterial infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Hiasa for the overexpressing constructs of E. coli Topo IV subunits, P. K. Chakraborty for the drug-resistant M. smegmatis mc2 155 strain, and A. Maxwell and K. Madhusudan for the overexpressing constructs of E. coli and M. tuberculosis gyrase subunits, respectively. We thank D. N. Rao, J. Berger, and T. Blower for critical reading of the manuscript and helpful suggestions. We acknowledge the phosphorimager facility of Indian Institute of Science, supported by the Department of Biotechnology, Government of India.
R. K. was a recipient of a senior research fellowship and B.S.M. is a recipient of postdoctoral fellowship from the Department of Biotechnology, Government of India. V. N. is a J. C. Bose fellow of Department of Science and Technology, Government of India.
Footnotes
Published ahead of print 13 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01958-13.
REFERENCES
- 1.Gellert M, Mizuuchi K, O'Dea MH, Nash HA. 1976. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. U. S. A. 73:3872–3876. 10.1073/pnas.73.11.3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang JC. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65:635–692. 10.1146/annurev.bi.65.070196.003223 [DOI] [PubMed] [Google Scholar]
- 3.Champoux JJ. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369–413. 10.1146/annurev.biochem.70.1.369 [DOI] [PubMed] [Google Scholar]
- 4.Berger JM, Gamblin SJ, Harrison SC, Wang JC. 1996. Structure and mechanism of DNA topoisomerase II. Nature 379:225–232. 10.1038/379225a0 [DOI] [PubMed] [Google Scholar]
- 5.Maxwell A. 1993. The interaction between coumarin drugs and DNA gyrase. Mol. Microbiol. 9:681–686. 10.1111/j.1365-2958.1993.tb01728.x [DOI] [PubMed] [Google Scholar]
- 6.Goetschi E, Angehrn P, Gmuender H, Hebeisen P, Link H, Masciadri R, Nielsen J. 1993. Cyclothialidine and its congeners: a new class of DNA gyrase inhibitors. Pharmacol. Ther. 60:367–380. 10.1016/0163-7258(93)90017-8 [DOI] [PubMed] [Google Scholar]
- 7.Drlica K. 1999. Mechanism of fluoroquinolone action. Curr. Opin. Microbiol. 2:504–508. 10.1016/S1369-5274(99)00008-9 [DOI] [PubMed] [Google Scholar]
- 8.Bahassi EM, O'Dea MH, Allali N, Messens J, Gellert M, Couturier M. 1999. Interactions of CcdB with DNA gyrase: inactivation of GyrA, poisoning of the gyrase-DNA complex, and the antidote action of CcdA. J. Biol. Chem. 274:10936–10944 [DOI] [PubMed] [Google Scholar]
- 9.Heddle JG, Blance SJ, Zamble DB, Hollfelder F, Miller DA, Wentzell LM, Walsh CT, Maxwell A. 2001. The antibiotic microcin B17 is a DNA gyrase poison: characterisation of the mode of inhibition. J. Mol. Biol. 307:1223–1234. 10.1006/jmbi.2001.4562 [DOI] [PubMed] [Google Scholar]
- 10.Bax BD, Chan PF, Eggleston DS, Fosberry A, Gentry DR, Gorrec F, Giordano I, Hann MM, Hennessy A, Hibbs M, Huang J, Jones E, Jones J, Brown KK, Lewis CJ, May EW, Saunders MR, Singh O, Spitzfaden CE, Shen C, Shillings A, Theobald AJ, Wohlkonig A, Pearson ND, Gwynn MN. 2010. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 466:935–940. 10.1038/nature09197 [DOI] [PubMed] [Google Scholar]
- 11.Flatman RH, Howells AJ, Heide L, Fiedler HP, Maxwell A. 2005. Simocyclinone D8, an inhibitor of DNA gyrase with a novel mode of action. Antimicrob. Agents Chemother. 49:1093–1100. 10.1128/AAC.49.3.1093-1100.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashimi SM, Wall MK, Smith AB, Maxwell A, Birch RG. 2007. The phytotoxin albicidin is a novel inhibitor of DNA gyrase. Antimicrob. Agents Chemother. 51:181–187. 10.1128/AAC.00918-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karkare S, Chung TT, Collin F, Mitchenall LA, McKay AR, Greive SJ, Meyer JJ, Lall N, Maxwell A. 2012. The naphthoquinone diospyrin is an inhibitor of DNA gyrase with a novel mechanism of action. J. Biol. Chem. 288:5149–5156. 10.1074/jbc.M112.419069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCullough JE, Muller MT, Howells AJ, Maxwell A, O'Sullivan J, Summerill RS, Parker WL, Wells JS, Bonner DP, Fernandes PB. 1993. Clerocidin, a terpenoid antibiotic, inhibits bacterial DNA gyrase. J. Antibiot. (Tokyo) 46:526–530. 10.7164/antibiotics.46.526 [DOI] [PubMed] [Google Scholar]
- 15.Tari LW, Trzoss M, Bensen DC, Li X, Chen Z, Lam T, Zhang J, Creighton CJ, Cunningham ML, Kwan B, Stidham M, Shaw KJ, Lightstone FC, Wong SE, Nguyen TB, Nix J, Finn J. 2012. Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). I. Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. Bioorg. Med. Chem. Lett. 23:1529–1536. 10.1016/j.bmcl.2012.11.032 [DOI] [PubMed] [Google Scholar]
- 16.Drlica K, Malik M, Kerns RJ, Zhao X. 2008. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 52:385–392. 10.1128/AAC.01617-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. 10.1016/j.cell.2007.06.049 [DOI] [PubMed] [Google Scholar]
- 18.Oliphant CM, Green GM. 2002. Quinolones: a comprehensive review. Am. Fam. Physician 65:455–464 [PubMed] [Google Scholar]
- 19.Blumberg HM, Burman WJ, Chaisson RE, Daley CL, Etkind SC, Friedman LN, Fujiwara P, Grzemska M, Hopewell PC, Iseman MD, Jasmer RM, Koppaka V, Menzies RI, O'Brien RJ, Reves RR, Reichman LB, Simone PM, Starke JR, Vernon AA. 2003. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 167:603–662. 10.1164/rccm.167.4.603 [DOI] [PubMed] [Google Scholar]
- 20.Aubry A, Pan XS, Fisher LM, Jarlier V, Cambau E. 2004. Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob. Agents Chemother. 48:1281–1288. 10.1128/AAC.48.4.1281-1288.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manjunatha UH, Dalal M, Chatterji M, Radha DR, Visweswariah SS, Nagaraja V. 2002. Functional characterisation of mycobacterial DNA gyrase: an efficient decatenase. Nucleic Acids Res. 30:2144–2153. 10.1093/nar/30.10.2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen LL, Pernet AG. 1985. Mechanism of inhibition of DNA gyrase by analogues of nalidixic acid: the target of the drugs is DNA. Proc. Natl. Acad. Sci. U. S. A. 82:307–311. 10.1073/pnas.82.2.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen LL. 2001. Quinolone interactions with DNA and DNA gyrase. Methods Mol. Biol. 95:171–184 [DOI] [PubMed] [Google Scholar]
- 24.Shen LL, Kohlbrenner WE, Weigl D, Baranowski J. 1989. Mechanism of quinolone inhibition of DNA gyrase. Appearance of unique norfloxacin binding sites in enzyme-DNA complexes. J. Biol. Chem. 264:2973–2978 [PubMed] [Google Scholar]
- 25.Shen LL, Mitscher LA, Sharma PN, O'Donnell TJ, Chu DW, Cooper CS, Rosen T, Pernet AG. 1989. Mechanism of inhibition of DNA gyrase by quinolone antibacterials: a cooperative drug-DNA binding model. Biochemistry 28:3886–3894. 10.1021/bi00435a039 [DOI] [PubMed] [Google Scholar]
- 26.Yoshida H, Nakamura M, Bogaki M, Ito H, Kojima T, Hattori H, Nakamura S. 1993. Mechanism of action of quinolones against Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 37:839–845. 10.1128/AAC.37.4.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heddle J, Maxwell A. 2002. Quinolone-binding pocket of DNA gyrase: role of GyrB. Antimicrob. Agents Chemother. 46:1805–1815. 10.1128/AAC.46.6.1805-1815.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar R, Riley JE, Parry D, Bates AD, Nagaraja V. 2012. Binding of two DNA molecules by type II topoisomerases for decatenation. Nucleic Acids Res. 40:10904–10915. 10.1093/nar/gks843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maxwell A, Howells AJ. 1999. Overexpression and purification of bacterial DNA gyrase. Methods Mol. Biol. 94:135–144 [DOI] [PubMed] [Google Scholar]
- 30.Peng H, Marians KJ. 1993. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 268:24481–24490 [PubMed] [Google Scholar]
- 31.Banerjee SK, Bhatt K, Misra P, Chakraborti PK. 2000. Involvement of a natural transport system in the process of efflux-mediated drug resistance in Mycobacterium smegmatis. Mol. Gen. Genet. 262:949–956. 10.1007/PL00008663 [DOI] [PubMed] [Google Scholar]
- 32.Favicchio R, Dragan AI, Kneale GG, Read CM. 2009. Fluorescence spectroscopy and anisotropy in the analysis of DNA-protein interactions. Methods Mol. Biol. 543:589–611. 10.1007/978-1-60327-015-1_35 [DOI] [PubMed] [Google Scholar]
- 33.Bougie I, Charpentier S, Bisaillon M. 2003. Characterization of the metal ion binding properties of the hepatitis C virus RNA polymerase. J. Biol. Chem. 278:3868–3875. 10.1074/jbc.M209785200 [DOI] [PubMed] [Google Scholar]
- 34.Hooper DC. 2001. Mechanisms of action of antimicrobials: focus on fluoroquinolones. Clin. Infect. Dis. 32(Suppl 1):S9–S15 [DOI] [PubMed] [Google Scholar]
- 35.Cambau E, Sougakoff W, Besson M, Truffot-Pernot C, Grosset J, Jarlier V. 1994. Selection of a gyrA mutant of Mycobacterium tuberculosis resistant to fluoroquinolones during treatment with ofloxacin. J. Infect. Dis. 170:479–483. 10.1093/infdis/170.2.479 [DOI] [PubMed] [Google Scholar]
- 36.Grosset JH. 1992. Treatment of tuberculosis in HIV infection. Tuberc. Lung Dis. 73:378–383. 10.1016/0962-8479(92)90044-K [DOI] [PubMed] [Google Scholar]
- 37.Tsukamura M, Nakamura E, Yoshii S, Amano H. 1985. Therapeutic effect of a new antibacterial substance ofloxacin (DL8280) on pulmonary tuberculosis. Am. Rev. Respir. Dis. 131:352–356 [DOI] [PubMed] [Google Scholar]
- 38.Dong Y, Xu C, Zhao X, Domagala J, Drlica K. 1998. Fluoroquinolone action against mycobacteria: effects of C-8 substituents on growth, survival, and resistance. Antimicrob. Agents Chemother. 42:2978–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corbett KD, Berger JM. 2004. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33:95–118. 10.1146/annurev.biophys.33.110502.140357 [DOI] [PubMed] [Google Scholar]
- 40.Noble CG, Barnard FM, Maxwell A. 2003. Quinolone-DNA interaction: sequence-dependent binding to single-stranded DNA reflects the interaction within the gyrase-DNA complex. Antimicrob. Agents Chemother. 47:854–862. 10.1128/AAC.47.3.854-862.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR. 2009. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 16:667–669. 10.1038/nsmb.1604 [DOI] [PubMed] [Google Scholar]
- 42.Wohlkonig A, Chan PF, Fosberry AP, Homes P, Huang J, Kranz M, Leydon VR, Miles TJ, Pearson ND, Perera RL, Shillings AJ, Gwynn MN, Bax BD. 2010. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struct. Mol. Biol. 17:1152–1153. 10.1038/nsmb.1892 [DOI] [PubMed] [Google Scholar]
- 43.Yoshida H, Bogaki M, Nakamura M, Nakamura S. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271–1272. 10.1128/AAC.34.6.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng AF, Yew WW, Chan EW, Chin ML, Hui MM, Chan RC. 2004. Multiplex PCR amplimer conformation analysis for rapid detection of gyrA mutations in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 48:596–601. 10.1128/AAC.48.2.596-601.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Critchlow SE, Maxwell A. 1996. DNA cleavage is not required for the binding of quinolone drugs to the DNA gyrase-DNA complex. Biochemistry 35:7387–7393. 10.1021/bi9603175 [DOI] [PubMed] [Google Scholar]
- 46.Palu G, Valisena S, Ciarrocchi G, Gatto B, Palumbo M. 1992. Quinolone binding to DNA is mediated by magnesium ions. Proc. Natl. Acad. Sci. U. S. A. 89:9671–9675. 10.1073/pnas.89.20.9671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aldred KJ, McPherson SA, Turnbough CL, Jr, Kerns RJ, Osheroff N. 2013. Topoisomerase IV-quinolone interactions are mediated through a water-metal ion bridge: mechanistic basis of quinolone resistance. Nucleic Acids Res. 41:4628–4639. 10.1093/nar/gkt124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aldred KJ, McPherson SA, Wang P, Kerns RJ, Graves DE, Turnbough CL, Jr, Osheroff N. 2012. Drug interactions with Bacillus anthracis topoisomerase IV: biochemical basis for quinolone action and resistance. Biochemistry 51:370–381. 10.1021/bi2013905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kampranis SC, Maxwell A. 1998. The DNA gyrase-quinolone complex: ATP hydrolysis and the mechanism of DNA cleavage. J. Biol. Chem. 273:22615–22626 [DOI] [PubMed] [Google Scholar]
- 50.Sengupta S, Nagaraja V. 2008. Inhibition of DNA gyrase activity by Mycobacterium smegmatis MurI. FEMS Microbiol. Lett. 279:40–47. 10.1111/j.1574-6968.2007.01005.x [DOI] [PubMed] [Google Scholar]
- 51.Sengupta S, Nagaraja V. 2008. YacG from Escherichia coli is a specific endogenous inhibitor of DNA gyrase. Nucleic Acids Res. 36:4310–4316. 10.1093/nar/gkn355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tran JH, Jacoby GA, Hooper DC. 2005. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49:118–125. 10.1128/AAC.49.1.118-125.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eaves DJ, Randall L, Gray DT, Buckley A, Woodward MJ, White AP, Piddock LJ. 2004. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob. Agents Chemother. 48:4012–4015. 10.1128/AAC.48.10.4012-4015.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamura S, Nakamura M, Kojima T, Yoshida H. 1989. gyrA and gyrB mutations in quinolone-resistant strains of Escherichia coli. Antimicrob. Agents Chemother. 33:254–255. 10.1128/AAC.33.2.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maruri F, Sterling TR, Kaiga AW, Blackman A, van der Heijden YF, Mayer C, Cambau E, Aubry A. 2012. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J. Antimicrob. Chemother. 67:819–831. 10.1093/jac/dkr566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou J, Dong Y, Zhao X, Lee S, Amin A, Ramaswamy S, Domagala J, Musser JM, Drlica K. 2000. Selection of antibiotic-resistant bacterial mutants: allelic diversity among fluoroquinolone-resistant mutations. J. Infect. Dis. 182:517–525. 10.1086/315708 [DOI] [PubMed] [Google Scholar]
- 57.Piton J, Petrella S, Delarue M, Andre-Leroux G, Jarlier V, Aubry A, Mayer C. 2010. Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase. PLoS One 5:e12245. 10.1371/journal.pone.0012245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sissi C, Perdona E, Domenici E, Feriani A, Howells AJ, Maxwell A, Palumbo M. 2001. Ciprofloxacin affects conformational equilibria of DNA gyrase A in the presence of magnesium ions. J. Mol. Biol. 311:195–203. 10.1006/jmbi.2001.4838 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.