Abstract
With 29 individual antiretroviral drugs available from six classes that are approved for the treatment of HIV-1 infection, a combination of different phenotypic and genotypic tests is currently needed to monitor HIV-infected individuals. In this study, we developed a novel HIV-1 genotypic assay based on deep sequencing (DeepGen HIV) to simultaneously assess HIV-1 susceptibilities to all drugs targeting the three viral enzymes and to predict HIV-1 coreceptor tropism. Patient-derived gag-p2/NCp7/p1/p6/pol-PR/RT/IN- and env-C2V3 PCR products were sequenced using the Ion Torrent Personal Genome Machine. Reads spanning the 3′ end of the Gag, protease (PR), reverse transcriptase (RT), integrase (IN), and V3 regions were extracted, truncated, translated, and assembled for genotype and HIV-1 coreceptor tropism determination. DeepGen HIV consistently detected both minority drug-resistant viruses and non-R5 HIV-1 variants from clinical specimens with viral loads of ≥1,000 copies/ml and from B and non-B subtypes. Additional mutations associated with resistance to PR, RT, and IN inhibitors, previously undetected by standard (Sanger) population sequencing, were reliably identified at frequencies as low as 1%. DeepGen HIV results correlated with phenotypic (original Trofile, 92%; enhanced-sensitivity Trofile assay [ESTA], 80%; TROCAI, 81%; and VeriTrop, 80%) and genotypic (population sequencing/Geno2Pheno with a 10% false-positive rate [FPR], 84%) HIV-1 tropism test results. DeepGen HIV (83%) and Trofile (85%) showed similar concordances with the clinical response following an 8-day course of maraviroc monotherapy (MCT). In summary, this novel all-inclusive HIV-1 genotypic and coreceptor tropism assay, based on deep sequencing of the PR, RT, IN, and V3 regions, permits simultaneous multiplex detection of low-level drug-resistant and/or non-R5 viruses in up to 96 clinical samples. This comprehensive test, the first of its class, will be instrumental in the development of new antiretroviral drugs and, more importantly, will aid in the treatment and management of HIV-infected individuals.
INTRODUCTION
According to the Joint United Nations Programme on HIV/AIDS (UNAIDS), at the end of 2012, close to 10 million of the approximately 34 million people living with HIV had access to antiretroviral therapy, with the goal being to reach 15 million people with HIV treatment by 2015 (1). Not only has this broader access to antiretroviral drugs led to considerable reductions in morbidity and mortality (2, 3), but unfortunately, it has increased the risk of virologic failure due to the emergence (3, 4) and potential transmission (5) of drug-resistant viruses. Therefore, detecting and quantifying drug resistance, and in the case of CCR5 receptor antagonists, determining HIV-1 coreceptor tropism, have become the standard of care prior to designing new antiretroviral regimens (3, 6–8).
To date, 29 individual antiretroviral drugs from six drug classes have been approved by the U.S. Food and Drug Administration (FDA) to be used in the treatment of HIV-1 infection, including protease inhibitors (PI), nucleoside/nucleotide reverse transcriptase inhibitors (NRTI), nonnucleoside reverse transcriptase inhibitors (NNRTI), integrase inhibitors (INI), fusion inhibitors (FI), and entry inhibitors (EI). HIV-1 resistance to PI, NRTI, NNRTI, and INI can be determined using (i) indirect methods based on the detection of specific amino acid substitutions (due to underlying nucleotide mutations) in the respective coding regions previously associated with resistance to specific antiretroviral drugs (i.e., genotyping) (6, 9, 10), (ii) more direct methods that test the ability of a patient-derived virus to replicate in the presence of antiretroviral drugs in a cell-based assay (i.e., phenotyping) (11–13), or (iii) a combination of the two approaches that takes advantage of a large database in order to infer the level of HIV-1 drug resistance based on genotyping and its relationship with matched phenotypic data (14). Similarly, since treatment with CCR5 antagonists requires prior knowledge of the HIV-1 coreceptor tropism in the patient, i.e., CCR5- or CXCR4-tropic viruses (R5 and X4, respectively), dual tropism (R5/X4), or a mixture of both R5 and X4 viruses (7, 15), a multitude of phenotypic and genotypic approaches to determine HIV-1 coreceptor tropism have been developed (8, 16, 17). Phenotypic assays to determine HIV-1 drug resistance or tropism usually involve the generation of patient-derived pol (11–13) or env recombinant (18–20) viruses, respectively, to quantify their ability to infect susceptible cell lines expressing the appropriate HIV-1 receptors and coreceptors; in the case of HIV-1 tropism, these measures may also be based on the quantification of cell-to-cell fusion events (21–23). Conversely, genotypic HIV-1 tropism tests (8, 15, 16, 24–26) take advantage of the properties of specific regions in the env gene as determinants of CCR5 or CXCR4 tropism, mainly the V3 region of gp120, and their interpretations are based on a series of bioinformatics methods to infer the ability of HIV-1 to use either or both coreceptors to enter host cells (27–30).
As expected, phenotypic (experimental) and genotypic (computational) approaches to determining HIV-1 drug resistance or HIV-1 coreceptor tropism have some disadvantages, including the longer turnaround times and higher cost of the phenotypic assays or the intrinsic predictive nature of the genotypic tests. Particular emphasis has been made on the limited sensitivities of genotypic HIV-1 tropism assays to detect minor non-R5 variants (16, 31), and to a lesser extent on the ability of genotypic HIV-1 drug resistance tests to detect minority drug-resistant variants (32–34). In the case of HIV-1 drug resistance, the vast amount of information accumulated during the last 2 decades by correlating mutations with phenotypic data has led to the almost exclusive use of genotypic antiretroviral testing based on population (Sanger) sequencing to manage patients infected with HIV-1 (2, 35). In contrast, although several studies have shown significant concordance and similar predictive values (36–40), genotypic HIV-1 tropism assays based on population sequencing seem to be less sensitive and specific than phenotypic assays (8, 16, 17, 41). Thus, a cell-based assay (Trofile; Monogram Biosciences) (19, 42) is currently the standard method in the United States for determining HIV-1 coreceptor tropism, while genotypic HIV-1 tropism tests are largely used in Europe (16, 31).
To date, all current commercial genotypic HIV-1 drug resistance assays are based on population sequencing (10, 43, 44), which can detect only minority variants that are present in >20% of the viral population (44–48). However, and although this is still uncertain, drug-resistant HIV-1 minority variants (i.e., those present in as low as 1% of the viral population) have been suggested to be clinically relevant, as they have a high chance of selection under antiretroviral drug pressure conditions (49–57). For that reason, a series of ultrasensitive assays have been developed to detect drug-resistant HIV-1 minority variants, e.g., allele-specific PCR (49, 58), oligonucleotide ligation assays (33, 59), and deep (next-generation) sequencing (60–62). On the other hand, as described above, the adoption of genotypic HIV-1 tropism assays in the clinical setting has been hampered by the limited sensitivities of the population-based sequencing assays to detect minor non-R5 variants. Therefore, more sensitive genotypic HIV-1 tropism assays based on deep sequencing have been developed to detect non-R5 variants present at frequencies of <20% of the population, and these have been shown to correlate well with both phenotypic assays (36, 63–67) and the virological response to CCR5-receptor antagonists, such as maraviroc (Selzentry/Celsentri, Pfizer, NY) (36, 63, 66). Nevertheless, a combination of at least two different genotypic assays is still needed to assess the susceptibility of a patient-derived HIV-1 infection to all FDA-approved antiretroviral drugs, including CCR5 antagonists. Therefore, in this study, we have developed, characterized, and validated a novel HIV-1 genotyping assay based on deep sequencing to simplify the monitoring of patients infected with HIV-1. This all-inclusive sensitive methodology accurately provides drug resistance information for all protease, reverse transcriptase, integrase, and maturation inhibitors, as well as HIV-1 coreceptor tropism, in a single, more efficient, rapid, and affordable clinical assay.
(This research was presented in part at the International HIV & Hepatitis Virus Drug Resistance Workshop and Curative Strategies, Toronto, Ontario, Canada, 4 to 8 June 2013.)
MATERIALS AND METHODS
Viruses and plasmids.
The following viruses were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1A-92RW009, HIV-1A-93RW020, HIV-1A-92UG029, HIV-1B-92BR014, HIV-1B-92TH593, HIV-1B-US714, HIV-1B-92US727, HIV-1B-92US076, HIV-1C-92BR025, HIV-1D-94UG108, HIV-1D-92UG038, HIV-1D-93UG065, HIV-1F-93BR029, HIV-1F-93BR020, HIV-1G-RU570, HIV-1G-RU132, HIV-1AE-CMU02, HIV-1AE-CMU06, HIV-1AE-92TH021, HIV-1BF-93BR029, and HIV-2CBL-20. Other viruses were from Eric J. Arts' laboratory at Case Western Reserve University (CWRU), Cleveland, OH: HIV-1A-V115, HIV-1A-V120, HIV-1C-C18, HIV-1C-C20, HIV-1C-C21, HIV-1C-C22, HIV-1D-V89, HIV-1D-V122, HIV-1D-V126, HIV-1F-VI820, HIV-1F-V164, HIV-1F-CA16, and HIV-1F-CA20. Aliquots of additional RNA or DNA viruses were obtained from the Molecular Diagnostics or Medical Microbiology laboratories at University Hospitals Case Medical Center (UHCMC), Cleveland, OH (BK virus [BKV], cytomegalovirus [CMV], herpes simplex virus 1 and 2 [HSV-1 and HSV-2], and varicella zoster virus [VZV]) or the Division of Infectious Diseases, School of Medicine at CWRU (hepatitis B virus [HBV], hepatitis C virus [HCV], and Epstein-Barr virus [EBV]). Plasmids containing patient-derived HIV-1 gag-p2/NCp7/p1/p6/pol-PR/RT/IN-coding sequences from multidrug-resistant viruses, i.e., 08-180 and 08-194, have been described (68), along with a pNL4-3 plasmid containing the env gene from the R5 HIV-1YU2 virus (69). Five plasmid mixtures containing drug resistance mutations in the HIV-1 pol gene, i.e., K65R (5%) plus wild type (95%), K103N (5%) plus wild type (95%), K101E (5%) plus E138K (5%) plus wild type (90%), K101E plus E138K (10%) plus wild type (90%), and M184V (RT) plus E92Q (IN) (10%) plus wild type (90%), were obtained from Gilead Sciences, Inc. (Foster City, CA).
Clinical samples.
Blood plasma samples for the characterization and verification of the novel HIV-1 genotypic and coreceptor tropism assay were obtained during routine patient monitoring from a well-characterized cohort of HIV-infected individuals at the AIDS Clinical Trials Unit (ACTU) at CWRU/UHCMC, with the understanding and written consent of each participant. RNA specimens, derived from plasma samples collected from HIV-infected individuals enrolled in the (i) maraviroc expanded-access program in Europe or (ii) the ALLEGRO trial, were obtained from the Hospital Carlos III (Madrid, Spain) (40). Written informed consent was obtained from the patients before participation in the study, as previously described (40, 70). HIV-1 coreceptor tropism was determined at baseline using two phenotypic assays, i.e., the original version of the Trofile (19) and VeriTrop (23), and by population sequencing analyzed with Geno2Pheno (27), with false-positive rates (FPR) (predicted frequency of classifying an R5 sequence as a non-R5 virus) based on optimized cutoffs associated with the analysis of clinical data from MOTIVATE (2.5% and 5.75%) (63). Finally, plasma samples were obtained from HIV-infected individuals at the Infectious Diseases Unit Virgen del Rocio University Hospital (Seville, Spain) participating in a study to evaluate the use of an 8-day maraviroc monotherapy clinical test (MCT) (71, 72). Patients provided written informed consent, and the ethics committee of the hospital approved the study (72). HIV-1 coreceptor tropism in these samples was determined at baseline using two different phenotypic assays, i.e., the enhanced-sensitivity Trofile assay (ESTA) (42) and TROCAI (73), and by population sequencing analyzed with Geno2Pheno (27) with an FPR of 10%, according to the recommendations from the European Consensus Group on the clinical management of HIV-1 tropism testing, as described on the Geno2Pheno website (http://coreceptor.bioinf.mpi-inf.mpg.de/index.php).
Reverse transcription-PCR amplification of gag-p2/NCp7/p1/p6/pol-PR/RT/IN- and env-C2V3-coding regions.
Plasma viral RNA was purified from pelleted virus particles by centrifuging 1 ml of plasma at 18,000 × g for 60 min at 4°C, removing 860 μl of cell-free supernatant, resuspending the pellet in the remaining 140 μl, and finally extracting viral RNA using the QIAamp viral RNA minikit (Qiagen; Valencia, CA). Viral RNA was reverse transcribed using AccuScript high-fidelity reverse transcriptase (Stratagene Agilent; Santa Clara, CA) and the corresponding antisense external primers in a 20-μl reaction mixture containing 1 mM deoxynucleoside triphosphates (dNTPs), 10 mM dithiothreitol (DTT), and 10 units of RNase inhibitor. The HIV-1 genomic region encoding the Gag proteins p2, p7, p1, and p6 and the protease, reverse transcriptase, and integrase enzymes was amplified as two overlapping fragments (1,657 nucleotides [nt] and 2,002 nt corresponding to the p2-5′half RT and 3′half RT-INT, respectively) using a series of external and nested primers with defined cycling conditions (13). External PCRs were carried out in a 50-μl mixture containing 0.2 mM dNTPs, 1 mM MgCl2, and 2.5 units of Pfu Turbo DNA polymerase (Stratagene). Nested PCRs were carried out in a 50-μl mixture containing 0.2 mM dNTPs, 0.3 units of Pfu Turbo DNA polymerase, and 1.9 units of Taq polymerase (Denville Scientific; Metuchen, NJ). A fragment corresponding to the C2V3 region (480 nt) of the surface glycoprotein (gp120) in the envelope gene was amplified using a series of external and nested primers with defined cycling conditions, as previously described (74).
Population (Sanger) sequencing analysis.
PCR products corresponding to the gag-p2/NCp7/p1/p6/pol-PR/RT/IN- and env-C2V3-coding regions of HIV-1 were purified with the QIAquick PCR purification kit (Qiagen) and sequenced (Sanger, population, or global sequence) using AP Biotech DYEnamic ET Terminator cycle with Thermosequenase II (Davis Sequencing LLC, Davis, CA). Nucleotide sequences were analyzed using DNAStar Lasergene Software Suite version 10.0.1 (Madison, WI).
Deep sequencing of gag-p2/NCp7/p1/p6/pol-PR/RT/IN- and env-C2V3-coding regions.
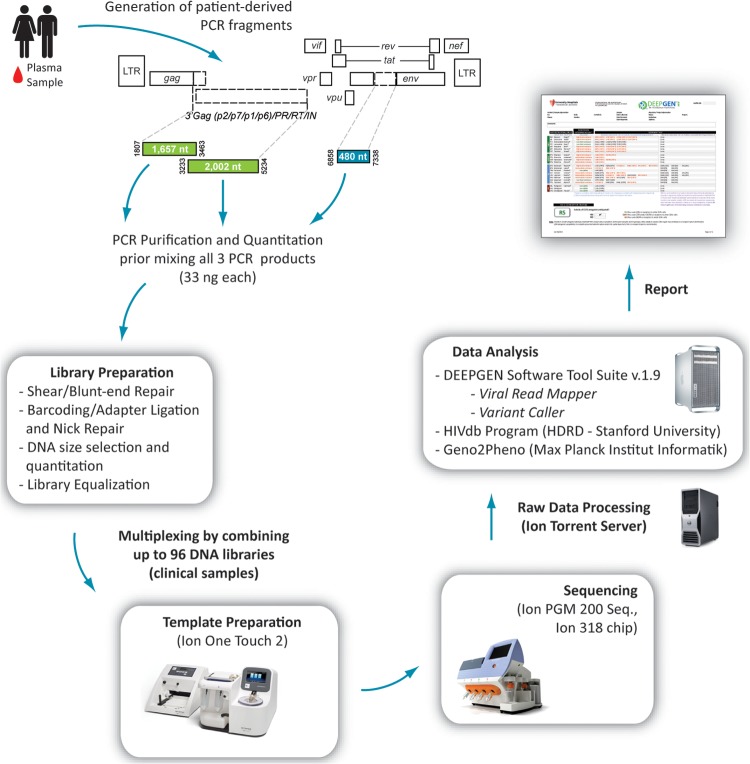
The three PCR products corresponding to the gag-p2/NCp7/p1/p6/pol-PR/RT/IN- (1,657-nt and 2,002-nt fragments) and env-C2V3- (480-nt fragment) coding regions of HIV-1 were purified (Agencourt AMPure XP; Beckman Coulter) and quantified (2100 Bioanalyzer DNA 7500; Agilent Technologies) prior to using the Ion Xpress fragment library kit (Life Technologies, Carlsbad CA) to construct a multiplexed library for shotgun sequencing on the Ion Personal Genome Machine (PGM) (Life Technologies) (Fig. 1). Briefly, a mixture of all three purified DNA amplicons (33 ng each) was randomly fragmented and the blunt ends repaired using the Ion Shear Plus reagent (Life Technologies), followed by DNA purification (Agencourt AMPure XP). The P1 adapter (5′-CCA CTA CGC CTC CGC TTT CCT CTC TAT GGG CAG TCG GTG AT, 5′-ATC ACC GAC TGC CCA TAG AGA GGA AAG CGG AGG CGT AGT GG*T*T [asterisks indicate phosphorothioate bonds]) and one of 96 barcodes were ligated to the repaired fragment ends prior to DNA purification (Agencourt AMPure XP). DNA fragments were then selected by size (i.e., 300 bp; Pippin Prep; Life Technologies) and each bar-coded library, i.e., a mixture of all three amplicons per sample, was purified (Agencourt AMPure XP) and normalized using the Ion Library Equalizer kit (Life Technologies). All bar-coded DNA libraries corresponding to patient-derived amplicons plus the HIV-1NL4-3 control were pooled in equimolar concentrations and the templates prepared and enriched for sequencing on the Ion Sphere particles (ISPs) using the Ion OneTouch 200 template kit version 2 (Life Technologies) in the Ion OneTouch 2 system (Life Technologies). Templated ISPs were quantified (Qubit 2.0; Life Technologies) and loaded into an Ion 318 Chip (Life Technologies) to be sequenced on the Ion PGM using the Ion PGM sequencing 200 kit version 2 (Life Technologies). Following a 4-h-and-20-min sequencing run, signal processing and base calling were performed with the Torrent Analysis Suite version 3.4.2.
FIG 1.
Overview of the protocol for the novel HIV-1 genotyping and coreceptor tropism assay (DeepGen HIV). Three PCR products corresponding to the gag-p2/NCp7/p1/p6/pol-PR/RT/IN- (1,657 bp and 2,002 bp) and env-C2V3- (480 bp) coding regions of HIV-1 were used to construct a multiplexed library for shotgun sequencing on the Ion PGM. Signal processing and base calling were performed with the Torrent analysis suite version 3.4.2 and sequences analyzed using DEEPGEN Software Tool Suite. The HIVdb Program Genotypic Resistance Interpretation Algorithm from the Stanford University HIV Drug Resistance Database (see http://hivdb.stanford.edu) and Geno2Pheno (27) were used to infer the levels of susceptibility to PI, RTI, and INI and for HIV-1 coreceptor tropism determination, respectively.
Read mapping, variant calling, and phylogenetic analysis.
As part of the novel HIV-1 genotypic and coreceptor tropism assay, we developed the DeepGen Software Tool Suite for the processing of HIV-1 deep sequencing data and HIV-1 drug resistance determination. DeepGen uses two main tools: Viral Read Mapper and Variant Caller.
(i) Viral Read Mapper.
To minimize the amount of data loss during mapping due to the high HIV-1 sequence variability and to allow for interpatient indel variation across the gag-p2/NCp7/p1/p6/pol-PR/RT/IN- and env-C2V3-coding regions, sample-specific reference sequences were constructed for each of these two genomic regions, i.e., positions 1807 to 5096 and 6900 to 7400 in the HXB2 reference strain (GenBank accession no. K03455), respectively. The mapping of reads from each sample/region occurred in three stages. First, a guide template for mapping was selected from the Los Alamos HIV Sequence Database (see http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html) by comparing 100 randomly selected reads to the corresponding region within all full-length sequences present within the HIV Sequence Database. This comparison was performed using a k-mer approach that rapidly identifies regions of similarity between any two sequences to select a guide sequence for mapping with minimal divergence from the read data, as such divergence is the primary cause of biased data loss (75). Following the selection of a guide sequence, the reads were mapped and aligned using the mapping algorithm described previously (76). During mapping, site indexes in relation to HXB2 were also maintained. Next, to reduce the diversity between the reads and reference sequence, a consensus was generated across each site of the guide sequence, and the reads were remapped to this final consensus template. Reads spanning the 3′ end of Gag, PR, RT, and INT were then translated and assembled for genotyping.
(ii) Variant Caller.
Variant calling, which is the identification and calculation of the frequency of each amino acid present in each genomic position, was calculated using a table generated by the Viral Read Mapper as input, which includes the nucleotide frequencies at each position relative to the reference sequence and numbering relative to the HIV-1B-HXB2 reference strain. The coverage, indel, codon, and residue frequencies at each position were also listed. Variant Caller summarized the results in a graphical interface, with a particular focus on the sites of known drug resistance based on the latest edition of the International AIDS Society-USA (IAS-USA) HIV drug resistance mutations list (77). A list of the amino acids at these positions and their frequencies was exported as a tabulated text file and used with the HIVdb Program Genotypic Resistance Interpretation Algorithm from the Stanford University HIV Drug Resistance Database (see http://hivdb.stanford.edu) to infer the levels of susceptibility to protease, reverse transcriptase, and integrase inhibitors.
In addition, for each data set, reads spanning amino acid positions (i) 50 to 85 in the protease (HXB2 2400 to 2508), (ii) 180 to 215 in the RT (HXB2 3087 to 3195), (iii) 130 to 165 in the integrase (HXB2 4617 to 4725), and (iv) 1 to 35 in the V3 region (HXB2 7110 to 7217) were extracted, truncated, and translated for phylogenetic analysis and HIV-1 coreceptor tropism prediction as described below. Within each data set, only one representative of any identical variant was maintained, but the overall frequency was stored. All variants with a frequency of >1 within the population were aligned using ClustalW (78) and the phylogeny reconstructed using the neighbor-joining statistical method as implemented within MEGA 5.05 (79). In this study, a minority variant was defined as a variation detected at >1% (based on the intrinsic error rate of the system as described below) and <20% of the virus population, corresponding to those mutations that cannot be determined using population sequencing (44–48).
Genotypic HIV-1 coreceptor tropism determination.
HIV-1 coreceptor tropism was predicted from population and deep sequencing V3 sequences using Geno2Pheno (27). Regarding the population V3 sequences, nucleotide mixtures were considered when the second highest peak in the electropherogram was >25% and the nucleotide mixtures translated into all possible permutations. Geno2Pheno with FPR of 2.5% and 5.75%, based on optimized cutoffs associated with the analysis of clinical data from MOTIVATE (2.5% and 5.75%) (63), or an FPR of 10%, according to the recommendations from the European Consensus Group on the clinical management of HIV-1 tropism testing as described on the Geno2Pheno website (http://coreceptor.bioinf.mpi-inf.mpg.de/index.php), was used for the clinical samples obtained from the Madrid and Seville cohorts, respectively. In the case of deep sequencing V3 sequences, reads spanning amino acid positions 1 to 35 in the V3 region (HXB2 7110 to 7217) were extracted and truncated for HIV-1 coreceptor tropism determination using Geno2Pheno (27) with an FPR of 3.5%, based on optimized cutoffs for determining HIV-1 coreceptor usage, as previously described (36, 63, 80). Deep sequencing V3 sequences usually spanned 105 nucleotides (35 amino acids), with some minor discrepancies associated with natural HIV-1 variation (81, 82), which led to V3 sequences with an open reading frame of 96, 99, 102, 108, or 111 nucleotides, all starting and ending with a cysteine codon, i.e., TG(T/C). V3 reads with stop codons (TGA, TAA, or TAG) and/or where the nucleotide length was not a multiple of 3 (e.g., 101, 103, 104, and 106), mostly associated with naturally or methodology (PCR or sequencing)-induced insertions and/or deletions, were not included in the analysis. Deep sequencing of the V3 region was considered unsuccessful if reads from the majority variants had to be omitted from the analysis. Finally, blood plasma samples were classified as containing non-R5 viruses if ≥2% of the individual sequences, as determined by deep sequencing, were predicted to be non-R5 (36, 63).
Statistical analyses.
The descriptive results are expressed as median values, interquartile ranges, standard deviations, and confidence intervals. Pearson's correlation coefficient was used to determine the strength of the associations between categorical variables. A paired t test was used to compare the number of drug resistance mutations detected by population and deep sequencing in the same sample. All differences with a P value of <0.05 were considered statistically significant. The kappa coefficient, which assesses a chance-adjusted measure of the agreement between any number of categories, was calculated using ComKappa3 version 3.0.1 (83) to quantify the concordance among the different HIV-1 tropism determinations. All statistical analyses were performed using GraphPad Prism version 6.0b (GraphPad Software, La Jolla, CA), unless otherwise specified. The gag-p2/NCp7/p1/p6/pol-PR/RT/IN and/or env-V3 nucleotide sequences obtained in this study by deep sequencing have been submitted to the Los Alamos National Laboratory HIV-DB Next-Generation Sequence Archive.
RESULTS
Characterization of the reverse transcription-PCR amplification step.
As described in Materials and Methods and as shown in Fig. 1, the novel HIV-1 genotyping and coreceptor tropism assay requires reverse transcription-PCR amplification of three amplicons covering the HIV-1 gag-p2/NCp7/p1/p6/pol-PR/RT/IN- and env-C2V3-coding regions. The characterization of the two overlapping PCR products (1,657 nt and 2,002 nt) comprising the subgenomic HIV-1 region spanning the Gag proteins p2, p7, p1, and p6 and the protease, reverse transcriptase, and integrase coding regions was detailed previously as part of a study describing ViralARTS HIV, an HIV-1 phenotyping assay (13). Thus, here, we focused in the characterization of the 480-nt fragment of the HIV-1 env gene, corresponding to the C2V3 region of the surface glycoprotein (gp120). The sensitivity of the reverse transcription-PCR amplification step was tested by analyzing 79 plasma samples obtained from the ACTU (Cleveland, OH). Blood samples from HIV-infected individuals with plasma viral loads ranging from 1,000 to >10,000 copies of viral RNA/ml were used to PCR amplify the C2V3 fragment. Similar to the results observed with the two overlapping fragments spanning the gag-p2/NCp7/p1/p6/pol-PR/RT/IN-coding region (13), reverse transcription-PCR products of the correct size were consistently obtained (92% [73/79]) in plasma samples with ≥1,000 copies/ml of HIV RNA (Table 1).
TABLE 1.
Sensitivity of reverse transcription-PCR amplification of the C2V3 region of the HIV-1 env gene (480 nt)
| Viral load (copies/ml) | % positive samples by RT-PCR (no. of positive samples/total no. of samples tested)a |
|---|---|
| 1,001–5,000 | 95 (19/20) |
| 5,001–10,000 | 85 (17/20) |
| >10,000 | 95 (37/39) |
Reverse transcription-PCR amplification of patient-derived env fragments was performed with plasma samples (n = 79) from HIV-infected individuals with viral loads ranging from 1,000 to >10,000 copies of viral RNA/ml, as described in Materials and Methods.
Highly reproducible success in reverse transcription-PCR amplification of the specific HIV-1 gag-p2/NCp7/p1/p6/pol-PR/RT/IN- and env-C2V3 products was obtained when testing 15 plasma samples with different viral loads. The details of these tests using two different operators with different lots of critical reagents and over a 7-day period are described in Weber et al. (13) for the gag-p2/NCp7/p1/p6/pol-PR/RT/IN fragments, and in Table S1 in the supplemental material for the env-C2V3 fragment. The specificities of the reverse transcription-PCR primers and reactions for the env-C2V3 fragment were analyzed using nucleic acids from a series of RNA and DNA viruses (i.e., BKV, CMV, HSV-1, HSV-2, VZV, HBV, HCV, and EBV). As expected, no cross-reactivity was observed with any of these viruses, as all reverse transcription-PCRs failed to generate any detectable amplicons (see Fig. S1 in the supplemental material). Similar results were obtained for the gag-p2/NCp7/p1/p6/pol-PR/RT/IN fragments, as previously described (13).
Finally, although most of the HIV-1 genotyping and coreceptor tropism determinations are performed in North America, Europe, and Australia, where subtype HIV-1 strains are predominant (see http://www.who.int/hiv/pub/global_report2010/en/index.html), it was important to test the ability of the assay to work with more worldwide prevalent non-B HIV-1 variants. For that, the env-C2V3 fragment was reverse transcription-PCR amplified from 33 diverse HIV-1 isolates, including five subtype A (HIV-1A-92RW009, HIV-1A-93RW020, HIV-1A-92UG029, HIV-1A-V115, and HIV-1A-V120,), five subtype B (HIV-1B-92BR014, HIV-1B-92TH593, HIV-1B-US714, HIV-1B-92US727, and HIV-1B-92US076), five subtype C (HIV-1C-92BR025, HIV-1C-C18, HIV-1C-C20, HIV-1C-C21, and HIV-1C-C22,), six subtype D (HIV-1D-94UG108, HIV-1D-92UG038, HIV-1D-93UG065, HIV-1D-V89, HIV-1D-V122, and HIV-1D-V126,), six subtype F (HIV-1F-93BR029, HIV-1F-93BR020, HIV-1F-VI820, HIV-1F-V164, HIV-1F-CA16, and HIV-1F-CA20), two subtype G (HIV-1G-RU570 and HIV-1G-RU132,), and four circulating recombinant forms (HIV-1AE-CMU02, HIV-1AE-CMU06, HIV-1AE-92TH021, and HIV-1BF-93BR029). Amplicons of the correct sizes were obtained for the env-C2V3 (see Table S2 in the supplemental material) and gag-p2/NCp7/p1/p6/pol-PR/RT/IN fragments (13) from all HIV-1 group M isolates analyzed, while negative or inconclusive results were obtained with the HIV-2CBL-20 strain (data not shown).
Estimation of the intrinsic error rate of the assay.
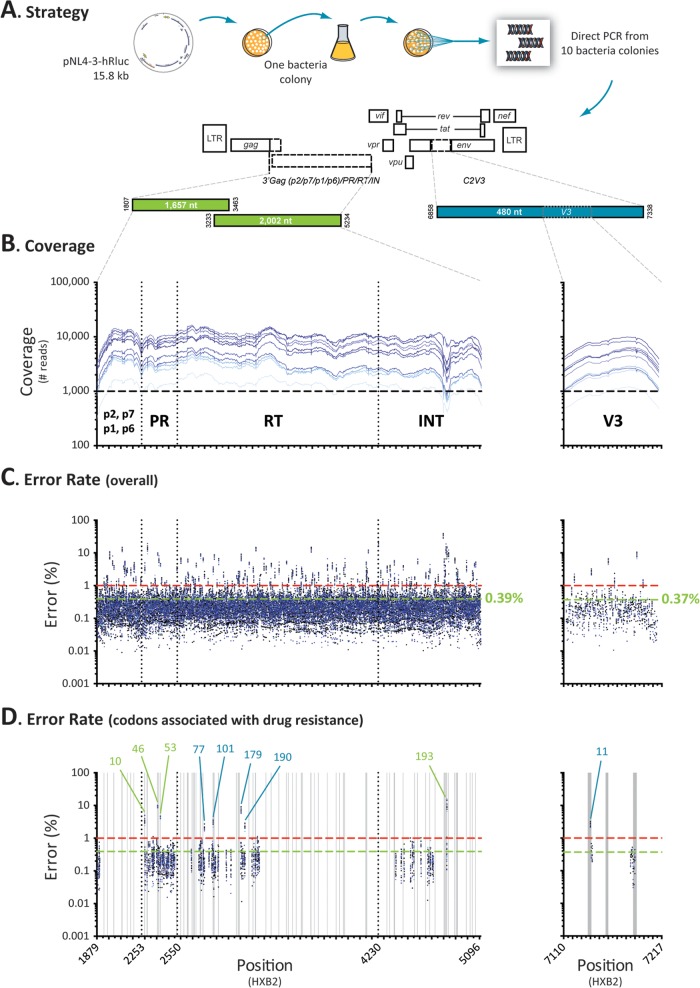
Point mutations, insertions, and deletions (indels) can be introduced in the PCR amplification and sequencing steps of any deep-sequencing-based assay (84). Therefore, it was important to calculate the intrinsic (combined) error rate of our novel HIV-1 genotyping and coreceptor tropism assay, since this value might affect the practical limit of detection of the assay. For that, the pNL4-3-hRluc plasmid containing the entire genome of the wild-type HIV-1NL4-3 strain (69) was transformed into Electrocomp TOP10 bacteria (Invitrogen). One bacterial colony was grown overnight in 10 ml of bacterial culture, and the plasmid DNA was purified and transformed again into bacteria. Ten individual and theoretically identical bacterial colonies were used for direct PCR amplification of the gag-p2/NCp7/p1/p6/pol-PR/RT/IN and env-C2V3 fragments (Fig. 2A), and these were sequenced using the same protocol utilized with the clinical samples. The quality of the DNA sequences was analyzed and the reads were filtered in the Ion Torrent server using a Phred quality score of 20 (Q20), which provides a base call accuracy of 99% (i.e., a 1 in 100 probability of an incorrect base call). The average coverages (sequencing depth) per nucleotide position for the 10 clones were 5,750 (range, 681 to 15,614) and 3,797 (range, 942 to 9,981) for the gag-p2/NCp7/p1/p6/pol-PR/RT/IN and env-V3 regions, respectively (Fig. 2B). For each individual NL4-3 clone, the reads were independently mapped to the pNL4-3-hRluc reference sequence (13, 69), and all point mutation and indel information in relation to the reference was analyzed using Segminator II (76).
FIG 2.
Error rate determination. (A) The pNL4-3-hRluc plasmid containing the entire genome of the wild-type HIV-1NL4-3 strain (69) was transformed into bacteria and the gag-p2/NCp7/p1/p6/pol-PR/RT/IN and env-C2V3 fragments were PCR amplified and deeply sequenced from 10 individual colonies. The reads from each individual NL4-3 clone were independently mapped to the pNL4-3-hRluc reference sequence (13, 69) using Segminator II (76). (B) Coverage, i.e., number of reads per nucleotide position, for the 10 NL4-3 clones. (C) Overall (point mutation, insertions, and deletions) error rate per nucleotide position calculated using a Phred quality score of 20. The mean error rates for the gag-p2/NCp7/p1/p6/pol-PR/RT/IN and env-V3 regions are indicated in green, while the minimum thresholds to detect mutations in the minority HIV-1 variants with this novel assay (1%) are indicated by dashed red lines. (D) Overall error rate in positions associated with drug resistance. Only codon changes with error rates >1% are indicated, i.e., L10, M46, and F53 in the protease region, F77, K101, V179, and G190 in the RT region, G193 in the integrase region, and amino acid 11 in the V3 region. Homopolymeric regions, defined as four or more identical consecutive nucleotides, are indicated as vertical gray bars.
Although all 10 NL4-3 clones were expected to have no mutations (i.e., point mutation and/or indels) relative to the pNL4-3-hRluc reference sequence, a number of errors were observed throughout the p2/NCp7/p1/p6/pol-PR/RT/IN and V3 regions, with error rates ranging from 0% to 29% (mean, 0.39%) and 0% to 9.5% (mean, 0.37%), respectively (Fig. 2C). The average error frequencies due to point mutations were 0.17% (range, 0% to 2.5%) and 0.12% (0% to 0.3%) for the p2/NCp7/p1/p6/pol-PR/RT/IN and V3 regions, respectively, whereas the average error rates associated with indels were 0.22% (range, 0% to 28%) and 0.25% (0% to 9.2%), respectively. Most of the positions with a total (point mutation plus indels) error rate of >1% corresponded to the last nucleotide of a homopolymeric region, defined as four or more identical consecutive nucleotides (data not shown). Some of these nucleotide positions corresponded to codons that have been associated with resistance to antiretroviral drugs, e.g., L10 in the protease (3.5%), K101 in the RT (3%), and G193 in the integrase (10.5%), or with coreceptor tropism, e.g., position 11 in the V3 region (2.6%) (Fig. 2D, Table 2). Interestingly, most of the errors in these (homopolymeric) positions corresponded to indels, with a limited number of point mutation errors, e.g., L10 (3.3% versus 0.22%), K101 (2.7% versus 0.24%), G193 (9.6% versus 0.89%), and position 11 in the V3 (2.3% versus 0.25%), respectively (Table 2). Therefore, considering that (i) the overall error rates for the gag-p2/NCp7/p1/p6/pol-PR/RT/IN and env-V3 regions were 0.39% and 0.37%, respectively, (ii) the point mutation error rates were <1% for all the codons associated with drug resistance, and (iii) the Variant Caller in the DeepGen Software Tool Suite identifies and filters out the indels, it was reasonable to define a frequency of 1% as the minimum threshold to detect mutations in minority HIV-1 variants with this novel assay.
TABLE 2.
Error rate distribution
| Genomic regiona | Codonb | Sequencec | Error rate (mean ± SD)d |
||
|---|---|---|---|---|---|
| Total | Point mutations | Indels | |||
| gag-pol | NAe | NA | 0.0039 ± 0.0001 | 0.0017 ± 0.0001 | 0.0022 ± 0.0001 |
| env-V3 | NA | NA | 0.0037 ± 0.0002 | 0.0012 ± 0.0001 | 0.0025 ± 0.0002 |
| Protease | L10 | cgacccCTCgtc | 0.0353 ± 0.0086 | 0.0022 ± 0.0002 | 0.0331 ± 0.0051 |
| M46 | accaaaaATGata | 0.0920 ± 0.0056 | 0.0013 ± 0.0001 | 0.0907 ± 0.0043 | |
| F53 | aggtTTTatc | 0.0436 ± 0.0034 | 0.0021 ± 0.0002 | 0.0415 ± 0.0022 | |
| RT | F77 | agatTTCag | 0.0205 ± 0.0028 | 0.0018 ± 0.0001 | 0.0187 ± 0.0011 |
| K101 | gttaAAAcag | 0.0295 ± 0.0044 | 0.0024 ± 0.0002 | 0.0271 ± 0.0032 | |
| V179 | ataGTCatc | 0.0756 ± 0.0122 | 0.0036 ± 0.0002 | 0.0720 ± 0.0091 | |
| G190 | gtaGGAtct | 0.0221 ± 0.0036 | 0.0012 ± 0.0001 | 0.0209 ± 0.0021 | |
| Integrase | G193 | attgggGGGtac | 0.1048 ± 0.0277 | 0.0089 ± 0.0003 | 0.0959 ± 0.0145 |
| V3 | R11 | aagaaaaAGTatc | 0.0256 ± 0.0037 | 0.0025 ± 0.0002 | 0.0231 ± 0.0020 |
HIV-1 genomic region analyzed. gag-pol and env-V3 correspond to the gag-p2/NCp7/p1/p6/pol-PR/RT/IN and V3 region of the gp120 in the envelope gene, respectively.
Codons associated with resistance to antiretroviral drugs were determined to have total error rate values of >1%.
Nucleotide sequence based on the population sequencing of the HIV-1NL4-3 clone (13), which around these codons was identical to the HIV-1HXB2 reference sequence (GenBank accession no. K03455). The respective codons are indicated in uppercase letters, while the nucleotide positions associated with the elevated error rate (>1%) are shown in bold type. Position numbering is relative to the HIV-1HXB2 reference sequence.
Number of combined PCR and sequencing errors, i.e., point mutations, insertions, and deletions (indels), per read calculated using Segminator II (76). Mean and standard deviation values indicated were obtained from 10 independent sequences.
NA, not applicable.
Performance of the novel deep-sequencing-based HIV-1 genotypic and coreceptor tropism assay.
The measure of success for any deep-sequencing-based assay depends on its ability to generate the maximum number of reads per sequencing run (individual sequences), which then allows for the detection of minority variants within the HIV-1 population. This inherent quality is the sum of a series of metrics, including, but not limited to (i) the number of samples multiplexed and sequenced per run, (ii) chip-loading efficiency, (iii) the total number of quality reads, (iv) mean read length, and (v) the sequencing coverage at each nucleotide position. Most of the deep sequencing runs described in this study involved multiplexing up to 96 individual samples per sequencing reaction, a number that ensured a minimum coverage of 1,000 per nucleotide position sequenced that was required to secure the detection of a minor variant present in at least 1% of the population (85). Efficient loading of Ion Sphere particles into the Ion 318 Chip proved to be user dependent (mean, 72%; range, 60% to 84%). The total number of quality reads was proportional to the chip-loading efficiency (empty wells), with other parameters, such as enrichment (no template), polyclonality (ISPs with excess DNA library), test fragments, and primer dimers, potentially affecting the final number of total reads in this study (mean, 3,827,323 reads; range, 3,051,463 to 4,936,375 reads). We used the Ion PGM sequencing 200 kit version 2 in all sequencing runs, generating an average read length of 147 bp (range, 119 bp to 178 bp). As expected, the average coverage varied with each sequencing run, correlating mostly with the number of multiplexed samples per sequencing reaction, e.g., 20 samples (mean, 9,008; range, 3,776 to 15,458 in the gag-p2/NCp7/p1/p6/pol-PR/RT/IN region and mean, 6,494; range, 2,322 to 8,599 in the env-V3 region) or 96 samples (mean, 4,485; range, 1,612 to 7,274 in the gag-p2/NCp7/p1/p6/pol-PR/RT/IN region and mean, 1,017; range, 966 to 1,070 in the env-V3 region).
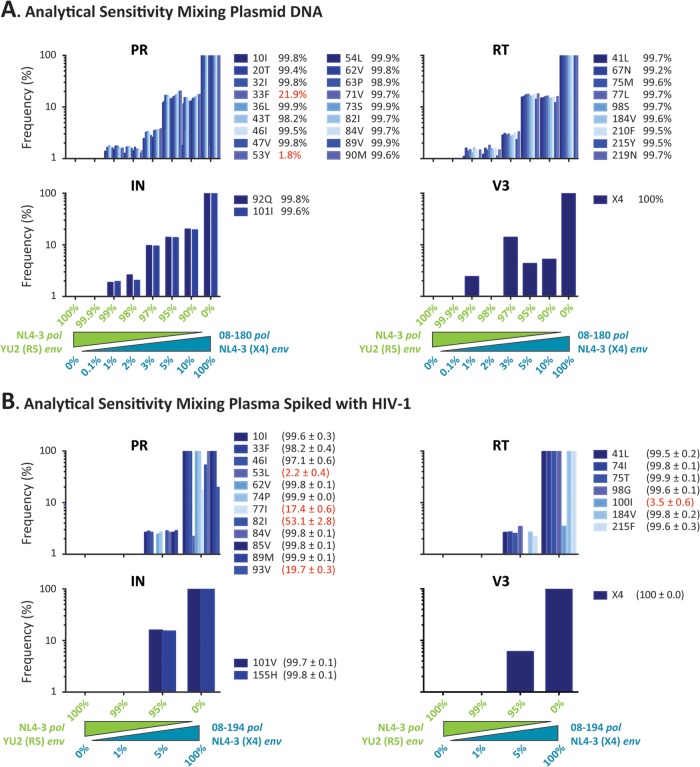
As described above, we calculated the error rate of the HIV-1 genotyping and coreceptor tropism assay to be <1%, and we incorporated this error-defined cutoff into the evaluation of the analytical sensitivity and the limit of detection for minority HIV-1 variants. For that, we evaluated extensively the analytical sensitivity of the test to detect and quantify drug resistance mutations (gag-p2/NCp7/p1/p6/pol-PR/RT/IN) and non-R5 variants (env-V3) within mixtures of viral populations. First, we sequenced five plasmid mixtures that contained one or two drug resistance mutations in the RT- and/or IN-coding regions at a frequency of 5% or 10% (i.e., mixture of plasmids containing the respective mutations with a plasmid comprising the wild-type HIV-1HXB2 sequence). As shown in Fig. S2 in the supplemental material, all mutations were detected and quantified at the expected proportions, including those at a frequency of 5% of the total population. Next, in order to quantify more accurately the analytical sensitivity of the assay, we mixed DNA from a plasmid containing a patient-derived multidrug-resistant gag-p2/NCp7/p1/p6/pol-PR/RT/IN fragment in the X4 HIV-1NL4-3 backbone (08-180) (68) with DNA from a plasmid containing the genome of the wild-type HIV-1NL4-3 virus carrying the env gene from the R5 HIV-1YU2 virus (69). Plasmid DNA was quantified and dilutions were used to prepare eight mixtures containing the X4 multidrug-resistant 08-180 plasmid at 0%, 0.1%, 1%, 2%, 3%, 5%, 10%, and 100% concentrations at a final concentration of 0.1 ng/ml. This total plasmid concentration (0.1 ng/ml or 100,000 fg/ml) theoretically allowed the detection of 100 fg of the plasmid when diluted to 0.1% of the population using nested PCR (86). Plasmid 08-180pol/NL43-(X4)env was generated by the yeast cloning method, which allows for a better representation of the in vivo HIV-1 quasispecies (13). It contained numerous drug resistance mutations in the protease, RT, and integrase, most of them as majority members of the quasispecies (>99% of the population); however, two amino acid substitutions in the protease were present as minority variants, i.e., L33F at 21.9% and F53Y at 1.7%. Interestingly, most drug resistance mutations were detected in the plasmid mixtures containing approximately 1% of the 08-180pol/NL43-(X4) plasmid, with the exception of substitution T215Y in the RT, which was identified at 0.95% (Fig. 3A; see also Fig. S3 in the supplemental material). As expected, the detection of minority mutations leading to two amino acid substitutions (L33F and F53Y) faded quickly and in proportion to their frequencies in the original population. Similar results were observed during the detection of X4 (NL4-3) V3 sequences, which were detected in a plasmid mixture containing approximately 1% of the 08-180pol/NL43-(X4) env plasmid (Fig. 3A; see also Fig. S3). Unfortunately, and most likely due to the need to remove V3 sequences with odd open reading frames (as described in Materials and Methods), the quantification of X4 sequences in the mixtures containing the 08-180pol/NL43-(X4) env plasmid at 2% and 3% of the population failed or was not accurate, respectively (Fig. 3A; see also Fig. S3).
FIG 3.
The ability of the novel HIV-1 genotyping and coreceptor tropism assay to detect drug resistance mutations (in the gag-p2/NCp7/p1/p6/pol-PR/RT/IN fragment) and non-R5 variants (in the env-V3 region) within mixtures of viral populations was used to calculate the analytical sensitivity of the test. (A) A gag-p2/NCp7/p1/p6/pol-PR/RT/IN PCR product was obtained from an antiretroviral-experienced patient (08-180) and used to construct p2-INT recombinant viruses based on the yeast cloning method to maintain the HIV-1 quasispecies (13, 68). This plasmid preparation contained the pol gene from the patient and the env gene from the CXCR4-tropic HIV-1NL4-3 strain, i.e., 08-180 pol/NL43-(X4)env. Plasmid NL4-3 pol/YU2(R5)env contains the genome of the wild-type HIV-1NL4-3 virus carrying the env gene from the R5 HIV-1YU2 virus (69). A series of plasmid mixtures were created by mixing 0.1%, 1%, 2%, 3%, 5%, and 10% of the 08-180pol/NL43-(X4)env plasmid with the corresponding amount of the NL4-3pol/YU2(R5)env plasmid at a final concentration of 0.1 ng/ml. DNA from the entire plasmid mixtures, together with the two individual plasmids as controls (100%), was purified and deeply sequenced as described in Materials and Methods. (B) Plasma containing a patient-derived multidrug-resistant gag-p2/NCp7/p1/p6/pol-PR/RT/IN recombinant virus constructed using the X4 HIV-1NL4-3 backbone (08-194) and a wild-type HIV-1NL4-3 virus carrying the env gene from the R5 HIV-1YU2 virus was mixed at 0%, 1%, 5%, and 100% of the 08-194 virus at a final concentration of 100,000 copies/ml. The frequency of each mutation detected in the original population at ≥1% of the population, the threshold calculated based on the intrinsic error rate of the assay, is indicated (mean ± standard deviation from quadruplicate experiments in the case of the experiment using mixtures of HIV-infected plasma). Amino acid substitutions detected at a frequency of <90% of the population are indicated in red.
Finally, in order to mimic the first steps of the assay (i.e., RNA purification and reverse transcription-PCR) under controlled conditions, HIV-1-seronegative plasma samples were spiked with two viruses, the first being a patient-derived multidrug-resistant gag-p2/NCp7/p1/p6/pol-PR/RT/IN recombinant virus constructed using the X4 HIV-1NL4-3 backbone (08-194) (68) and the second being a wild-type HIV-1NL4-3 virus carrying the env gene from the R5 HIV-1YU2 virus (69). The plasma HIV-1 RNA (viral) load was determined (Cobas AmpliPrep/Cobas TaqMan HIV-1 test version 2.0; Roche) and dilutions were used to prepare four mixtures containing the X4 multidrug-resistant 08-194 virus at 0%, 1%, 5%, and 100% concentrations in a final viral load of 100,000 copies/ml. This was the average viral load in plasma samples obtained from highly antiretroviral-experienced patients, usually carrying multidrug-resistant viruses, from recent previous studies in our laboratory (13, 68). The viral RNA was purified, reverse transcription-PCR amplified, bar coded in quadruplicate, and deeply sequenced as described in Materials and Methods. As expected, all drug resistance mutations from 08-194 were detected when the mixture contained 100% of this virus (Fig. 3B). Interestingly, a few amino acid substitutions were identified by deep sequencing that were not detected in the original study using Sanger sequencing (68), e.g., F53L (2.2%), V77I (17.4%), and I93V (19.7%) in the protease and L100I (3.5%) in the RT coding regions (Fig. 3B). All mutations present at a frequency of >50% in the original 08-194 virus were detected in the 5%:95% (08-194-to-wild type) mixture; however, none of these mutations were identified when the mixture included 1% 08-194 virus (Fig. 3B). Similar results were observed in the env gene, i.e., X4 sequences corresponding to the V3 region of the HIV-1NL4-3 were detected when present at a frequency of 100% and 5% in the viral mixture (Fig. 3B).
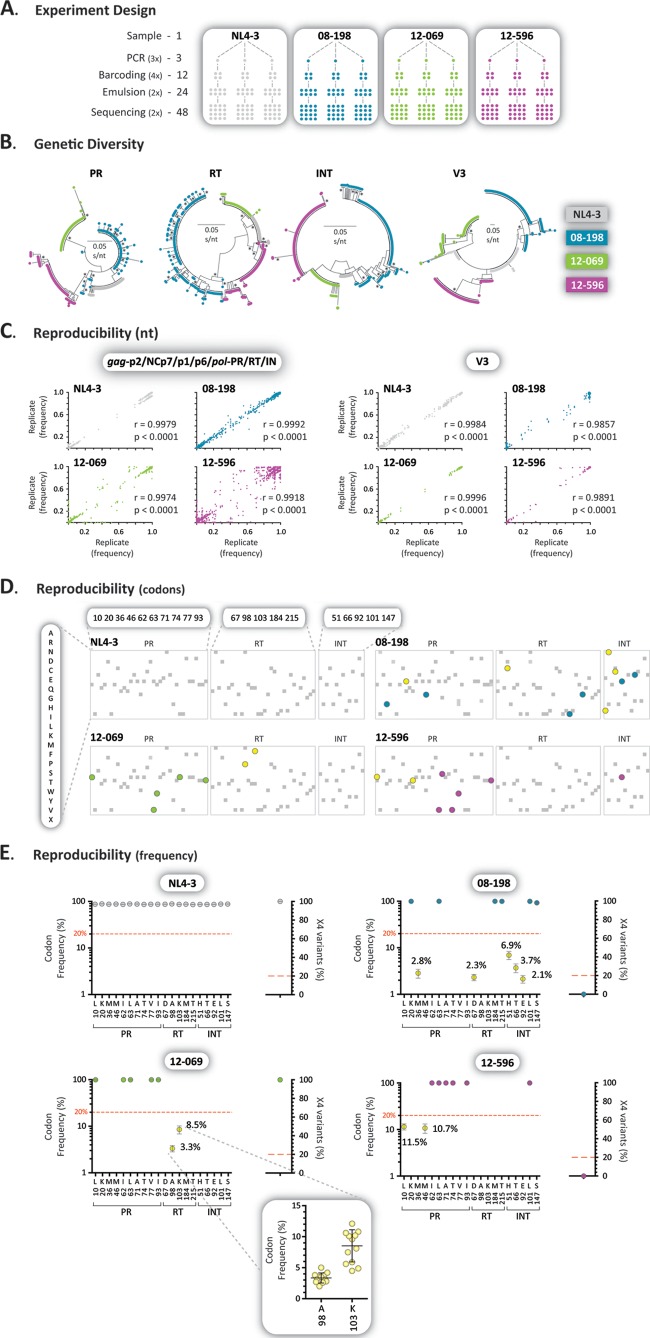
The reproducibility of the HIV-1 genotyping and coreceptor tropism assay was evaluated by testing samples from the wild-type HIV-1 control strain (NL4-3), an antiretroviral-naive individual (12-596), and two antiretroviral-experienced (08-198 and 12-069) individuals. The four samples were reverse transcription-PCR amplified in triplicate (3×), each amplicon was bar coded in quadruplicate (4×), and DNA libraries were prepared in duplicate (2×) and then sequenced twice (2×), for a total of 48 sequences per virus (Fig. 4A). First, reads with a frequency of >1 corresponding to 105-bp fragments from the protease, RT, integrase, and V3 regions were used to construct neighbor-joining phylogenetic trees to quantify intra- and interpatient genetic distances and rule out any potential cross-contamination. Figure 4B shows a clear virus-dependent clustering of sequences in all four HIV-1 regions. As expected, the interpatient genetic distances were larger than the range of intrapatient genetic diversity in the four HIV-1 regions, i.e., 0.0495 (range, 0.0023 to 0.0113), 0.0698 (range, 0.0019 to 0.0150), 0.0554 (range, 0.0001 to 0.0145), and 0.3671 (range, 0.0046 to 0.1067) substitutions per site in the protease, RT, integrase, and V3 regions, respectively. Next, the frequency of each nucleotide at each position was compared among the 16 sequences obtained for each one of the triplicate amplicons (n = 48) for all four viruses in the gag-p2/NCp7/p1/p6/pol-PR/RT/IN and env-V3 regions. Statistically significant correlations were observed when the three sets of 16 sequences were compared for each virus, with r values ranging from 0.9857 to 0.9996 (P < 0.0001, Pearson's coefficient correlation) (Fig. 4C). More important, all 48 sequences detected the same amino acids with similar frequencies in each position in all four viruses. This was evident when only positions associated with drug resistance in the protease, RT, and integrase regions were evaluated (Fig. 4D). Wild-type amino acids were basically the only ones identified in these positions (range, 99.6% to 100%) in the NL4-3 reference virus, while various mutations and different frequencies were detected in the patient-derived samples (Fig. 4D and E). Finally, a series of minor amino acid substitutions were identified repeatedly in the patient-derived samples (08-198, 12-069, and 12-596) at frequencies below the limit of detection of Sanger sequencing, e.g., A98G (mean ± standard deviation, 3.3% ± 0.8%) and K103N (8.5% ± 2.6%) in virus 12-069 (Fig. 4E).
FIG 4.
Assay reproducibility. (A) Samples from two antiretroviral-naive (NL4-3 and 12-596) and two antiretroviral-experienced (08-198 and 12-069) individuals were reverse transcription-PCR amplified in triplicate, each amplicon was bar coded four times, and two DNA libraries were prepared and sequenced in duplicate for a total of 48 sequences per sample. (B) Neighbor-joining phylogenetic trees constructed using reads with a frequency of >1 corresponding to 105-bp fragments from the protease, RT, integrase, and V3 regions. Each color-coded dot represents a unique variant; frequency is not depicted. Bootstrap resampling (1,000 data sets) of the multiple alignments tested the statistical robustness of the trees, with percentage values of >75% indicated by an asterisk. s/nt, substitutions per nucleotide. (C) Pearson's correlation coefficient was used to determine the strength of association between the frequency of each nucleotide at each position among the 16 sequences obtained for each one of triplicate amplicons (n = 48) for all four viruses in the gag-p2/NCp7/p1/p6/pol-PR/RT/IN and env-V3 regions. Over 135,000 and 5,000 points are included in each one of the gag-p2/NCp7/p1/p6/pol-PR/RT/IN and env-V3 plots, respectively. r, correlation coefficient; p, two-tailed P value. (D) Amino acids detected in codons associated with drug resistance in the protease, RT, and integrase regions according to the IAS-USA (77). Drug resistance mutations with a frequency of ≥20% (blue, green, or purple), <20% (yellow), or any other amino acid changes (gray) are indicated. Only amino acid substitutions with a frequency >1% are depicted. (E) Frequency of amino acids in positions associated with drug resistance (gag-p2/NCp7/p1/p6/pol-PR/RT/IN) or X4 variants (env-V3) found in any of the four samples. Each dot represents the mean and 95% confidence interval, with the exception of the inset (sample 12-069), in which each dot indicates the frequency of amino acids detected in each of the 48 replicates, including their mean ± standard deviation.
Comparison of drug susceptibility determination using deep sequencing (DeepGen HIV) with the current standard HIV-1 genotypic assays based on population sequencing.
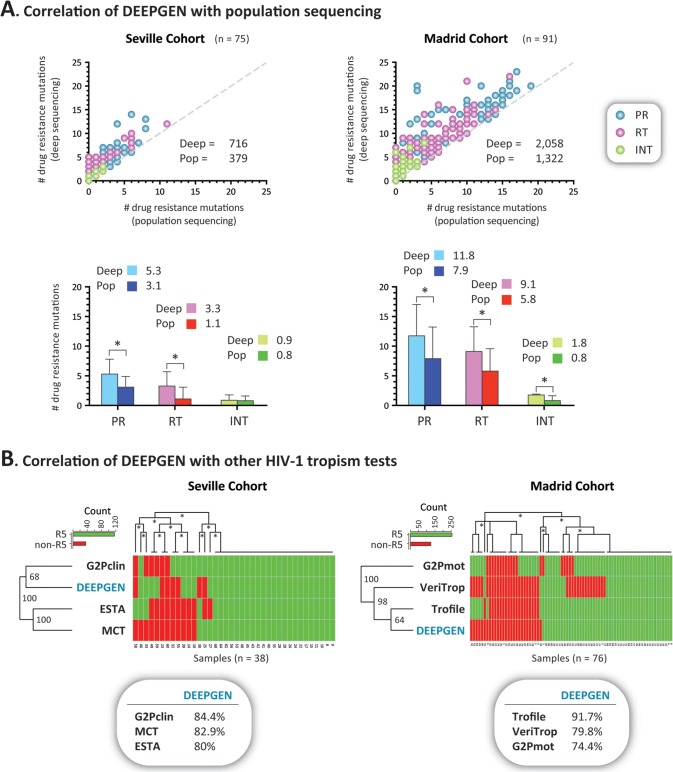
As described above, a multitude of HIV-1 drug resistance methods have been developed, but only a few have been deployed in the clinical setting, including several genotypic tests based on population sequencing (10, 43, 44). Here, blood plasma samples from 166 treatment-experienced HIV-infected individuals from two cohorts of patients (Seville and Madrid) were analyzed using standard population-based HIV-1 genotyping and the novel deep-sequencing assay. The mean CD4+ T-cell count in these patients was 353 cells/μl (interquartile range [IQR], 190 to 488) and their mean plasma viral load was 69,459 copies/ml (IQR, 5,218 to 87,000). The length of the antiretroviral treatment varied among individuals, averaging 8.2 years (between years 1989 and 2012), and included a diversity of treatment regimens using a multitude of PI, NRTI, NNRTI, raltegravir, and/or maraviroc. Altogether, a total of 1,701 mutations (379 and 1,322 in the Seville and Madrid cohorts, respectively) in positions associated with drug resistance were detected by both methodologies (i.e., 954 in the protease, 613 in the RT, and 134 in the integrase region) (Fig. 5A). As expected, all drug resistance mutations identified by population sequencing were also detected by deep sequencing, while an additional 1,073 drug resistance mutations (337 and 736 in the Seville and Madrid cohorts, respectively) were detected by deep sequencing only (i.e., 511 in the protease, 1,015 in the RT, and 97 in the integrase region) (Fig. 5A). Overall, the difference in the numbers of drug resistance mutations detected by the two methods was significant, even when the mutations were quantified by drug class; i.e., an average of 3.1, 2.8, and 0.6 additional mutations associated with PI, RTI, and INI, respectively, were detected by deep sequencing compared to population sequencing (paired t test, P < 0.0001) (Fig. 5A). Interestingly, additional PI (mean, 5.3 versus 3.1) and RTI (3.1 versus 1.1) but not INI (0.9 versus 0.8) resistance mutations were identified by deep sequencing in patients from the Seville cohort (Fig. 5A). Unlike some of the HIV-infected individuals from the Madrid cohort, these patients from Seville were not treated with raltegravir. The slight difference in the number of INI mutations detected by deep sequencing in the Seville patients corresponded to the identification of the L101I polymorphism, which has been associated with decreased susceptibility to the INI dolutegravir (87). Figure S4 in the supplemental material shows a comparison of the frequency of amino acids detected by population and deep sequencing in a virus carrying multiple mutations associated with resistance to PI, RTI, and INI.
FIG 5.
Comparison of the novel HIV-1 genotyping and coreceptor tropism assay with other HIV-1 genotypic phenotypic tests. Plasma samples from 166 treatment-experienced HIV-infected individuals from two cohorts of patients (Seville and Madrid) were analyzed as described in Materials and Methods. (A) Top two plots compare the number of drug resistance mutations detected by standard population (Sanger) and deep sequencing in each patient. The total numbers of drug resistance mutations identified by each sequencing method are indicated. The difference in the numbers of drug resistance mutations (mean ± standard deviation) detected by population and deep sequencing in the protease (PR), reverse transcriptase (RT), and integrase (INT) regions is indicated in the bottom two plots. The mean values are indicated. Statistically significant differences are marked with an asterisk (paired t test, P < 0.0001). Deep, deep sequencing; Pop, population sequencing. The dashed gray line reflects the exact number of mutations detected by deep and population sequencing. (B) Hierarchical clustering analysis was used to group the different HIV-1 coreceptor tropism determinations by similarity. Dendrograms (bootstrap values are indicated) were calculated using the Euclidean distance and complete cluster methods with 100 bootstrap iterations (as described at http://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html). Bootstrap values of >60% are indicated with an asterisk. Green and red blocks indicate the absence or presence of non-R5 (X4) viruses, respectively, as determined by each assay. Concordance between DeepGen HIV and the other HIV-1 coreceptor tropism assays is indicated in bubbles below the graphs. G2Pclin, Geno2Pheno with an FPR of 10%; MCT, 8-day maraviroc monotherapy clinical test (71, 72); ESTA, enhanced sensitivity Trofile assay (42); Trofile, the original version of the Trofile assay (19); VeriTrop, phenotypic HIV-1 tropism assay (23); G2Pmot, Geno2Pheno with a FPR of 2.5% and 5.75% based on optimized cutoffs associated with the analysis of clinical data from MOTIVATE (63).
Comparison of HIV-1 coreceptor tropism data obtained with DeepGen HIV to other phenotypic or genotypic HIV-1 coreceptor tropism assays.
Plasma samples from 114 HIV-infected individuals screened to be treated with a maraviroc-containing regimen, a subset of samples from the same Seville and Madrid cohorts of patients, were analyzed using the novel deep-sequencing-based HIV-1 coreceptor tropism assay. These results were compared with those from a series of genotypic (population sequencing) and phenotypic (ESTA [42], TROCAI [73], and VeriTrop [23]) HIV-1 tropism tests. Only samples with results from all the different tests (i.e., 38 and 76 from Seville and Madrid, respectively) were included. Hierarchical clustering analysis grouped the different HIV-1 coreceptor tropism determinations based on their ability to detect R5 and non-R5 (X4, dual tropic, and/or dual mixed [D/M]) sequences (Fig. 5B). The plasma samples from the Seville cohort were from patients participating in a study evaluating the use of an 8-day maraviroc monotherapy clinical test (MCT), the rationale being that the plasma viral load in patients carrying non-R5 viruses will fail to decrease at least one log10 or will be undetectable in subjects with <1,000 copies/ml after receiving maraviroc in this period of time (71, 72). Overall, in this cohort of patients, the concordance and agreement were high among the different HIV-1 tropism methods, with DeepGen HIV (Geno2Pheno with an FPR of 3.5%) showing good agreement with population sequencing analyzed using Geno2Pheno with an FPR of 10% (84.4%, kappa = 0.37), MCT (82.9%, kappa = 0.44), and ESTA (80%, kappa = 0.47) (Fig. 5B). Similar concordance was observed between ESTA and MCT (85%, kappa = 0.64) and between population sequencing/Geno2Pheno with an FPR of 10% and MCT (82.9%, kappa = 0.51). Interestingly, a perfect agreement (100%, kappa = 1) was observed between MCT and TROCAI, the phenotypic HIV-1 tropism assay performed in Seville (data not shown). Using slightly older samples from the Madrid cohort, DeepGen HIV showed excellent agreement with the original Trofile assay (91.7%, kappa = 0.79) and good concordance with VeriTrop (79.8%, kappa = 0.58) or population sequencing/Geno2Pheno with an FPR of 2.5% to 5.75% (74.4%, kappa = 0.37) (Fig. 5B). Concordance between the original Trofile assay and population sequencing/Geno2Pheno with an FPR of 2.5% to 5.75% was comparable (80.3%, kappa = 0.54), while a 73.7% (kappa = 0.5) agreement was observed between the two phenotypic tests (VeriTrop and Trofile) in baseline samples from these patients (Fig. 5B).
DISCUSSION
Almost 25 years since drug-resistant HIV-1 strains were first reported following initial treatments with zidovudine (AZT) (88), and 28 additional antiretroviral drugs later, the detection and characterization of drug-resistant variants have become essential in the clinical care of HIV-infected patients (3, 6–8). A multitude of genotypic (sequencing-based) and phenotypic (cell-based) assays have been developed to identify and quantify HIV-1 drug resistance, and in the case of CCR5 receptor antagonists, to determine HIV-1 coreceptor tropism (8, 34, 35, 89). Most of these HIV-1 drug resistance or tropism tests correspond to methods developed and used in research settings; however, a few selected assays are commercially available. All current commercial genotypic HIV-1 drug resistance assays are based on population (Sanger) sequencing (10, 43, 44), limiting the detection of minority variants to those in >20% of the viral population (44–48). In the case of HIV-1 tropism determination, only a couple of ultrasensitive deep-sequencing-based genotypic tests are currently used in the clinical setting (63, 66). Here, we have developed, characterized, and validated the first all-inclusive HIV-1 genotyping and tropism test (DeepGen HIV) based on deep sequencing, which is able to detect minority drug-resistant and non-R5 HIV-1 variants in a single assay.
Deep sequencing is here to stay. The efficiency, sensitivity, and cost-effectiveness of deep-sequencing-based methodologies have allowed their use in a multitude of biological fields, including the sequencing of whole genomes of animals and plants (90, 91), targeted studies on polymorphisms related to various genetic disorders (92) and cancer (93, 94), and research aimed at decoding the genomes of bacteria (95, 96), fungi (97), and viruses (98, 99). We have also observed an exponential increase during the last 5 years in the number of HIV studies based on deep sequencing. Many of them are related to HIV-1 pathogenesis, transmission, and evolution (98, 100), but the majority are aimed at detecting minority non-R5 HIV-1 variants (24, 26, 36, 63, 65–67, 75, 101–104) or low-frequency drug-resistant variants that might lead to treatment failure (51, 60–62, 84, 105–108). However, none of the methods used in these studies have been able to simultaneously evaluate resistance to antiretroviral drugs targeting Gag, protease, reverse transcriptase, and integrase and the ability of the virus to use the CCR5 coreceptor to enter the host cell all in a single assay. DeepGen HIV was designed to use the same two overlapping amplicons covering the 3′ end of Gag and the entire pol gene used in the phenotypic HIV-1 assay ViralARTS HIV (13), with the addition of the V3 region of the gp120 in the env gene, which allows the possibility to reflex from the ultrasensitive genotypic test (DeepGen HIV) to the comprehensive cell-based ViralARTS HIV assay using the same PCR products. The 97% amplification success of the two overlapping fragments covering the gag-p2/NCp7/p1/p6/pol-PR/RT/IN region from plasma samples with ≥1,000 copies/ml of HIV RNA (13) was matched here by the successful amplification (92%) of the env-V3 fragment from plasma samples with a similar viral load, which is comparable to the success rates observed in other studies (23, 62, 109). Moreover, we have been able to PCR amplify intermittently these long fragments from plasma samples with viral loads between 50 and 1,000 copies/ml, depending on the quality of the clinical specimen (data not shown). The reverse transcription-PCR step was not only reproducible but ensured successful amplification of the samples from diverse HIV-1 subtypes while avoiding amplification of nonspecific products from endogenous or any of the related viruses tested.
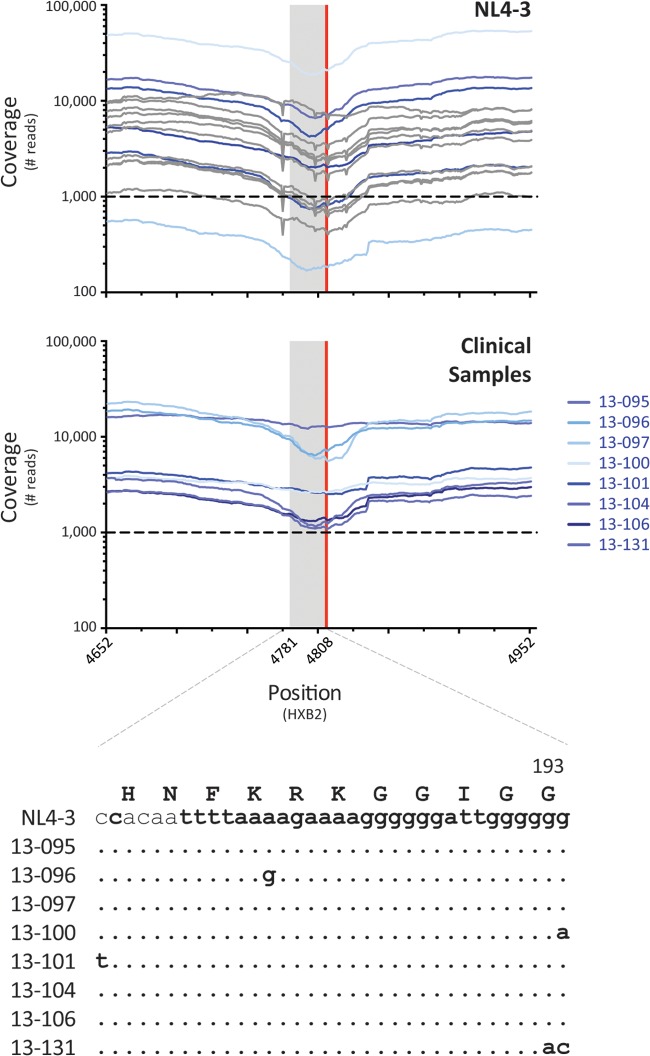
Any methodology based on PCR amplification and deep sequencing endures the same fundamental problem, that is, errors are introduced during the process (67, 84, 85, 110, 111). In fact, a limited number of errors are introduced during the PCR step, with most of the errors produced during deep sequencing, mainly insertions and deletions (84). Thus, it was important here to calculate the intrinsic error rate of the entire system since it undoubtedly affects the limit of detection of the assay. The combined error rates, i.e., those for point mutations, insertions, and deletions, of our new HIV-1 genotyping and coreceptor tropism assay were 0.39% and 0.37% for the gag-p2/NCp7/p1/p6/pol-PR/RT/IN and env-V3 regions, respectively. These values were similar to the average error rates previously reported in the HIV-1 pol or env genes using other deep-sequencing platforms, ranging from 0.3% to 0.98% (62, 66, 67, 84, 85, 110, 112–115). Approximately a 10-fold-higher combined error rate was observed in nucleotide positions associated with homopolymeric regions, resembling findings from other studies describing similar difficulties when sequencing regions with identical consecutive nucleotides (61, 62, 84, 85, 113). More importantly, some of these homopolymeric regions encompass positions associated with resistance to antiretroviral drugs, which might represent a challenge during the interpretation of minority mutations detected at these positions. Using computational methods, we were able to discern between genuine genetic variation and errors introduced during the sequencing process. For example, codon 193 in the integrase region showed the highest error rate in the entire HIV-1 genomic region analyzed, i.e., 9.6% for indels and 10.5% overall. In HIV-1NL4-3, the 25 nucleotides upstream of this position correspond to a series of identical consecutive nucleotides (Fig. 6), which might explain the elevated error rate in this particular region. However, the Ion Torrent software was able to filter most of the reads with sequencing errors at the expense of reducing approximately 5-fold the coverage around this region (Fig. 2B and 6) but still above the 1,000 reads required to guarantee the detection of a minor variant present in ≥1% of the population (85). Moreover, the Variant Caller in the DeepGen Software Tool Suite filtered out all the indels at position 193, and as a consequence, we were able to accurately and repeatedly determine the correct amino acids in all wild-type HIV-1NL4-3 sequences and the different variants from the patient-derived samples (Fig. 6).
FIG 6.
Coverage around codon 193 of the HIV-1 integrase region. Top two panels summarize the coverage of the 10 HIV-1NL4-3 sequences (top) obtained from the clones used to calculate the intrinsic error rate of the assay (gray lines) and 6 HIV-1NL4-3 sequences obtained from the positive controls in six regular sequencing runs (blue lines). The middle panel includes the coverage of 8 patient-derived HIV-1 sequences (clinical samples labeled 13-xxx). Gray bar indicates the homopolymeric region at and upstream of codon 193, i.e., 25 nucleotides in bold in the nucleotide alignment. Red vertical line depicts codon 193. The bottom panel shows the nucleotide sequences of the 8 patient-derived HIV-1 sequences.
The approach of using a deep-sequencing-based assay to detect minority HIV-1 variants relies on the capacity of the methodology to truthfully detect low-frequency sequences from usually complex virus populations. The limits of detection of these assays are a combination of the intrinsic error rate of the method and the analytical sensitivity of the test. As described above, the ability of our HIV-1 genotyping and coreceptor tropism assay to discern between genetic variation and sequencing error was calculated to be 1%. The analytical sensitivity of the assay, determined using mixtures of plasmid DNA, seems to suggest that the deep-sequencing-based test is able to detect minority variants when present in as low as 1% of the population; however, mutations were detected in the HIV-infected plasma mixtures at a 5% frequency but not at 1%. Five percent is the limit of detection previously calculated when studying HIV using 454 pyrosequencing (62, 66). It is possible that although sequences at a frequency of 1% theoretically can be identified, the bias for the majority members of the population during reverse transcription-PCR amplification, library preparation, and/or sequencing might limit the ability of the assay to efficiently detect minority variants of <5%. Additional studies using a larger number of clinical samples will be needed to verify this limit of detection. Nevertheless, our novel assay was able to reproducibly detect HIV-1 minority variants at frequencies at least 4-fold lower than the 20% reported for the standard Sanger-based sequencing (16, 44–48).
A multitude of studies have compared the efficacies of phenotypic and genotypic assays to detect and quantify HIV-1 drug resistance (116–120) or coreceptor tropism (38, 121–124). While there is usually high concordance between drug resistance methodologies (13, 119, 120), in general, population-based sequencing HIV-1 tropism tests are less sensitive and less specific than phenotypic assays (8, 41). Here, DeepGen HIV identified all the drug resistance mutations detected by the standard population sequencing-based HIV-1 genotyping tests used to monitor 166 individuals in two well-established cohorts of patients. More importantly, >1,000 additional minor drug resistance mutations at frequencies between 1% and 20% were detected using the novel deep-sequencing-based assay. In the cases of detection of minority non-R5 variants and determination of HIV-1 coreceptor tropism, DeepGen HIV showed good concordance with both ESTA and the original Trofile assay (80% and 92%, respectively), similar to the agreement of these phenotypic tests with 454-based sequencing systems used to determine HIV-1 tropism (80% to 87%) (24, 36, 63–66, 104, 125, 126). Deep-sequencing-based HIV-1 coreceptor tropism assays have shown good concordance with virological responses to CCR5 antagonists, such as maraviroc (36, 63); thus, more sensitive HIV-1 genotyping methods based on deep sequencing may also help detect minority drug-resistant variants earlier, possibly improving the design of antiretroviral treatment schemes.
As described above, DeepGen HIV detected additional (minority) HIV-1 drug resistance mutations and non-R5 variants compared to population sequencing; however, the clinical relevance of these minority members of the viral population is still under debate (33, 51, 55, 57, 127–129). It is reasonable to assume that under appropriate selection (i.e., drug pressure), minority variants carrying drug resistance mutations will eventually outcompete other members of the viral population, and the earlier these mutations are detected, the sooner the proper strategy can be defined to control the growth of these viruses. However, the threshold for the identification of significant low-level variants may be mutation and/or antiretroviral drug dependent (32); therefore, early detection of drug-resistant HIV-1 minority variants, or non-R5 variants in the case of CCR5 antagonists, may contribute to the prediction of clinical or treatment outcome (32, 36, 57, 63, 127). Additional effort, most likely in the form of longitudinal studies, is needed to better understand the clinical relevance of these minority HIV-1 variants. Here, we have developed, characterized, and validated a novel all-inclusive and ultrasensitive HIV-1 genotyping assay based on deep sequencing to simplify the monitoring of patients infected with HIV-1. This first-in-class methodology accurately provides drug resistance information for all protease, reverse transcriptase, integrase, and maturation inhibitors, as well as HIV-1 coreceptor tropism, in a single, more efficient, rapid, and affordable test. This unique assay may provide the platform not only for predicting clinical outcome in HIV-infected individuals—who otherwise might fail treatment regimens—but in the development and clinical assessment of novel antiretroviral drugs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eric J. Arts (HIV-1 isolates), Michael R. Jacobs (BKV, CMV, HSV-1, HSV-2, and VZV), David Canaday (EBV), and Donald Anthony (HBV and HCV) from Case Western Reserve University (Cleveland, OH) for providing access to the respective RNA or DNA viruses. We also thank Benigno Rodriguez and the Case Western Reserve University/University Hospitals Center for AIDS Research (grant no. NIH P30 AI036219) and Vicente Soriano (Hospital Carlos III, Madrid, Spain) for providing clinical samples for the characterization and validation studies. We are grateful to Michael D. Miller and Kirsten L. White (Gilead Sciences, Inc., Foster City, CA) for providing some of the plasmid mixtures used in the analytical sensitivity experiments.
E.R.-M. and M.L. were supported by Redes Telemáticas de Investigación Cooperativa en Salud (RETICS), Red de Investigación en SIDA (no. RIS RD12/0017/0029).
M.E.Q.-M. designed the study, collected and assembled the data, and wrote and drafted the manuscript. R.M.G., A.M.M., and D.W. performed all cell culture, molecular, and sequencing experiments. J.A., F.F., M.E.Q.-M. and D.L.R. designed the DEEPGEN Software Tool Suite, while J.A. and F.F. developed the suite. E.R.-M. and M.L. performed the studies associated with the Seville cohort, including population sequencing, MCT, and TROCAI analyses. R.M.G., C.L.S., and M.E.Q.-M. contributed to the overall analysis of the data. All the authors read and approved the final manuscript.
Footnotes
Published ahead of print 27 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02710-13.
REFERENCES
- 1.WHO. 2013. Global update on HIV treatment 2013: results, impact, and opportunities. World Health Organizzation, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/85327/1/WHO_HIV_2013.9_eng.pdf [Google Scholar]
- 2.Paredes R, Clotet B. 2010. Clinical management of HIV-1 resistance. Antiviral Res. 85:245–265. 10.1016/j.antiviral.2009.09.015 [DOI] [PubMed] [Google Scholar]
- 3.Zolopa AR. 2010. The evolution of HIV treatment guidelines: current state-of-the-art of ART. Antiviral Res. 85:241–244. 10.1016/j.antiviral.2009.10.018 [DOI] [PubMed] [Google Scholar]
- 4.Deeks SG. 2003. Treatment of antiretroviral-drug-resistant HIV-1 infection. Lancet 362:2002–2011. 10.1016/S0140-6736(03)15022-2 [DOI] [PubMed] [Google Scholar]
- 5.Tang JW, Pillay D. 2004. Transmission of HIV-1 drug resistance. J. Clin. Virol. 30:1–10. 10.1016/j.jcv.2003.12.002 [DOI] [PubMed] [Google Scholar]
- 6.Johnson VA, Calvez V, Günthard HF, Paredes R, Pillay D, Shafer R, Wensing AM, Richman DD. 2011. 2011 update of the drug resistance mutations in HIV-1. Top. Antivir. Med. 19:156–164 [PMC free article] [PubMed] [Google Scholar]
- 7.Weber J, Piontkivska H, Quiñones-Mateu ME. 2006. HIV type 1 tropism and inhibitors of viral entry: clinical implications. AIDS Rev. 8:60–77 [PubMed] [Google Scholar]
- 8.Rose JD, Rhea AM, Weber J, Quiñones-Mateu ME. 2009. Current tests to evaluate HIV-1 coreceptor tropism. Curr. Opin. HIV AIDS 4:136–142. 10.1097/COH.0b013e328322f973 [DOI] [PubMed] [Google Scholar]
- 9.Rhee SY, Taylor J, Wadhera G, Ben-Hur A, Brutlag DL, Shafer RW. 2006. Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc. Natl. Acad. Sci. U. S. A. 103:17355–17360. 10.1073/pnas.0607274103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paar C, Palmetshofer C, Flieger K, Geit M, Kaiser R, Stekel H, Berg J. 2008. Genotypic antiretroviral resistance testing for human immunodeficiency virus type 1 integrase inhibitors by use of the TruGene sequencing system. J. Clin. Microbiol. 46:4087–4090. 10.1128/JCM.01246-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hertogs K, de Béthune MP, Miller V, Ivens T, Schel P, Van Cauwenberge A, Van Den Eynde C, Van Gerwen V, Azijn H, Van Houtte M, Peeters F, Staszewski S, Conant M, Bloor S, Kemp S, Larder B, Pauwels R. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269–276. 10.1093/jac/42.2.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, Tian H, Smith D, Winslow GA, Capon DJ, Whitcomb JM. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920–928. 10.1128/AAC.44.4.920-928.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber J, Vazquez AC, Winner D, Rose JD, Wylie D, Rhea AM, Henry K, Pappas J, Wright A, Mohamed N, Gibson R, Rodriguez B, Soriano V, King K, Arts EJ, Olivo PD, Quiñones-Mateu ME. 2011. Novel method for simultaneous quantification of phenotypic resistance to maturation, protease, reverse transcriptase, and integrase HIV inhibitors based on 3′Gag(p2/p7/p1/p6)/PR/RT/INT-recombinant viruses: a useful tool in the multitarget era of antiretroviral therapy. Antimicrob. Agents Chemother. 55:3729–3742. 10.1128/AAC.00396-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vermeiren H, Van Craenenbroeck E, Alen P, Bacheler L, Picchio G, Lecocq P, Virco Clinical Response Collaborative Team 2007. Prediction of HIV-1 drug susceptibility phenotype from the viral genotype using linear regression modeling. J. Virol. Methods 145:47–55. 10.1016/j.jviromet.2007.05.009 [DOI] [PubMed] [Google Scholar]
- 15.Poveda E, Briz V, Quiñones-Mateu M, Soriano V. 2006. HIV tropism: diagnostic tools and implications for disease progression and treatment with entry inhibitors. AIDS 20:1359–1367. 10.1097/01.aids.0000233569.74769.69 [DOI] [PubMed] [Google Scholar]
- 16.Obermeier M, Symons J, Wensing AM. 2012. HIV population genotypic tropism testing and its clinical significance. Curr. Opin. HIV AIDS 7:470–477. 10.1097/COH.0b013e328356eaa7 [DOI] [PubMed] [Google Scholar]
- 17.Raymond S, Delobel P, Izopet J. 2012. Phenotyping methods for determining HIV tropism and applications in clinical settings. Curr. Opin. HIV AIDS 7:463–469. 10.1097/COH.0b013e328356f6d7 [DOI] [PubMed] [Google Scholar]
- 18.Trouplin V, Salvatori F, Cappello F, Obry V, Brelot A, Heveker N, Alizon M, Scarlatti G, Clavel F, Mammano F. 2001. Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay. J. Virol. 75:251–259. 10.1128/JVI.75.1.251-259.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitcomb JM, Huang W, Fransen S, Limoli K, Toma J, Wrin T, Chappey C, Kiss LD, Paxinos EE, Petropoulos CJ. 2007. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob. Agents Chemother. 51:566–575. 10.1128/AAC.00853-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raymond S, Delobel P, Mavigner M, Cazabat M, Souyris C, Encinas S, Bruel P, Sandres-Sauné K, Marchou B, Massip P, Izopet J. 2010. Development and performance of a new recombinant virus phenotypic entry assay to determine HIV-1 coreceptor usage. J. Clin. Virol. 47:126–130. 10.1016/j.jcv.2009.11.018 [DOI] [PubMed] [Google Scholar]
- 21.Holland AU, Munk C, Lucero GR, Nguyen LD, Landau NR. 2004. Alpha-complementation assay for HIV envelope glycoprotein-mediated fusion. Virology 319:343–352. 10.1016/j.virol.2003.11.012 [DOI] [PubMed] [Google Scholar]
- 22.Grund S, Klein A, Adams O. 2010. Expression plasmids are only useful for the investigation of co-receptor tropism and fusion capacity of short HIV-1 envelope domains. J. Virol. Methods 166:106–109. 10.1016/j.jviromet.2010.03.015 [DOI] [PubMed] [Google Scholar]
- 23.Weber J, Vazquez AC, Winner D, Gibson RM, Rhea AM, Rose JD, Wylie D, Henry K, Wright A, King K, Archer J, Poveda E, Soriano V, Robertson DL, Olivo PD, Arts EJ, Quiñones-Mateu ME. 2013. Sensitive cell-based assay for determination of human immunodeficiency virus type 1 coreceptor tropism. J. Clin. Microbiol. 51:1517–1527. 10.1128/JCM.00092-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swenson LC, Moores A, Low AJ, Thielen A, Dong W, Woods C, Jensen MA, Wynhoven B, Chan D, Glascock C, Harrigan PR. 2010. Improved detection of CXCR4-using HIV by V3 genotyping: application of population-based and “deep” sequencing to plasma RNA and proviral DNA. J. Acquir. Immune Defic. Syndr. 54:506–510. 10.1097/QAI.0b013e3181d0558f [DOI] [PubMed] [Google Scholar]
- 25.Swenson LC, Däumer M, Paredes R. 2012. Next-generation sequencing to assess HIV tropism. Curr. Opin. HIV AIDS 7:478–485. 10.1097/COH.0b013e328356e9da [DOI] [PubMed] [Google Scholar]
- 26.Archer J, Braverman MS, Taillon BE, Desany B, James I, Harrigan PR, Lewis M, Robertson DL. 2009. Detection of low-frequency pretherapy chemokine (CXC motif) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. AIDS 23:1209–1218. 10.1097/QAD.0b013e32832b4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sing T, Low AJ, Beerenwinkel N, Sander O, Cheung PK, Domingues FS, Büch J, Däumer M, Kaiser R, Lengauer T, Harrigan PR. 2007. Predicting HIV coreceptor usage on the basis of genetic and clinical covariates. Antivir. Ther. 12:1097–1106 [PubMed] [Google Scholar]
- 28.Jensen MA, Li FS, van ‘t Wout AB, Nickle DC, Shriner D, He HX, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376–13388. 10.1128/JVI.77.24.13376-13388.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardozo T, Kimura T, Philpott S, Weiser B, Burger H, Zolla-Pazner S. 2007. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res. Hum. Retroviruses 23:415–426. 10.1089/aid.2006.0130 [DOI] [PubMed] [Google Scholar]
- 30.Rosen O, Sharon M, Quadt-Akabayov SR, Anglister J. 2006. Molecular switch for alternative conformations of the HIV-1 V3 region: implications for phenotype conversion. Proc. Natl. Acad. Sci. U. S. A. 103:13950–13955. 10.1073/pnas.0606312103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poveda E, Alcamí J, Paredes R, Córdoba J, Gutiérrez F, Llibre JM, Delgado R, Pulido F, Iribarren JA, García Deltoro M, Hernández Quero J, Moreno S, García F. 2010. Genotypic determination of HIV tropism - clinical and methodological recommendations to guide the therapeutic use of CCR5 antagonists. AIDS Rev. 12:135–148 [PubMed] [Google Scholar]
- 32.Gianella S, Richman DD. 2010. Minority variants of drug-resistant HIV. J. Infect. Dis. 202:657–666. 10.1086/655397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stekler JD, Ellis GM, Carlsson J, Eilers B, Holte S, Maenza J, Stevens CE, Collier AC, Frenkel LM. 2011. Prevalence and impact of minority variant drug resistance mutations in primary HIV-1 infection. PLoS One 6:e28952. 10.1371/journal.pone.0028952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunn DT, Coughlin K, Cane PA. 2011. Genotypic resistance testing in routine clinical care. Curr. Opin. HIV AIDS 6:251–257. 10.1097/COH.0b013e32834732e8 [DOI] [PubMed] [Google Scholar]
- 35.Grant PM, Zolopa AR. 2009. The use of resistance testing in the management of HIV-1-infected patients. Curr. Opin. HIV AIDS 4:474–480. 10.1097/COH.0b013e328331c14f [DOI] [PubMed] [Google Scholar]
- 36.Swenson LC, Mo T, Dong WW, Zhong X, Woods CK, Thielen A, Jensen MA, Knapp DJ, Chapman D, Portsmouth S, Lewis M, James I, Heera J, Valdez H, Harrigan PR. 2011. Deep V3 sequencing for HIV type 1 tropism in treatment-naive patients: a reanalysis of the MERIT trial of maraviroc. Clin. Infect. Dis. 53:732–742. 10.1093/cid/cir493 [DOI] [PubMed] [Google Scholar]
- 37.Seclén E, Soriano V, González MM, Gómez S, Thielen A, Poveda E. 2011. High concordance between the position-specific scoring matrix and Geno2Pheno algorithms for genotypic interpretation of HIV-1 tropism: V3 length as the major cause of disagreement. J. Clin. Microbiol. 49:3380–3382. 10.1128/JCM.00908-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Recordon-Pinson P, Soulié C, Flandre P, Descamps D, Lazrek M, Charpentier C, Montes B, Trabaud MA, Cottalorda J, Schneider V, Morand-Joubert L, Tamalet C, Desbois D, Macé M, Ferré V, Vabret A, Ruffault A, Pallier C, Raymond S, Izopet J, Reynes J, Marcelin AG, Masquelier B, ANRS AC11 Resistance Study Group 2010. Evaluation of the genotypic prediction of HIV-1 coreceptor use versus a phenotypic assay and correlation with the virological response to maraviroc: the ANRS GenoTropism study. Antimicrob. Agents Chemother. 54:3335–3340. 10.1128/AAC.00148-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGovern RA, Thielen A, Mo T, Dong W, Woods CK, Chapman D, Lewis M, James I, Heera J, Valdez H, Harrigan PR. 2010. Population-based V3 genotypic tropism assay: a retrospective analysis using screening samples from the A4001029 and MOTIVATE studies. AIDS 24:2517–2525. 10.1097/QAD.0b013e32833e6cfb [DOI] [PubMed] [Google Scholar]
- 40.Poveda E, Seclén E, González Mdel M, García F, Chueca N, Aguilera A, Rodríguez JJ, González-Lahoz J, Soriano V. 2009. Design and validation of new genotypic tools for easy and reliable estimation of HIV tropism before using CCR5 antagonists. J. Antimicrob. Chemother. 63:1006–1010. 10.1093/jac/dkp063 [DOI] [PubMed] [Google Scholar]
- 41.Lin NH, Kuritzkes DR. 2009. Tropism testing in the clinical management of HIV-1 infection. Curr. Opin. HIV AIDS 4:481–487. 10.1097/COH.0b013e328331b929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reeves JD, Coakley E, Petropoulos CJ, Whitcomb JM. 2009. An enhanced-sensitivity Trofile HIV coreceptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5: a review of analytical and clinical studies. J. Viral Entry 3:94–102 [Google Scholar]
- 43.Cunningham S, Ank B, Lewis D, Lu W, Wantman M, Dileanis JA, Jackson JB, Palumbo P, Krogstad P, Eshleman SH. 2001. Performance of the applied Biosystems ViroSeq human immunodeficiency virus type 1 (HIV-1) genotyping system for sequence-based analysis of HIV-1 in pediatric plasma samples. J. Clin. Microbiol. 39:1254–1257. 10.1128/JCM.39.4.1254-1257.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grant RM, Kuritzkes DR, Johnson VA, Mellors JW, Sullivan JL, Swanstrom R, D'Aquila RT, Van Gorder M, Holodniy M, Lloyd RM, Jr, Reid C, Morgan GF, Winslow DL. 2003. Accuracy of the Trugene HIV-1 genotyping kit. J. Clin. Microbiol. 41:1586–1593. 10.1128/JCM.41.4.1586-1593.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larder BA, Kohli A, Kellam P, Kemp SD, Kronick M, Henfrey RD. 1993. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature 365:671–673. 10.1038/365671a0 [DOI] [PubMed] [Google Scholar]
- 46.Church JD, Jones D, Flys T, Hoover D, Marlowe N, Chen S, Shi C, Eshleman JR, Guay LA, Jackson JB, Kumwenda N, Taha TE, Eshleman SH. 2006. Sensitivity of the ViroSeq HIV-1 genotyping system for detection of the K103N resistance mutation in HIV-1 subtypes A, C, and D. J. Mol. Diagn. 8:430–432; quiz 527. 10.2353/jmoldx.2006.050148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halvas EK, Aldrovandi GM, Balfe P, Beck IA, Boltz VF, Coffin JM, Frenkel LM, Hazelwood JD, Johnson VA, Kearney M, Kovacs A, Kuritzkes DR, Metzner KJ, Nissley DV, Nowicki M, Palmer S, Ziermann R, Zhao RY, Jennings CL, Bremer J, Brambilla D, Mellors JW. 2006. Blinded, multicenter comparison of methods to detect a drug-resistant mutant of human immunodeficiency virus type 1 at low frequency. J. Clin. Microbiol. 44:2612–2614. 10.1128/JCM.00449-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leitner T, Halapi E, Scarlatti G, Rossi P, Albert J, Fenyö EM, Uhlén M. 1993. Analysis of heterogeneous viral populations by direct DNA sequencing. Biotechniques 15:120–127 [PubMed] [Google Scholar]
- 49.Paredes R, Lalama CM, Ribaudo HJ, Schackman BR, Shikuma C, Giguel F, Meyer WA, III, Johnson VA, Fiscus SA, D'Aquila RT, Gulick RM, Kuritzkes DR, AIDS Clinical Trials Group (ACTG) A5095 Study Team 2010. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J. Infect. Dis. 201:662–671. 10.1086/650543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson JA, Li JF, Wei X, Lipscomb J, Irlbeck D, Craig C, Smith A, Bennett DE, Monsour M, Sandstrom P, Lanier ER, Heneine W. 2008. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 5:e158. 10.1371/journal.pmed.0050158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simen BB, Simons JF, Hullsiek KH, Novak RM, Macarthur RD, Baxter JD, Huang C, Lubeski C, Turenchalk GS, Braverman MS, Desany B, Rothberg JM, Egholm M, Kozal MJ, Terry Beirn Community Programs for Clinical Research on AIDS 2009. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 199:693–701. 10.1086/596736 [DOI] [PubMed] [Google Scholar]
- 52.Metzner KJ, Giulieri SG, Knoepfel SA, Rauch P, Burgisser P, Yerly S, Gunthard HF, Cavassini M. 2009. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin. Infect. Dis. 48:239–247. 10.1086/595703 [DOI] [PubMed] [Google Scholar]
- 53.Kuritzkes DR, Lalama CM, Ribaudo HJ, Marcial M, Meyer WA, III, Shikuma C, Johnson VA, Fiscus SA, D'Aquila RT, Schackman BR, Acosta EP, Gulick RM. 2008. Preexisting resistance to nonnucleoside reverse-transcriptase inhibitors predicts virologic failure of an efavirenz-based regimen in treatment-naive HIV-1-infected subjects. J. Infect. Dis. 197:867–870. 10.1086/528802 [DOI] [PubMed] [Google Scholar]
- 54.Halvas EK, Wiegand A, Boltz VF, Kearney M, Nissley D, Wantman M, Hammer SM, Palmer S, Vaida F, Coffin JM, Mellors JW. 2010. Low frequency nonnucleoside reverse-transcriptase inhibitor-resistant variants contribute to failure of efavirenz-containing regimens in treatment- experienced patients. J. Infect. Dis. 201:672–680. 10.1086/650542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li JZ, Paredes R, Ribaudo HJ, Svarovskaia ES, Kozal MJ, Hullsiek KH, Miller MD, Bangsberg DR, Kuritzkes DR. 2012. Relationship between minority nonnucleoside reverse transcriptase inhibitor resistance mutations, adherence, and the risk of virologic failure. AIDS 26:185–192. 10.1097/QAD.0b013e32834e9d7d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fisher R, van Zyl GU, Travers SA, Kosakovsky Pond SL, Engelbrech S, Murrell B, Scheffler K, Smith D. 2012. Deep sequencing reveals minor protease resistance mutations in patients failing a protease inhibitor regimen. J. Virol. 86:6231–6237. 10.1128/JVI.06541-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li JZ, Kuritzkes DR. 2013. Clinical implications of HIV-1 minority variants. Clin. Infect. Dis. 56:1667–1674. 10.1093/cid/cit125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmer S, Boltz V, Maldarelli F, Kearney M, Halvas EK, Rock D, Falloon J, Davey RT, Jr, Dewar RL, Metcalf JA, Mellors JW, Coffin JM. 2006. Selection and persistence of non-nucleoside reverse transcriptase inhibitor-resistant HIV-1 in patients starting and stopping non-nucleoside therapy. AIDS 20:701–710. 10.1097/01.aids.0000216370.69066.7f [DOI] [PubMed] [Google Scholar]
- 59.Lalonde MS, Troyer RM, Syed AR, Bulime S, Demers K, Bajunirwe F, Arts EJ. 2007. Sensitive oligonucleotide ligation assay for low-level detection of nevirapine resistance mutations in human immunodeficiency virus type 1 quasispecies. J. Clin. Microbiol. 45:2604–2615. 10.1128/JCM.00431-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Avidor B, Girshengorn S, Matus N, Talio H, Achsanov S, Zeldis I, Fratty IS, Katchman E, Brosh-Nissimov T, Hassin D, Alon D, Bentwich Z, Yust I, Amit S, Forer R, Vulih Shultsman I, Turner D. 2013. Evaluation of a benchtop HIV ultradeep pyrosequencing drug resistance assay in the clinical laboratory. J. Clin. Microbiol. 51:880–886. 10.1128/JCM.02652-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang MW, Oliveira G, Yuan J, Okulicz JF, Levy S, Torbett BE. 2013. Rapid deep sequencing of patient-derived HIV with ion semiconductor technology. J. Virol. Methods 189:232–234. 10.1016/j.jviromet.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dudley DM, Chin EN, Bimber BN, Sanabani SS, Tarosso LF, Costa PR, Sauer MM, Kallas EG, O'Connor DH. 2012. Low-cost ultra-wide genotyping using Roche/454 pyrosequencing for surveillance of HIV drug resistance. PLoS One 7:e36494. 10.1371/journal.pone.0036494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swenson LC, Mo T, Dong WW, Zhong X, Woods CK, Jensen MA, Thielen A, Chapman D, Lewis M, James I, Heera J, Valdez H, Harrigan PR. 2011. Deep sequencing to infer HIV-1 co-receptor usage: application to three clinical trials of maraviroc in treatment-experienced patients. J. Infect. Dis. 203:237–245. 10.1093/infdis/jiq030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saliou A, Delobel P, Dubois M, Nicot F, Raymond S, Calvez V, Masquelier B, Izopet J, ANRS AC11 Resistance Study Group 2011. Concordance between two phenotypic assays and ultradeep pyrosequencing for determining HIV-1 tropism. Antimicrob. Agents Chemother. 55:2831–2836. 10.1128/AAC.00091-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kagan RM, Johnson EP, Siaw MF, Van Baelen B, Ogden R, Platt JL, Pesano RL, Lefebvre E. 2014. Comparison of genotypic and phenotypic HIV type 1 tropism assay: results from the screening samples of cenicriviroc study 202, a randomized phase II trial in treatment-naive subjects. AIDS Res. Hum. Retroviruses 30:151–159. 10.1089/aid.2013.0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kagan RM, Johnson EP, Siaw M, Biswas P, Chapman DS, Su Z, Platt JL, Pesano RL. 2012. A genotypic test for HIV-1 tropism combining Sanger sequencing with ultradeep sequencing predicts virologic response in treatment-experienced patients. PLoS One 7:e46334. 10.1371/journal.pone.0046334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Archer J, Weber J, Henry K, Winner D, Gibson R, Lee L, Paxinos E, Arts EJ, Robertson DL, Mimms L, Quiñones-Mateu ME. 2012. Use of four next-generation sequencing platforms to determine HIV-1 coreceptor tropism. PLoS One 7:e49602. 10.1371/journal.pone.0049602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber J, Rose JD, Vazquez AC, Winner D, Margot N, McColl DJ, Miller MD, Quiñones-Mateu ME. 2013. Resistance mutations outside the integrase coding region have an effect on human immunodeficiency virus replicative fitness but do not affect its susceptibility to integrase strand transfer inhibitors. PLoS One 8:e65631. 10.1371/journal.pone.0065631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weber J, Quiñones-Mateu ME. 2007. Novel anti-HIV-1 screening system based on intact recombinant viruses expressing synthetic firefly and Renilla luminescent proteins. Antivir. Ther. 12:S155 [Google Scholar]
- 70.Moreno S, Clotet B, Sarria C, Ortega E, Leal M, Rodríguez-Arrondo F, Sánchez-de la Rosa R, Allegro Study Group 2009. Prevalence of CCR5-tropic HIV-1 among treatment-experienced individuals in Spain. HIV Clin. Trials 10:394–402. 10.1310/hct1006-394 [DOI] [PubMed] [Google Scholar]
- 71.Gonzalez-Serna A, Romero-Sánchez MC, Ferrando-Martinez S, Genebat M, Vidal F, Muñoz-Fernández MA, Abad MA, Leal M, Ruiz-Mateos E. 2012. HIV-1 tropism evolution after short-term maraviroc monotherapy in HIV-1-infected patients. Antimicrob. Agents Chemother. 56:3981–3983. 10.1128/AAC.00507-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Romero-Sánchez MC, Machmach K, Gonzalez-Serna A, Genebat M, Pulido I, García-García M, Alvarez-Ríos AI, Ferrando-Martinez S, Ruiz-Mateos E, Leal M. 2012. Effect of maraviroc on HIV disease progression-related biomarkers. Antimicrob. Agents Chemother. 56:5858–5864. 10.1128/AAC.01406-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonzalez-Serna A, Leal M, Genebat M, Abad MA, Garcia-Perganeda A, Ferrando-Martinez S, Ruiz-Mateos E. 2010. TROCAI (tropism coreceptor assay information): a new phenotypic tropism test and its correlation with Trofile enhanced sensitivity and genotypic approaches. J. Clin. Microbiol. 48:4453–4458. 10.1128/JCM.00953-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Quiñones-Mateu ME, Domingo E. 1999. Nucleotide diversity in three different genomic regions of Venezuelan HIV type 1 isolates: a subtyping update. AIDS Res. Hum. Retroviruses 15:73–79 [DOI] [PubMed] [Google Scholar]
- 75.Archer J, Rambaut A, Taillon BE, Harrigan PR, Lewis M, Robertson DL. 2010. The evolutionary analysis of emerging low frequency HIV-1 CXCR4 using variants through time–an ultra-deep approach. PLoS Comput. Biol. 6:e1001022. 10.1371/journal.pcbi.1001022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Archer J, Baillie G, Watson SJ, Kellam P, Rambaut A, Robertson DL. 2012. Analysis of high-depth sequence data for studying viral diversity: a comparison of next generation sequencing platforms using Segminator II. BMC Bioinformatics 13:47. 10.1186/1471-2105-13-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, Shafer RW, Wensing AM, Richman DD. 2013. Update of the drug resistance mutations in HIV-1: March 2013. Top. Antivir. Med. 21:6–14 [PMC free article] [PubMed] [Google Scholar]
- 78.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 79.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harrigan PR. 2009. MOTIVATE tropism study group. Optimization of clinical cutoffs for determining HIV co-receptor use by population and “deep” sequencing methods. Poster 297 Infectious Diseases Society of America, Philadelphia, PA, 30 October 2009 [Google Scholar]
- 81.Bunnik EM, Swenson LC, Edo-Matas D, Huang W, Dong W, Frantzell A, Petropoulos CJ, Coakley E, Schuitemaker H, Harrigan PR, van 't Wout AB. 2011. Detection of inferred CCR5- and CXCR4-using HIV-1 variants and evolutionary intermediates using ultra-deep pyrosequencing. PLoS Pathog. 7:e1002106. 10.1371/journal.ppat.1002106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lobritz MA, Marozsan AJ, Troyer RM, Arts EJ. 2007. Natural variation in the V3 crown of human immunodeficiency virus type 1 affects replicative fitness and entry inhibitor sensitivity. J. Virol. 81:8258–8269. 10.1128/JVI.02739-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson BF, Bakeman R. 1998. ComKappa: a Windows 95 program for calculating kappa and related statistics. Behav. Res. Methods Instrum. Comput. 30:731–732. 10.3758/BF03209495 [DOI] [Google Scholar]
- 84.Shao W, Boltz VF, Spindler JE, Kearney MF, Maldarelli F, Mellors JW, Stewart C, Volfovsky N, Levitsky A, Stephens RM, Coffin JM. 2013. Analysis of 454 sequencing error rate, error sources, and artifact recombination for detection of low-frequency drug resistance mutations in HIV-1 DNA. Retrovirology 10:18. 10.1186/1742-4690-10-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. 2007. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 17:1195–1201. 10.1101/gr.6468307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sato T, Matsuyama J, Kumagai T, Mayanagi G, Yamaura M, Washio J, Takahashi N. 2003. Nested PCR for detection of mutans streptococci in dental plaque. Lett. Appl. Microbiol. 37:66–69. 10.1046/j.1472-765X.2003.01359.x [DOI] [PubMed] [Google Scholar]
- 87.Garrido C, Soriano V, Geretti AM, Zahonero N, Garcia S, Booth C, Gutierrez F, Viciana I, de Mendoza C. 2011. Resistance associated mutations to dolutegravir (S/GSK1349572) in HIV-infected patients–impact of HIV subtypes and prior raltegravir experience. Antiviral Res. 90:164–167. 10.1016/j.antiviral.2011.03.178 [DOI] [PubMed] [Google Scholar]
- 88.Marx JL. 1989. Drug-resistant strains of AIDS virus found. Science 243:1551–1552. 10.1126/science.2928791 [DOI] [PubMed] [Google Scholar]
- 89.MacArthur RD. 2009. Understanding HIV phenotypic resistance testing: usefulness in managing treatment-experienced patients. AIDS Rev. 11:223–230 [PubMed] [Google Scholar]
- 90.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53–59. 10.1038/nature07517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, Hobolth A, Lappalainen T, Mailund T, Marques-Bonet T, McCarthy S, Montgomery SH, Schwalie PC, Tang YA, Ward MC, Xue Y, Yngvadottir B, Alkan C, Andersen LN, Ayub Q, Ball EV, Beal K, Bradley BJ, Chen Y, Clee CM, Fitzgerald S, Graves TA, Gu Y, Heath P, Heger A, Karakoc E, Kolb-Kokocinski A, Laird GK, Lunter G, Meader S, Mort M, Mullikin JC, Munch K, O'Connor TD, Phillips AD, Prado-Martinez J, Rogers AS, Sajjadian S, Schmidt D, Shaw K, Simpson JT, Stenson PD, Turner DJ, Vigilant L, Vilella AJ, et al. 2012. Insights into hominid evolution from the gorilla genome sequence. Nature 483:169–175. 10.1038/nature10842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chatterjee R, Hoffman M, Cliften P, Seshan S, Liapis H, Jain S. 2013. Targeted exome sequencing integrated with clinicopathological information reveals novel and rare mutations in atypical, suspected and unknown cases of Alport syndrome or proteinuria. PLoS One 8:e76360. 10.1371/journal.pone.0076360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, Hundal J, Wendl MC, Demeter R, Wylie T, Allison JP, Smyth MJ, Old LJ, Mardis ER, Schreiber RD. 2012. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 482:400–404. 10.1038/nature10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, Kalyana-Sundaram S, Sam L, Balbin OA, Quist MJ, Barrette T, Everett J, Siddiqui J, Kunju LP, Navone N, Araujo JC, Troncoso P, Logothetis CJ, Innis JW, Smith DC, Lao CD, Kim SY, Roberts JS, Gruber SB, Pienta KJ, Talpaz M, Chinnaiyan AM. 2011. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci. Transl. Med. 3:111ra121. 10.1126/scitranslmed.3003161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Howden BP, McEvoy CR, Allen DL, Chua K, Gao W, Harrison PF, Bell J, Coombs G, Bennett-Wood V, Porter JL, Robins-Browne R, Davies JK, Seemann T, Stinear TP. 2011. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog. 7:e1002359. 10.1371/journal.ppat.1002359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. 2011. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364:33–42. 10.1056/NEJMoa1012928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Umemura M, Koyama Y, Takeda I, Hagiwara H, Ikegami T, Koike H, Machida M. 2013. Fine de novo sequencing of a fungal genome using only SOLiD short read data: verification on Aspergillus oryzae RIB40. PLoS One 8:e63673. 10.1371/journal.pone.0063673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, NISC Comparative Sequencing Program. Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602. 10.1126/science.1207532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ, Ellison JA, Gilbert AT, Kuzmin IV, Niezgoda M, Osinubi MO, Recuenco S, Markotter W, Breiman RF, Kalemba L, Malekani J, Lindblade KA, Rostal MK, Ojeda-Flores R, Suzan G, Davis LB, Blau DM, Ogunkoya AB, Alvarez Castillo DA, Moran D, Ngam S, Akaibe D, Agwanda B, Briese T, Epstein JH, Daszak P, Rupprecht CE, Holmes EC, Lipkin WI. 2013. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. U. S. A. 110:8194–8199. 10.1073/pnas.1303037110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, Berlin AM, Malboeuf CM, Ryan EM, Gnerre S, Zody MC, Erlich RL, Green LM, Berical A, Wang Y, Casali M, Streeck H, Bloom AK, Dudek T, Tully D, Newman R, Axten KL, Gladden AD, Battis L, Kemper M, Zeng Q, Shea TP, Gujja S, Zedlack C, Gasser O, Brander C, Hess C, Günthard HF, Brumme ZL, Brumme CJ, Bazner S, Rychert J, Tinsley JP, Mayer KH, Rosenberg E, Pereyra F, Levin JZ, Young SK, Jessen H, Altfeld M, Birren BW, Walker BD, Allen TM. 2012. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 8:e1002529. 10.1371/journal.ppat.1002529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vandenbroucke I, Van Marck H, Mostmans W, Van Eygen V, Rondelez E, Thys K, Van Baelen K, Fransen K, Vaira D, Kabeya K, De Wit S, Florence E, Moutschen M, Vandekerckhove L, Verhofstede C, Stuyver LJ. 2010. HIV-1 V3 envelope deep sequencing for clinical plasma specimens failing in phenotypic tropism assays. AIDS Res. Ther. 7:4. 10.1186/1742-6405-7-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Raymond S, Saliou A, Nicot F, Delobel P, Dubois M, Cazabat M, Sandres-Sauné K, Marchou B, Massip P, Izopet J. 2011. Frequency of CXCR4-using viruses in primary HIV-1 infections using ultra-deep pyrosequencing. AIDS 25:1668–1670. 10.1097/QAD.0b013e3283498305 [DOI] [PubMed] [Google Scholar]
- 103.Abbate I, Vlassi C, Rozera G, Bruselles A, Bartolini B, Giombini E, Corpolongo A, D'Offizi G, Narciso P, Desideri A, Ippolito G, Capobianchi MR. 2011. Detection of quasispecies variants predicted to use CXCR4 by ultra-deep pyrosequencing during early HIV infection. AIDS 25:611–617. 10.1097/QAD.0b013e328343489e [DOI] [PubMed] [Google Scholar]
- 104.Lee GQ, Harrigan PR, Dong W, Poon AF, Heera J, Demarest J, Rinehart A, Chapman D, Valdez H, Portsmouth S. 2013. Comparison of population and 454 “deep” sequence analysis for HIV type 1 tropism versus the original Trofile assay in non-B subtypes. AIDS Res. Hum. Retroviruses 29:979–984. 10.1089/aid.2012.0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Messiaen P, Verhofstede C, Vandenbroucke I, Dinakis S, Van Eygen V, Thys K, Winters B, Aerssens J, Vogelaers D, Stuyver LJ, Vandekerckhove L. 2012. Ultra-deep sequencing of HIV-1 reverse transcriptase before start of an NNRTI-based regimen in treatment-naive patients. Virology 426:7–11. 10.1016/j.virol.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 106.Armenia D, Vandenbroucke I, Fabeni L, Van Marck H, Cento V, D'Arrigo R, Van Wesenbeeck L, Scopelliti F, Micheli V, Bruzzone B, Lo Caputo S, Aerssens J, Rizzardini G, Tozzi V, Narciso P, Antinori A, Stuyver L, Perno CF, Ceccherini-Silberstein F. 2012. Study of genotypic and phenotypic HIV-1 dynamics of integrase mutations during raltegravir treatment: a refined analysis by ultra-deep 454 pyrosequencing. J. Infect. Dis. 205:557–567. 10.1093/infdis/jir821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mukherjee R, Jensen ST, Male F, Bittinger K, Hodinka RL, Miller MD, Bushman FD. 2011. Switching between raltegravir resistance pathways analyzed by deep sequencing. AIDS 25:1951–1959. 10.1097/QAD.0b013e32834b34de [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu J, Miller MD, Danovich RM, Vandergrift N, Cai F, Hicks CB, Hazuda DJ, Gao F. 2011. Analysis of low-frequency mutations associated with drug resistance to raltegravir before antiretroviral treatment. Antimicrob. Agents Chemother. 55:1114–1119. 10.1128/AAC.01492-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gall A, Ferns B, Morris C, Watson S, Cotten M, Robinson M, Berry N, Pillay D, Kellam P. 2012. Universal amplification, next-generation sequencing, and assembly of HIV-1 genomes. J. Clin. Microbiol. 50:3838–3844. 10.1128/JCM.01516-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. 10.1186/gb-2007-8-7-r143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. 2012. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 30:434–439. 10.1038/nbt.2198 [DOI] [PubMed] [Google Scholar]
- 112.Brodin J, Mild M, Hedskog C, Sherwood E, Leitner T, Andersson B, Albert J. 2013. PCR-induced transitions are the major source of error in cleaned ultra-deep pyrosequencing data. PLoS One 8:e70388. 10.1371/journal.pone.0070388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rozera G, Abbate I, Bruselles A, Vlassi C, D'Offizi G, Narciso P, Chillemi G, Prosperi M, Ippolito G, Capobianchi MR. 2009. Massively parallel pyrosequencing highlights minority variants in the HIV-1 env quasispecies deriving from lymphomonocyte sub-populations. Retrovirology 6:15. 10.1186/1742-4690-6-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zagordi O, Däumer M, Beisel C, Beerenwinkel N. 2012. Read length versus depth of coverage for viral quasispecies reconstruction. PLoS One 7:e47046. 10.1371/journal.pone.0047046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Willerth SM, Pedro HA, Pachter L, Humeau LM, Arkin AP, Schaffer DV. 2010. Development of a low bias method for characterizing viral populations using next generation sequencing technology. PLoS One 5:e13564. 10.1371/journal.pone.0013564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Church JD, Huang W, Parkin N, Marlowe N, Guay LA, Omer SB, Musoke P, Jackson JB, Eshleman SH. 2009. Comparison of laboratory methods for analysis of non-nucleoside reverse transcriptase inhibitor resistance in Ugandan infants. AIDS Res. Hum. Retroviruses 25:657–663. 10.1089/aid.2008.0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gallego O, Martin-Carbonero L, Aguero J, de Mendoza C, Soriano V. 2004. Correlation between rules-based interpretation and virtual phenotype interpretation of HIV-1 genotypes for predicting drug resistance in HIV-infected individuals. J. Virol. Methods 121:115–118 [DOI] [PubMed] [Google Scholar]
- 118.Dunne AL, Mitchell FM, Coberly SK, Hellmann NS, Hoy J, Mijch A, Petropoulos CJ, Mills J, Crowe SM. 2001. Comparison of genotyping and phenotyping methods for determining susceptibility of HIV-1 to antiretroviral drugs. AIDS 15:1471–1475. 10.1097/00002030-200108170-00003 [DOI] [PubMed] [Google Scholar]
- 119.Qari SH, Respess R, Weinstock H, Beltrami EM, Hertogs K, Larder BA, Petropoulos CJ, Hellmann N, Heneine W. 2002. Comparative analysis of two commercial phenotypic assays for drug susceptibility testing of human immunodeficiency virus type 1. J. Clin. Microbiol. 40:31–35. 10.1128/JCM.40.1.31-35.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang K, Samudrala R, Mittler JE. 2004. Antivirogram or PhenoSense: a comparison of their reproducibility and an analysis of their correlation. Antivir. Ther. 9:703–712 [PubMed] [Google Scholar]
- 121.Skrabal K, Low AJ, Dong W, Sing T, Cheung PK, Mammano F, Harrigan PR. 2007. Determining human immunodeficiency virus coreceptor use in a clinical setting: degree of correlation between two phenotypic assays and a bioinformatic model. J. Clin. Microbiol. 45:279–284. 10.1128/jcm.01118-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.de Mendoza C, Van Baelen K, Poveda E, Rondelez E, Zahonero N, Stuyver L, Garrido C, Villacian J, Soriano V. 2008. Performance of a population-based HIV-1 tropism phenotypic assay and correlation with V3 genotypic prediction tools in recent HIV-1 seroconverters. J. Acquir. Immune Defic. Syndr. 48:241–244. 10.1097/QAI.0b013e3181734f03 [DOI] [PubMed] [Google Scholar]
- 123.Raymond S, Delobel P, Mavigner M, Cazabat M, Souyris C, Sandres-Sauné K, Cuzin L, Marchou B, Massip P, Izopet J. 2008. Correlation between genotypic predictions based on V3 sequences and phenotypic determination of HIV-1 tropism. AIDS 22:F11–F16. 10.1097/QAD.0b013e32830ebcd4 [DOI] [PubMed] [Google Scholar]
- 124.Trabaud MA, Icard V, Scholtes C, Perpoint T, Koffi J, Cotte L, Makhloufi D, Tardy JC, André P. 2012. Discordance in HIV-1 co-receptor use prediction by different genotypic algorithms and phenotype assay: intermediate profile in relation to concordant predictions. J. Med. Virol. 84:402–413. 10.1002/jmv.23209 [DOI] [PubMed] [Google Scholar]
- 125.Pou C, Codoñer FM, Thielen A, Bellido R, Pérez-Álvarez S, Cabrera C, Dalmau J, Curriu M, Lie Y, Noguera-Julian M, Puig J, Martínez-Picado J, Blanco J, Coakley E, Däumer M, Clotet B, Paredes R. 2013. HIV-1 tropism testing in subjects achieving undetectable HIV-1 RNA: diagnostic accuracy, viral evolution and compartmentalization. PLoS One 8:e67085. 10.1371/journal.pone.0067085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gonzalez-Serna A, McGovern RA, Harrigan PR, Vidal F, Poon AF, Ferrando-Martinez S, Abad MA, Genebat M, Leal M, Ruiz-Mateos E. 2012. Correlation of the virological response to short-term maraviroc monotherapy with standard and deep-sequencing-based genotypic tropism prediction methods. Antimicrob. Agents Chemother. 56:1202–1207. 10.1128/AAC.05857-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li JZ, Paredes R, Ribaudo HJ, Svarovskaia ES, Metzner KJ, Kozal MJ, Hullsiek KH, Balduin M, Jakobsen MR, Geretti AM, Thiebaut R, Ostergaard L, Masquelier B, Johnson JA, Miller MD, Kuritzkes DR. 2011. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA 305:1327–1335. 10.1001/jama.2011.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li JZ, Paredes R, Ribaudo HJ, Kozal MJ, Svarovskaia ES, Johnson JA, Geretti AM, Metzner KJ, Jakobsen MR, Hullsiek KH, Ostergaard L, Miller MD, Kuritzkes DR. 2013. Impact of minority nonnucleoside reverse transcriptase inhibitor resistance mutations on resistance genotype after virologic failure. J. Infect. Dis. 207:893–897. 10.1093/infdis/jis925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Codoñer FM, Pou C, Thielen A, García F, Delgado R, Dalmau D, Álvarez-Tejado M, Ruiz L, Clotet B, Paredes R. 2011. Added value of deep sequencing relative to population sequencing in heavily pre-treated HIV-1-infected subjects. PLoS One 6:e19461. 10.1371/journal.pone.0019461 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.