Abstract
Capreomycin is a second-line drug for multiple-drug-resistant tuberculosis (TB). However, with increased use in clinics, the therapeutic efficiency of capreomycin is decreasing. To better understand TB resistance to capreomycin, we have done research to identify the molecular target of capreomycin. Mycobacterium tuberculosis ribosomal proteins L12 and L10 interact with each other and constitute the stalk of the 50S ribosomal subunit, which recruits initiation and elongation factors during translation. Hence, the L12-L10 interaction is considered to be essential for ribosomal function and protein synthesis. Here we provide evidence showing that capreomycin inhibits the L12-L10 interaction by using an established L12-L10 interaction assay. Overexpression of L12 and/or L10 in M. smegmatis, a species close to M. tuberculosis, increases the MIC of capreomycin. Moreover, both elongation factor G-dependent GTPase activity and ribosome-mediated protein synthesis are inhibited by capreomycin. When protein synthesis was blocked with thiostrepton, however, the bactericidal activity of capreomycin was restrained. All of these results suggest that capreomycin seems to inhibit TB by interrupting the L12-L10 interaction. This finding might provide novel clues for anti-TB drug discovery.
INTRODUCTION
Infections caused by Mycobacterium tuberculosis are common and in many cases lethal. The emergence of multidrug-resistant (MDR) strains and compromised immunity due to the epidemic of HIV/AIDS have increased the chance of infection with M. tuberculosis (1). The first-line antituberculosis (anti-TB) drugs are not helpful for infections caused by MDR strains; thus, new anti-TB drugs are badly needed for clinical use. Capreomycin (CAP) is a cyclic peptide antibiotic that is particularly active against mycobacteria and is regarded as the ideal second-line anti-TB drug for MDR and persistent infections (2). Unfortunately, the efficiency of CAP is reducing along with its increased use in patients (3). Therefore, understanding of the molecular mechanism of CAP might play a significant role in the discovery of novel anti-TB drugs.
A previous study indicated that CAP may bind both the 30S and 50S ribosomal subunits and interfere with their functions (4). CAP prevents the deacylated tRNA from moving from the A site to the P site, vacating the A site for the next aminoacylated tRNA (5). Recently, CAP was shown to bind to the bacterial ribosome where interbridge B2a extends across the subunit interface and links the decoding region of the 16S rRNA with the highly conserved 23S rRNA (6). Microarray assay results indicate that the treatment of cells with CAP results in the expression of ribosomal proteins L12 and L10, which might be the complementary response to the compromised ribosomal function caused by CAP treatment (7, 8). Although accumulating evidence indicates that the ribosome is likely to be the target of CAP, the ribosomal protein that CAP directly interacts with remains unidentified.
M. tuberculosis ribosomal proteins L12 (encoded by the rplL gene) and L10 (encoded by the rplJ gene) interact with each other, and this interaction plays an important role in ribosomal function (9). L12 and L10 belong to the stalk of the large ribosomal subunit (50S). It has been shown that the stalk interacts with elongation factors EF-G and EF-Tu, and their GTPase activity is enhanced upon binding to the stalk (10). The L12 N-terminal domain interacts with the C-terminal domain of L10. This complex binds to the conserved GTPase-association region of the 23S rRNA (helices 42 to 44) through the N-terminal domain of L10. Disruption of the L12-L10 interaction abolishes the binding of EF-G and EF-Tu to the stalk and hence impairs GTPase activity and undermines ribosome-dependent protein synthesis. Therefore, the L12-L10 interaction is a novel drug target for anti-TB drugs (11).
A previous report showed that CAP interacts with the 23S rRNA, which is a component of the large subunit (50S) (12). Because ribosomal proteins L12 and L10 are the important stalk of 50S and they are physically close to the 23S rRNA (13), an untested possibility is that CAP associates with the L12-L10 complex to disrupt ribosomal function. In this study, we used an established L12-L10 interaction assay to demonstrate that CAP inhibits the L12-L10 interaction. It provoked our interest to investigate the compound further. Our observation that overexpression of L12 and/or L10 in M. smegmatis increases the MIC of CAP indicates that L12 and L10 are likely the targets. We further found that both EF-G-dependent GTPase activity and ribosome-mediated protein synthesis were inhibited by CAP. We speculated that disruption of the L12-L10 interaction by CAP impairs EF-G-dependent GTPase activity and protein synthesis subsequently. Moreover, CAP inhibits the growth of M. smegmatis through a bactericidal effect. Interestingly, the bactericidal effect of CAP is restrained when protein synthesis is blocked. All of these data indicate that CAP may inhibit mycobacterial growth by blocking protein synthesis on the ribosome through a novel target.
MATERIALS AND METHODS
The established assay.
Using the yeast two-hybrid system, we have demonstrated the interaction between L12 and L10 from M. tuberculosis (11). In this two-hybrid system, the L12-L10 interaction induces the expression of lacZ, which encodes the enzyme β-galactosidase (β-gal). To assess β-gal enzyme activity, 4.5 ml of yeast cultures in mid-log phase (optical density at 600 nm [OD600] of 0.5) were harvested by centrifugation and then resuspended in 300 μl of Z buffer. A 100-μl volume of the cell suspension was lysed by repeated freeze-thaw cycles, and then 160 μl of o-nitrophenyl-β-d-galactopyranoside (ONPG; 4 mg/ml in Z buffer) was added to the lysate and incubated at 30°C until we observed a color change. The reaction was stopped by the addition of 0.4 ml of 1 M Na2CO3. After centrifugation at 14,000 × g for 10 min, the A420 of the supernatant was measured. As a control, yeast cells with a control vector were used. The units of β-gal activity were calculated according to the instructions in the Clontech Yeast Protocols Handbook. The experiments were repeated three times, and three sets of readings were taken each time.
Determination of the MIC for M. smegmatis mc2155 overexpressing L12 and/or L10.
An L12-L10-enhanced green fluorescent protein (eGFP)/L10-L12-eGFP expression plasmid was constructed with the Gly-Ser-Gly4-Ser-Gly4-Ser flexible linker. M. smegmatis strains overexpressing the L12-eGFP, L10-eGFP, L12-L10-eGFP, L10-L12-eGFP, S12-eGFP, and Icl-eGFP fusion proteins were isolated by fluorescence microscopy, and the strains with an obvious GFP signal were used for the experiment. The L12-eGFP-, L10-eGFP-, L12-L10-eGFP-, and L10-L12-eGFP-overexpressing strains were further confirmed by Western blotting with anti-eGFP antibody. The MICs of CAP for M. smegmatis mc2155 strains with a control vector or strains overexpressing L12, L10, L12-L10, L10-L12, S12, and Icl were determined by the conventional plate dilution method.
Bacterial growth curves of M. smegmatis mc2155 overexpressing strains in the presence or absence of CAP.
The growth of M. smegmatis mc2155 carrying the vector control, L12-eGFP, L10-eGFP, L12-L10-eGFP, or L10-L12-eGFP was monitored by OD600 determination over a period of 72 h. M. smegmatis mc2155 strains were grown in 7H9 medium. At the same time, 0.5 μg/ml CAP (half of the MIC of the vector control) was added to each strain. An inoculum of 106 cells ml−1 was used, and growth (OD600) was measured three times a day at 0, 12, 24, 36, 48, 60, and 72 h.
Inhibition of GTPase activity.
As M. tuberculosis grows slowly, we use M. smegmatis, a nonpathogenic strain biologically close to M. tuberculosis, to examine ribosome-stimulated GTPase activity. The M. smegmatis ribosome extract was prepared as described by Kigawa et al. (14). M. smegmatis EF-G was purified according to Rodnina et al. (15). To measure ribosome-stimulated GTP hydrolysis by EF-G, ribosome extract (0.2 μM) was mixed with EF-G (0.04 μM) in the reaction buffer. The reaction buffer contained 50 mM Tris-HCl (pH 7.5), 70 mM NH4Cl, 30 mM KCl, and 7 mM MgCl2. The reaction was initiated by the addition of GTP (1 mM), and GTPase activity was assessed in a microtiter plate-based colorimetric assay by determining the ability of the enzyme to liberate Pi from GTP. Briefly, the enzyme reaction was terminated by the addition of an acid solution (200 μl) of malachite green, ammonium molybdate, and polyvinyl alcohol. The liberated Pi formed a phosphomolybdate-malachite complex that was detected at 650 nm. To detect inhibition of GTPase activity by CAP, various concentrations of CAP were added to the ribosome extract and incubated at 37°C for 20 min. One hundred percent activity corresponds to the hydrolysis of 0.2 μM ribosome extract and 1 mM GTP.
Protein synthesis inhibition in the ribosome.
Translation inhibition by CAP was assessed by using an in vitro cell-free translation system supplied with ribosomes from M. smegmatis, as well as a luciferase reporter. Translation assays were carried out according to the manufacturer's instructions (Promega, Madison, WI). The M. smegmatis ribosome extract was prepared as described before (13), followed by the addition of a mixture of nucleotide triphosphates, amino acids, and a luciferase reporter. Light emission was recorded with a luminescence counter (PerkinElmer, Wellesley, MA). CAP was added to final concentrations ranging from 0.4 to 250 μg/ml. The 50% inhibitory concentration (IC50) was determined on the basis of the ratio of light emission units to the concentration of compounds (log plots) that fits a variable-slope dose-response equation.
Determination of the mode of action.
M. smegmatis mc2155 is close to M. tuberculosis but exhibits fast growth and is noninfectious. This strain was grown to early log phase in Middlebrook 7H9 broth at 37°C. The culture was diluted to 5 × 105 CFU/ml in fresh medium containing various concentrations of CAP (1 to 64 μg/ml). The bacteria were collected at different time points, serially diluted, and plated. After incubation for 48 h at 37°C, the colonies were counted.
Ribosome-dependent bactericidal activity.
M. smegmatis mc2155 was grown in 7H9 broth to mid-log phase and then diluted 10,000-fold into fresh prewarmed medium. Prior to the addition of test compounds, the cultures were preincubated with 16 μg/ml of thiostrepton (Tsr) for 5 min at 37°C. CAP and isoniazid (INH) were added to the medium at 64 times their MICs. Aliquots of each culture were removed at the indicated times, serially diluted, plated, and incubated at 37°C for 48 h for colony counting.
RESULTS
CAP inhibits the interaction of M. tuberculosis proteins L12 and L10 in a yeast two-hybrid assay.
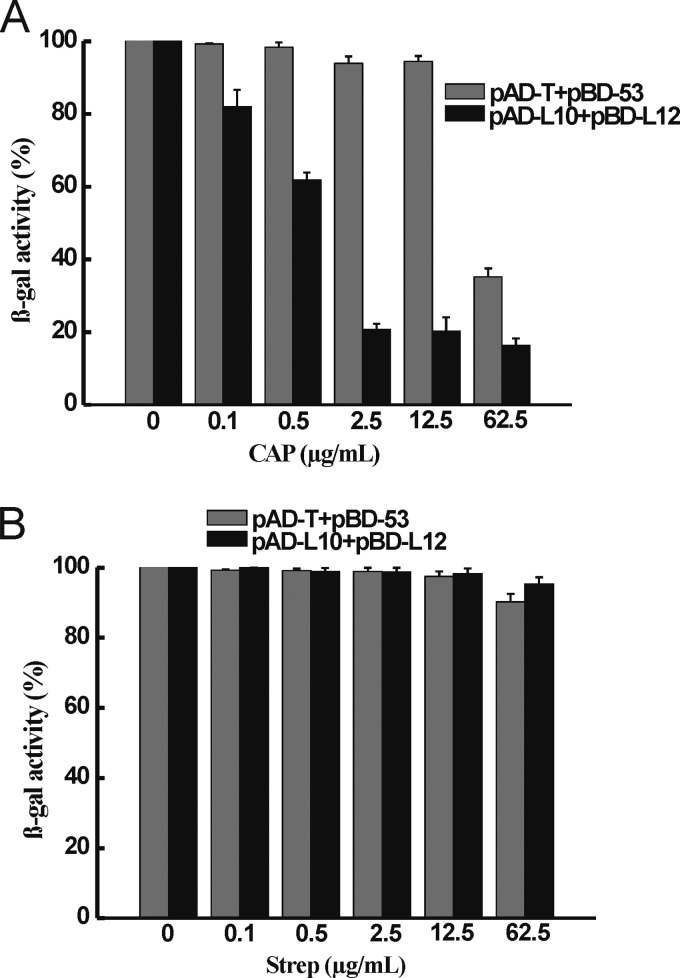
We have established a yeast two-hybrid assay system to detect the L12-L10 interaction. In this assay, DNA fragments of the genes for L12 and L10 of M. tuberculosis were fused in frame with the GAL4 transcription-activating domain and the DNA binding domain in the resulting plasmids pAD-L12 and pBD-L10. Then pAD-L12 and pBD-L10 were introduced into AH109 yeast cells to generate AH109(pAD-L12/pBD-L10). Strain AH109(pAD-T/pBD-53) (the tumor suppressor p53 protein binds to the simian virus 40 large T antigen) was used as a positive control. The L12-L10 interaction leads to the activation of reporter genes ADE2, HIS3, and LacZ in AH109 cells. Thus, the L12-L10 interaction can be determined by the growth of yeast cells on plates of synthetic defined medium lacking adenine and histidine (SD/-Ade-His) and strong β-gal activity can be determined by using ONPG as the LacZ substrate. Compounds that block the L12-L10 interaction can inhibit the growth of AH109(pAD-L12/pBD-L10) cells in SD/-Ade-His medium, as well as the β-gal activity of this strain. We found that CAP inhibited the growth of yeast strain AH109(pAD-L12/pBD-L10) in SD/-Ade-His medium but had no effect on the growth of yeast strain AH109(pAD-T/pBD-53) in SD/-Ade-His medium and AH109 yeast cells in yeast extract-peptone-dextrose medium (a rich medium for yeast cells) (data not shown). Strains AH109(pAD-L12/pBD-L10) and AH109(pAD-T/pBD-53) were subjected to a quantitative assay of β-gal activity in the presence of various concentrations of CAP. At 2.5 μg/ml, CAP inhibited the β-gal activity of AH109(pAD-L12/pBD-L10) almost completely on SD/-Leu-Trp but not that of AH109(pAD-T/pBD-53). Moreover, CAP inhibited the β-gal activity of AH109(pAD-L12/pBD-L10) in a dose-dependent manner, and it showed clear inhibition of β-gal activity at 0.5 μg/ml (Fig. 1A). Streptomycin was used as a control, and it had no effect on the β-gal activity of either strain AH109(pAD-L12/pBD-L10) or AH109(pAD-T/pBD-53) (Fig. 1B). All of these results indicate that CAP inhibits the interaction between ribosomal proteins L12 and L10 of M. tuberculosis.
FIG 1.
(A) CAP blocks the L12-L10 interaction in the yeast two-hybrid assay. Inhibition of the β-gal activity of AH109(pAD-L12/pBD-L10) cells by CAP at various concentrations is shown. Strain AH109(pAD-T/pBD-53) was used as a control. The percentage of β-gal activity in cells treated with CAP over that in untreated cells is shown. The activity of β-gal was determined in the presence of CAP at concentrations of 0.1, 0.5, 2.5, 12.5, and 62.5 μg/ml. Means and error bars (representing standard deviations) from three separate experiments are shown. (B) Streptomycin (Strep) was used as a control. It has no effect on the β-gal activities of strains AH109(pAD-L12/pBD-L10) and AH109(pAD-T/pBD-53).
Higher MICs for strains overexpressing L12 and L10.
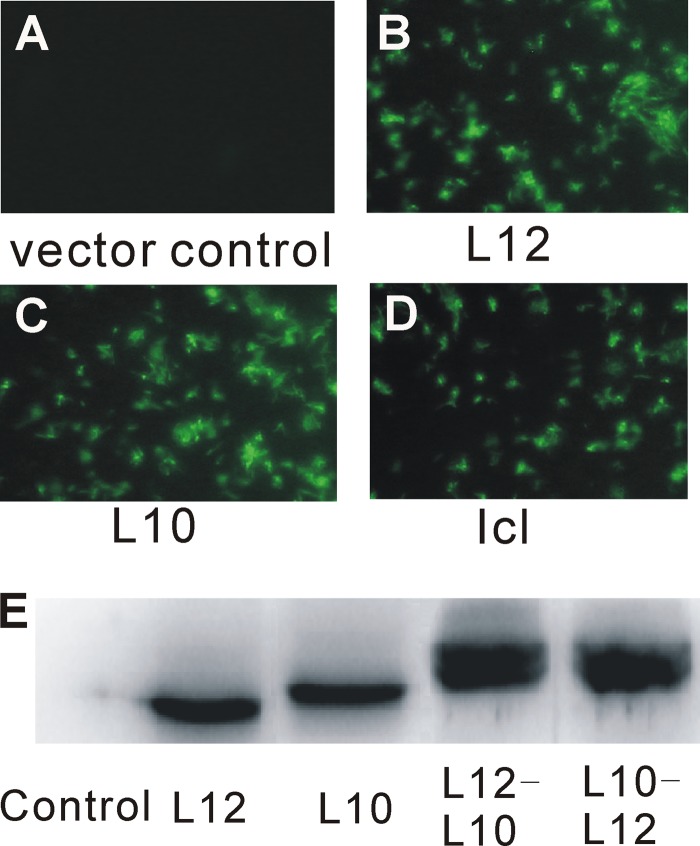
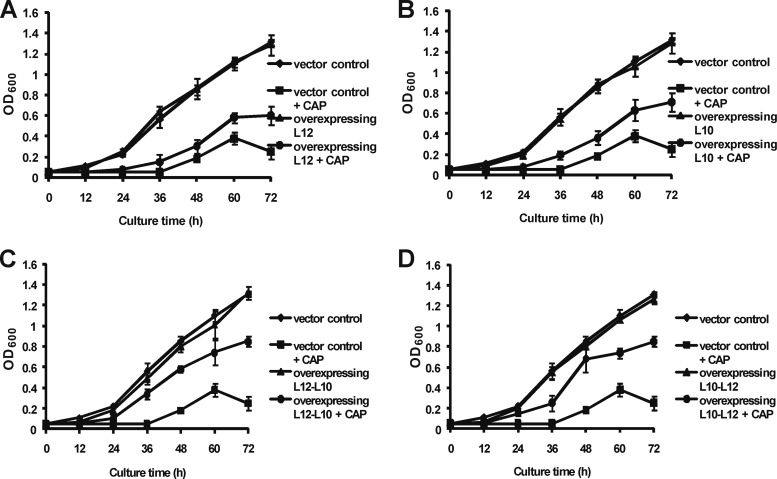
If CAP inhibits the growth of mycobacteria by disrupting the L12-L10 interaction, high levels of expression of L12 and/or L10 protein should lower its antibacterial activity. Therefore, we introduced expression plasmids containing the eGFP-tagged L12-, L10-, L12-L10-, L10-L12-, and 30S ribosomal protein S12 (streptomycin target)-encoding genes and the ribosome-irrelevant gene for isocitrase (Icl) into M. smegmatis and confirmed their expression by the appearance of a GFP signal (Fig. 2A to D). The expression of the L12, L10, L12-L10 fusion, and L10-L12 fusion proteins was also confirmed by Western blotting with anti-eGFP antibody (Fig. 2E). Interestingly, the MICs of CAP for the M. smegmatis strains overexpressing L12 and L10 were both 4 μg/ml, which was 4-fold higher than the MICs for the strain transfected with the vector control (Table 1). Furthermore, both of the strains overexpressing L12-L10 and L10-L12 showed an 8-fold CAP MIC increase (8 μg/ml). The MIC increased even more (2-fold) when L12 and L10 were overexpressed together than when they were expressed separately, while the expression of S12 and Icl in M. smegmatis did not increase the MIC of CAP. As a control, we examined the MICs of streptomycin for all of the overexpressing strains. These strains did not alter the MICs of streptomycin compared with the vector control, except for strains that overexpressed S12 (streptomycin target). For further proof, we examined the growth curves of all of the overexpressing strains in the presence or absence of CAP. The growth of each M. smegmatis strain overexpressing L12, L10, L12-L10, L10-L12, or the vector control did not show a significant difference in the absence of CAP. When induced with 0.5 μg/ml CAP (half of the MIC for the vector control), the growth rates of all of the strains decreased. However, the growth rate of the M. smegmatis strain overexpressing the vector control was lower than that of the strains overexpressing L12, L10, L12-L10, and L10-L12. Interestingly, the growth of M. smegmatis strains overexpressing L12 and L10 was slightly delayed with respect to that of strains overexpressing L12-L10 and L10-L12 (Fig. 3). Therefore, we speculate that the CAP sensitivity mediated by L12/L10 is likely to be a specific effect. Collectively, these data suggest that ribosomal proteins L12 and L10 are likely to be the in vivo targets of CAP. Because the C terminus of L10 anchors L12 by associating with the N-terminal domains of L12, we speculate that CAP may bind to the L12-L10 binding site.
FIG 2.
Selection of M. smegmatis mc2155 strains overexpressing L12-eGFP, L10-eGFP, L12-L10-eGFP, L10-L12-eGFP, and Icl-eGFP. M. smegmatis cells carrying plasmid pMV261 (A), pMV261:L12-eGFP (B), pMV261:L10-eGFP (C), or pMV261:Icl-eGFP (D) were visualized by fluorescence microscopy, and the expression of the L12 and/or L10 proteins in the selected strains was detected by Western blotting with anti-eGFP antibody (E).
TABLE 1.
Antibacterial activity of CAP against M. smegmatis strains with a vector control or plasmids overexpressing L10, L12, L12-L10, L10-L12, S12, or Icl
| Plasmid | MIC (μg/ml) |
|
|---|---|---|
| CAP | Streptomycin | |
| Vector control | 1 | 0.5 |
| L10 overexpression | 4 | 0.5 |
| L12 overexpression | 4 | 0.5 |
| L12-L10 overexpression | 8 | 0.5 |
| L10-L12 overexpression | 8 | 0.5 |
| S12 overexpression | 1 | 1 |
| Icl overexpression | 1 | 0.5 |
FIG 3.
Bacterial growth curves of M. smegmatis mc2155 overexpressing strains in the presence or absence of CAP. M. smegmatis cells carrying pMV261 (vector control) and pMV261:L12-eGFP (A), pMV261:L10-eGFP (B), pMV261:L12-L10-eGFP (C), or pMV261:L10-L12-eGFP (D) were grown in 7H9 broth. At the same time, CAP (at half the MIC of the vector control, 0.5 μg/ml) was added to all of the overexpressing strains. Bacterial populations were monitored by measuring OD600. Data are shown as mean values from three experiments ± standard deviations.
CAP blocks ribosomal GTPase activity and ribosome-dependent protein synthesis.
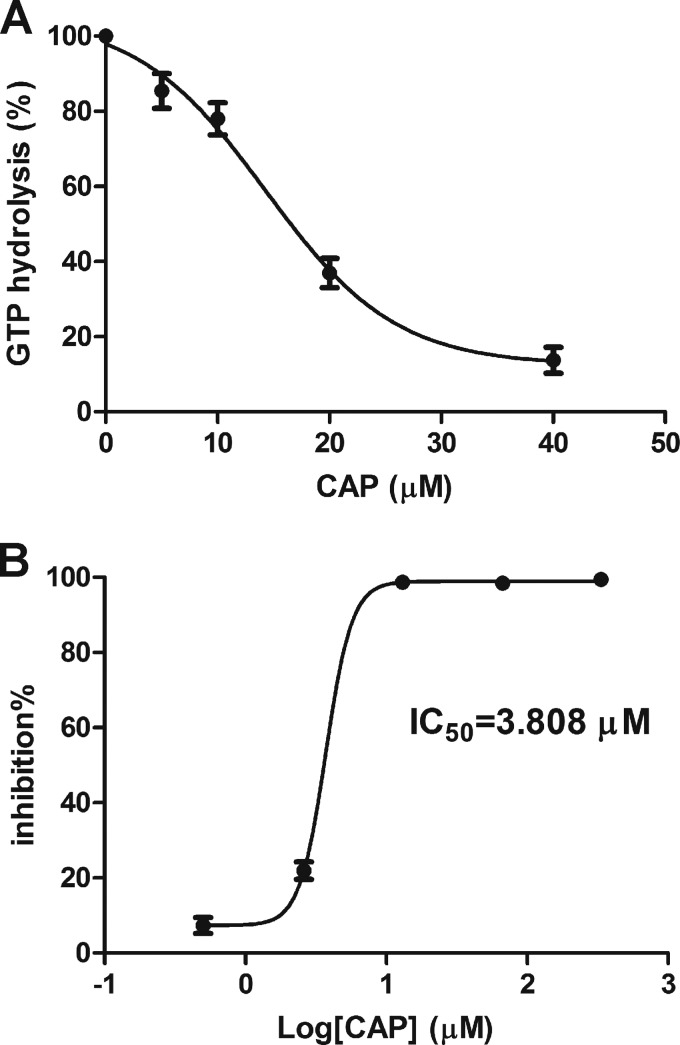
The L12 protein interacts with L10, and this interaction facilitates the recruitment of EF-G to the ribosome to enhance GTPase activity. As CAP blocks the L12-L10 interaction in the yeast two-hybrid assay, we speculate that CAP inhibits the GTPase activity that is essential for protein synthesis. Thus, we examined the effect of CAP on the GTPase activity of M. smegmatis ribosomes. In this assay, the release of Pi from ribosome-bound EF-G·GTP was monitored by measuring the phosphomolybdate-malachite complex, which increases significantly upon the binding of Pi. When CAP was added, the release of Pi from GTP was clearly reduced. As shown in Fig. 4A, the activity of EF-G-dependent GTPase was inhibited by CAP (5 to 40 μM) treatment with an IC50 of 14.31 μM.
FIG 4.
In vitro inhibition of GTPase activity and protein translation by CAP. (A) CAP reduced ribosome-stimulated GTP hydrolysis activity. Ribosome extract was incubated with different concentrations of CAP at 37°C for 20 min and mixed with EF-G and GTP. The reaction was quenched by the addition of an acid solution of malachite green, ammonium molybdate, and polyvinyl alcohol. The GTPase activity was assessed by determining the ability to liberate Pi from GTP. One hundred percent activity corresponds to the hydrolysis of 0.2 μM ribosome extract and 1 mM GTP. (B) Inhibition of protein translation by CAP. Inhibition was quantified with an in vitro cell-free translation system supplied with ribosomes from M. smegmatis, as well as a luciferase reporter. The experiment was repeated three times.
The L12-L10 interaction is essential for ribosome-dependent protein synthesis. Therefore, we took advantage of an in vitro system to determine whether CAP inhibits ribosome-mediated protein synthesis in M. smegmatis. For this purpose, we added CAP to a cell-free bacterial transcription-translation system containing M. smegmatis S30 extract and a luciferase reporter. Indeed, CAP inhibited protein translation with an IC50 of 3.808 μM (Fig. 4B), indicating the disruption of ribosomal function.
Bactericidal activity of CAP.
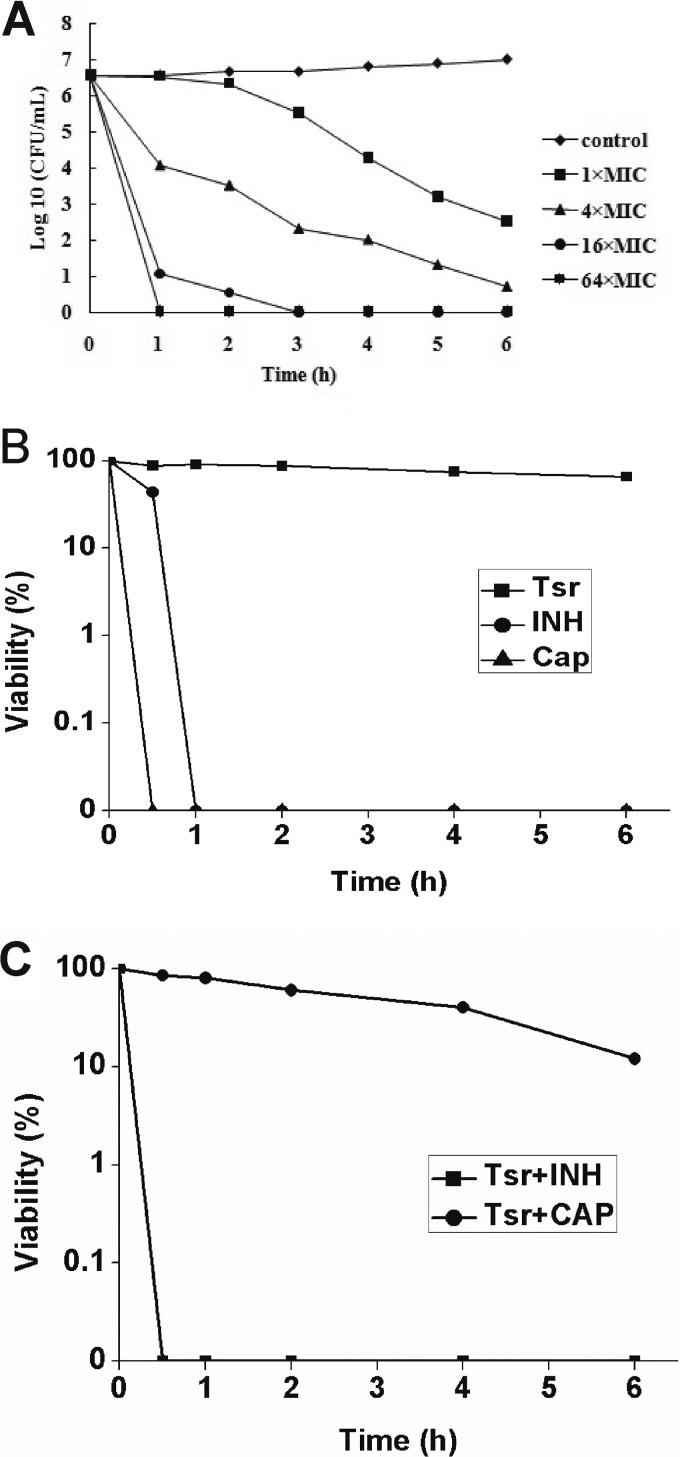
CAP may inhibit the growth of M. tuberculosis through a bacteriostatic or bactericidal effect. Because the growth of M. tuberculosis is extremely slow, we used M. smegmatis, to examine viability after treatment with the compound. The MIC of CAP for this nonpathogenic strain was 1 μg/ml. It was obvious that CAP showed bactericidal activity against this strain at its MIC, as evidenced by the viability loss. Moreover, the bactericidal activity increased dramatically when the concentrations of the compounds were 4, 16, or 64 times of the MIC. After 1 h of incubation, almost all of the bacteria were killed in the presence of CAP at 64 times the MIC (Fig. 5A). The results suggest a strong bactericidal activity of CAP against mycobacteria.
FIG 5.
Translation-dependent bactericidal activity of CAP. (A) Dose-dependent bactericidal activity of CAP against M. smegmatis mc2155. The MIC of CAP for this strain is 1 μg/ml. M. smegmatis mc2155 viability was determined in the presence of CAP concentrations ranging from 1 to 64 times the MIC. (B) The mode of action of the compounds tested against M. smegmatis mc2155. Growing bacteria in mid-log phase were incubated in the presence of Tsr (16 μg/ml), CAP (64 μg/ml), or INH (64 μg/ml). Viability was determined every hour for 6 h. (C) Bactericidal activity of CAP against M. smegmatis when preincubated with Tsr. Bacteria in the mid-log phase were pretreated with 16 μg/ml of Tsr for 5 min at 37°C before the test compounds (CAP, 64 μg/ml; INH, 64 μg/ml) were added to the cell cultures. Viability was determined every hour for 6 h.
Ribosome-dependent bactericidal activity.
Tsr exhibits bacteriostatic activity as it blocks protein synthesis by inhibiting the function of EF-G through the stalk of 50S (16). We found that Tsr inhibited the growth of M. smegmatis with a MIC of 2 μg/ml. Thus, we treated growing bacteria with 16 μg/ml Tsr for 5 min at 37°C before adding CAP. Although CAP alone killed M. smegmatis cells rapidly (Fig. 4B), many cells were viable when pretreated with Tsr (Fig. 5C). In contrast, INH, which targets the bacterial cell wall, maintained its bactericidal activity even when cells were pretreated with Tsr (Fig. 5C). One possibility is that disruption of protein synthesis in M. smegmatis by Tsr impairs the bactericidal activity of CAP. Therefore, we speculate that Tsr-dependent blockage of protein synthesis prevents CAP-induced ribosome damage, and this irreversible damage might contribute to the bactericidal activity of CAP. Alternatively, the growth inhibition by Tsr suppresses the bactericidal activity of CAP by targeting bacterial systems that remediate hydroxyl radical damage (17). Further investigation is needed to precisely demonstrate the primary target.
DISCUSSION
The emergence of MDR TB has significantly complicated TB treatment. CAP is particularly important for the treatment of TB caused by strains of M. tuberculosis that are resistant to first-line drugs. Previous data indicate that CAP binds across the ribosome interface involving the 23S and 16S rRNAs. One untested possibility is that CAP may also bind to ribosomal proteins adjacent to the 23S and 16S rRNAs to interfere with ribosomal function. The L12-L10 interaction is critical for ribosomal function, and our data indicate that CAP inhibits the L12-L10 interaction. Consistently, overexpression of L12 and/or L10 in M. smegmatis increases the MIC of CAP. Furthermore, CAP impairs EF-G-dependent GTPase activity and ribosome-mediated protein synthesis. Therefore, we inferred that the anti-TB antibiotic CAP inhibits protein synthesis by disrupting the interaction between two ribosomal proteins, L12 and L10.
An assay that targets the M. tuberculosis L12-L10 interaction by using a yeast two-hybrid system was established by our group. With this system, we identified two compounds, T766 and T054, and further proved that these compounds bind to L12 to block the L12-L10 interaction and protein synthesis (11). Also, compounds T766 and T054 were confirmed to have anti-TB activity. These results showed that the assay is feasible. CAP was found to have strong selective inhibition in the assay, suggesting that it may be an inhibitor of the L12-L10 interaction. The increased MICs for L12-, L10-, L12-L10-, and L10-L12-overexpressing strains and decreased EF-G-dependent GTPase activity verified that CAP may bind to L12 and/or L10 and abolish the L12-L10 interaction. Besides, the inhibited protein synthesis further validated the damage of the L12-L10 interaction. These results explain, at least in part, the anti-TB mechanism of CAP.
The results of the yeast two-hybrid assay indicate that CAP blocks the L12-L10 interaction. In support of this possibility, the CAP MIC for M. smegmatis overexpressing L12 and/or L10 was higher than that for the control. To further determine if CAP binds to L12 or L10 to disrupt their interaction, we performed a surface plasmon resonance (SPR) experiment. Unfortunately, no interaction of protein and CAP was detected by the SPR assay. We speculated that the small-molecular compound could lead to a reduced signal (18). Additionally, as dimethyl sulfoxide (DMSO) has a much higher refractive index than the common buffer used, it is difficult to produce reliable readings from DMSO-containing compounds by SPR assay. Further studies are needed to determine the direct relationship between CAP and L12 or L10.
Previous studies found that CAP interacts with helix 69 of the 23S rRNA and its antibacterial activity was reduced by inactivating a 2′-O-methyltransferase (encoded by the tlyA gene) that modifies nucleotide C1920 in helix 69 of the 23S rRNA, suggesting that the direct target of CAP is domain IV of the 23S rRNA (19, 20). While ribosomal protein L12 binds to L10 and forms a prominent stalk of the 50S subunit, which binds to a region in domain II of the 23S rRNA through the C-terminal region of L10 (21). Although there is no evidence of direct binding of L12 to the 23S rRNA, L12 binds to L10, forming a compact and stable stalk, and appears to be a functional unit activating ribosomes by its binding to the GTP-associated RNA domain (22). Therefore, we speculate that L12-L10 in its interacting site might be functional in the binding of 23S rRNA domain II to the ribosome. A chemical footprinting experiment indicates that domains IV and II interact with domain IV in forming the three-dimensional structure of the peptidyl transferase center within the ribosome (23). In addition to the increased CAP susceptibility caused by L12/L10 overexpression and reduced EF-G GTPase and ribosome-mediated translation activity affected by CAP, we speculate that CAP binds to both the 23S rRNA and L10/L12, which blocks the formation of the stalk and inhibits protein synthesis. This is the first report indicating a possible interaction between CAP and a ribosomal protein, and this interaction may play a key role in the antibacterial activity of this antibiotic. Interestingly, a recent microarray assay showed upregulated expression of L12 and L10 in CAP-treated cells, which could be a transcriptional response to the failure to form the L12-L10 complex (7, 8).
It has been reported that the CAP MIC for strain H37Rv is 2 μg/ml (24). We noticed that the IC50 of CAP for protein inhibition (3.808 μM) is similar to its MIC for H37Rv, but the IC50 for EF-G-dependent GTPase activity (14.31 μM) is much higher than the MIC. In bacteria, EF-G and EF-Tu are the two GTP-binding proteins essential for protein synthesis, and the L12-L10 interaction enhances EF-G/EF-Tu-dependent GTPase activity by stimulating their recruitment to the ribosome. We speculate that the block of the L12-L10 interaction by CAP inhibits GTPase activity and protein synthesis. In the in vitro translation system, CAP inhibits both EF-G- and EF-Tu-dependent protein synthesis (25). With an in vitro GTPase-detecting system supplied with ribosomes and EF-G, CAP showed less dramatic inhibition of GTPase activity. One possibility is that the in vitro system is not sensitive. Alternatively, CAP inhibits both EF-G- and EF-Tu-dependent GTPase activity but the inhibition of either EF-G or EF-Tu is sufficient to block protein synthesis, which may contribute to the lower MIC.
However, a key experiment such as the isolation of a CAP-resistant mutant is necessary to identify the exact target. Unfortunately, these results are not yet available. All of the current conclusions that CAP inhibits protein synthesis by disrupting interactions between ribosomal proteins L12 and L10 are based on indirect evidence.
In summary, our data indicate that CAP inhibits the L12-L10 interaction. Consistently, ribosomal GTPase activity and ribosome-dependent protein synthesis are inhibited by CAP. The higher MIC for the M. smegmatis strain overexpressing L12 and/or L10 than for the control suggests that L12 and L10 are likely to be the in vivo targets of CAP. Thus, the identification of novel drug targets might help us to find new drug candidates against TB.
ACKNOWLEDGMENTS
This work was supported by the Basic Scientific Research Program of Materia Medica, CAMS (2013ZD05), State Mega Programs (2012ZX09301002-003/006), the Beijing Key Laboratory of New Drug Mechanisms and Pharmacological Evaluation Study (BZ0150), and grants from the National Natural Science Foundation of China (81302816, 81001461, and 81072672).
Footnotes
Published ahead of print 21 January 2014
REFERENCES
- 1.Dye C, Espinal MA, Watt CJ, Mbiaga C, Williams BG. 2002. Worldwide incidence of multidrug-resistant tuberculosis. J. Infect. Dis. 185:1197–1202. 10.1086/339818 [DOI] [PubMed] [Google Scholar]
- 2.Thomas MG, Chan YA, Ozanick SG. 2003. Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster. Antimicrob. Agents Chemother. 47:2823–2830. 10.1128/AAC.47.9.2823-2830.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Georghiou SB, Magana M, Garfein RS, Catanzaro DG, Catanzaro A, Rodwell TC. 2012. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS One 7:e33275. 10.1371/journal.pone.0033275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maus CE, Plikaytis BB, Shinnick TM. 2005. Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:571–577. 10.1128/AAC.49.2.571-577.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. 2010. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat. Struct. Mol. Biol. 17:289–293. 10.1038/nsmb.1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansen SK, Maus CE, Plikaytis BB, Douthwaite S. 2006. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol. Cell 23:173–182. 10.1016/j.molcel.2006.05.044 [DOI] [PubMed] [Google Scholar]
- 7.Fu LM. 2006. Exploring drug action on Mycobacterium tuberculosis using Affymetrix oligonucleotide GeneChips. Tuberculosis 86:134–143. 10.1016/j.tube.2005.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu LM, Shinnick TM. 2007. Genome-wide exploration of the drug action of capreomycin on Mycobacterium tuberculosis using Affymetrix oligonucleotide GeneChips. J. Infect. 54:277–284. 10.1016/j.jinf.2006.05.012 [DOI] [PubMed] [Google Scholar]
- 9.Bruell CM, Eichholz C, Kubarenko A, Post V, Katunin VI, Hobbie SN, Rodnina MV, Bottger EC. 2008. Conservation of bacterial protein synthesis machinery: initiation and elongation in Mycobacterium smegmatis. Biochemistry 47:8828–8839. 10.1021/bi800527k [DOI] [PubMed] [Google Scholar]
- 10.Savelsbergh A, Mohr D, Wilden B, Wintermeyer W, Rodnina MV. 2000. Stimulation of the GTPase activity of translation elongation factor G by ribosomal protein L7/12. J. Biol. Chem. 275:890–894. 10.1074/jbc.275.2.890 [DOI] [PubMed] [Google Scholar]
- 11.Lin Y, Li Y, Zhu Y, Zhang J, Liu X, Jiang W, Yu S, You XF, Xiao C, Hong B, Wang Y, Jiang JD, Si S. 2012. Identification of antituberculosis agents that target ribosomal protein interactions using a yeast two-hybrid system. Proc. Natl. Acad. Sci. U. S. A. 109:17412–17417. 10.1073/pnas.1110271109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monshupanee T, Gregory ST, Douthwaite S, Chungjatupornchai W, Dahlberg AE. 2008. Mutations in conserved helix 69 of 23S rRNA of Thermus thermophilus that affect capreomycin resistance but not posttranscriptional modifications. J. Bacteriol. 190:7754–7761. 10.1128/JB.00984-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalo P, Reboud JP. 2003. The puzzling lateral flexible stalk of the ribosome. Biol. Cell 95:179–193. 10.1016/S0248-4900(03)00034-0 [DOI] [PubMed] [Google Scholar]
- 14.Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S. 2004. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genomics 5:63–68. 10.1023/B:JSFG.0000029204.57846.7d [DOI] [PubMed] [Google Scholar]
- 15.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. 2005. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 24:4316–4323. 10.1038/sj.emboj.7600884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. 10.1016/j.cell.2007.06.049 [DOI] [PubMed] [Google Scholar]
- 17.Walter JD, Hunter M, Cobb M, Traeger G, Spiegel PC. 2012. Thiostrepton inhibits stable 70S ribosome binding and ribosome-dependent GTPase activation of elongation factor G and elongation factor 4. Nucleic Acids Res. 40:360–370. 10.1093/nar/gkr623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann T, Junker HD, Schmidt K, Sekul R. 2007. SPR-based fragment screening: advantages and applications. Curr. Top. Med. Chem. 7:1630–1642. 10.2174/156802607782341073 [DOI] [PubMed] [Google Scholar]
- 19.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905–920. 10.1126/science.289.5481.905 [DOI] [PubMed] [Google Scholar]
- 20.Akbergenov R, Shcherbakov D, Matt T, Duscha S, Meyer M, Wilson DN, Bottger EC. 2011. Molecular basis for the selectivity of antituberculosis compounds capreomycin and viomycin. Antimicrob. Agents Chemother. 55:4712–4717. 10.1128/AAC.00628-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimizu T, Nakagaki M, Nishi Y, Kobayashi Y, Hachimori A, Uchiumi T. 2002. Interaction among silkworm ribosomal proteins P1, P2 and P0 required for functional protein binding to the GTPase-associated domain of 28S rRNA. Nucleic Acids Res. 30:2620–2627. 10.1093/nar/gkf379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griaznova O, Traut RR. 2000. Deletion of C-terminal residues of Escherichia coli ribosomal protein L10 causes the loss of binding of one L7/L12 dimer: ribosomes with one L7/L12 dimer are active. Biochemistry 39:4075–4081. 10.1021/bi992621e [DOI] [PubMed] [Google Scholar]
- 23.Kowalak JA, Bruenger E, Hashizume T, Peltier JM, Ofengand J, McCloskey JA. 1996. Structural characterization of U*-1915 in domain IV from Escherichia coli 23S ribosomal RNA as 3-methylpseudouridine. Nucleic Acids Res. 24:688–693. 10.1093/nar/24.4.688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho YI, Chan CY, Cheng AF. 1997. In-vitro activities of aminoglycoside-aminocyclitols against mycobacteria. J. Antimicrob. Chemother. 40:27–32. 10.1093/jac/40.1.27 [DOI] [PubMed] [Google Scholar]
- 25.Helgstrand M, Mandava CS, Mulder FA, Liljas A, Sanyal S, Akke M. 2007. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J. Mol. Biol. 365:468–479. 10.1016/j.jmb.2006.10.025 [DOI] [PubMed] [Google Scholar]