Abstract
The discovery of new compounds that are able to inhibit the growth of Burkholderia cenocepacia is of primary importance for cystic fibrosis patients. Here, the mechanism of resistance to a new pyridine derivative already shown to be effective against Mycobacterium tuberculosis and to have good activity toward B. cenocepacia was investigated. Increased expression of a resistance-nodulation-cell division (RND) efflux system was detected in the resistant mutants, thus confirming their important roles in B. cenocepacia antibiotic resistance.
TEXT
Respiratory tract infections are the main cause of mortality in cystic fibrosis (CF) patients (1). Burkholderia cenocepacia is frequently associated with reduced survival and a high risk of developing cepacia syndrome, a fulminating pneumonia (2).
Up to now, a few antibiotics have been effective for treating B. cenocepacia infections, such as meropenem plus ceftazidime, chloramphenicol, trimethoprim-sulfamethoxazole, and aztreonam (3, 4). In particular, combination therapy is recommended, since the bacteria causing these infections show a high level of intrinsic resistance (5). These treatments have had a substantial impact on the quality of life and survival of CF patients; however, the increasing diffusion of resistant bacteria requires new and alternative drugs for treatment.
Our laboratory is focused on the mechanisms of resistance mediated by resistance-nodulation-cell division (RND) efflux systems. These proteins have been shown to play a prominent role in drug resistance in Pseudomonas aeruginosa and Burkholderia pseudomallei, while their role in B. cenocepacia drug resistance has been only partially described (6, 7). In particular, we demonstrated that the deletion of the RND-4 system (BCAL2820 to BCAL2822) led to increased sensitivities to different antimicrobial compounds, suggesting its involvement in intrinsic drug resistance (8, 9).
This work describes a promising new molecule, a 2-thiopyridine derivative named 1-oxido-5-(trifluoromethyl)pyridin-2-yl thiocyanate (11026103) (Fig. 1), which is particularly active against B. cenocepacia growth and was recently shown to be effective against Mycobacterium tuberculosis (10). This compound was selected after a screening of >60 compounds, which were tested against B. cenocepacia strain J2315. For each compound, the MIC was evaluated, but only 11026103 was found to have promising antibacterial activity, with an MIC of 16 μg/ml (Table 1).
FIG 1.
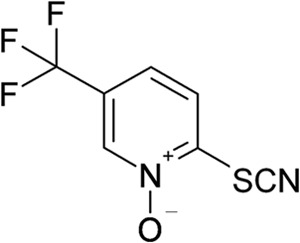
Chemical structure of compound 11026103.
TABLE 1.
MICs of 11026103
WT, wild type.
See Buroni et al. (8).
B. cenocepacia J2315 cells were spread on plates containing the 11026103 molecule at concentrations ranging from 4 to 10× the MIC. Two spontaneous resistant mutants were isolated through direct selection on solid medium containing 64 μg/ml of compound (4× the MIC) (Table 1). These mutant strains were named CU4 and CU14. The mutation frequency toward 11026103 was 10−10.
In order to understand which mutations were responsible for the resistance of these mutants, the genomic DNA of the mutant strains was extracted and sequenced by an Illumina Solexa method. The obtained reads were then mapped against the B. cenocepacia J2315 reference genome. The mutations with ≥60% frequency of variant alleles and leading to a change to a different amino acid or to the synthesis of a truncated protein were analyzed. In both mutants, one mutation was identified in the BCAL2823 gene. The BCAL2823 (bpeR) gene encodes a 213-amino acid protein that belongs to the TetR family of transcriptional regulators, localized next to the RND-4 efflux system (Fig. 1A). The mutation identified in the CU4 strain is the deletion of A250, which caused a frameshift in the nucleotide sequence, thus resulting in an early stop codon and a truncated protein of only 92 amino acids. In the CU14 mutant, the mutation was the substitution T458C, which led to the amino acid change L153P, with the two amino acids having different chemical properties.
In order to determine whether the resistance to 11026103 was associated with an upregulation of the RND-4 system due to mutations in its negative regulator, quantitative real-time PCR (RT-PCR) experiments were performed on both resistant strains. The results showed that compared to the wild type, the RND-4 (BCAL2821) gene was upregulated 4-fold in CU4 and 2-fold in CU14, respectively. As the overexpression detected in the CU14 mutant is very low, only CU4 was considered for further analyses.
Since the overexpression of efflux pumps can cause resistances to different compounds (11), the MICs of the most commonly used antibiotics were assessed for the CU4 strain. In particular, the MICs of chloramphenicol and nalidixic acid increased 8-fold in the mutant compared to the wild type, the MIC of levofloxacin increased 4-fold compared to the wild type, and the MICs of ciprofloxacin, norfloxacin, and sparfloxacin were 2-fold higher in the mutant. These results confirm that RND-4 overexpression is able to confer resistances to different compounds, as was already shown (8), and they suggest that this transporter is also involved in the 11026103 efflux.
To further confirm this hypothesis, the MIC of 11026103 was also assessed for the RND-4 deleted strain, which was 2-fold less resistant than the wild-type strain (Table 1).
To characterize the mechanism of resistance to the 11026103 drug, electrophoretic mobility shift assay (EMSA) experiments were performed using the TetR repressor (BCAL2823) purified from both B. cenocepacia J2315 and the CU4 mutant. The BCAL2823 coding region was cloned into the expression vector pET-28a (Novagen) and transformed in Escherichia coli BL21(DE3). Protein expression was obtained with the ZYP-5052 autoinducing medium (12). The recombinant proteins were purified by affinity chromatography on nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen).
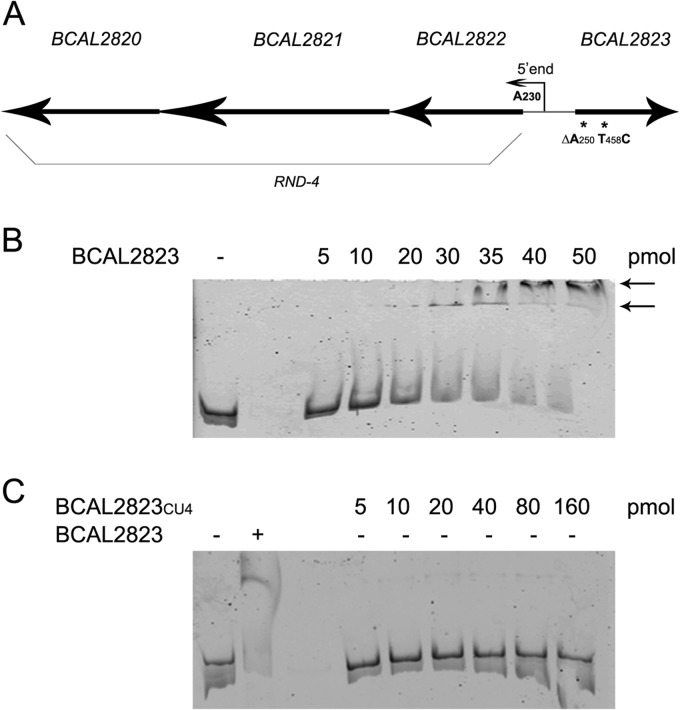
The 400-bp intergenic region located between the BCAL2822 and BCAL2823 genes (Fig. 2A) was used for DNA-binding assays and 5′-labeled with 6-carboxyfluorescein (6-FAM). The 20-μl binding reaction buffer was composed of 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1.25 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM EDTA, 3 μg bovine serum albumin, 1 μg salmon sperm DNA, and 10% glycerol. The labeled DNA region was used at 3.39 nM, with increasing concentrations of both BCAL2823 and BCAL2823CU4 (Fig. 2B and C). Specificity was demonstrated by performing the experiment with increasing concentrations of unlabeled probe (data not shown). The results showed that the BCAL2823 protein purified from the wild-type strain was able to bind to the 400-bp intergenic region at a concentration of 30 pmol (Fig. 2B, arrows), while the truncated protein purified from the CU4 mutant was unable to bind the same intergenic region even at higher concentrations (160 pmol) (Fig. 2C). Hence, as hypothesized, if BCAL2823 does not work correctly as a transcriptional repressor, an ineffective negative regulation might allow overexpression of the RND-4 operon, leading to drug resistance.
FIG 2.
Genetic organization and EMSAs with BCAL2823 and BCAL2823CU4. (A) Genetic organization of RND-4 operon and BCAL2823. RND-4 transcriptional start site position is indicated by the smaller arrow. The mutations in BCAL2823 are marked with asterisks. (B) A 3.39 nM 6-FAM-labeled probe was used, with increasing amounts of BCAL2823 (indicated above each lane). The bound BCAL2823 is indicated by arrows. −, control. (C) A 3.39 nM 6-FAM-labeled probe was used, with increasing amounts of BCAL2823CU4 (indicated above each lane) or 35 pmol BCAL2823. − and + indicate absence or presence, respectively, of BCAL2823 protein.
The transcriptional start site of the RND-4 operon was then mapped by 5′-Rapid amplification of cDNA Ends (RACE) technique. The reaction was made with ImProm-II reverse transcriptase (Promega) using an adaptor primer (5′-GACCACGCGTATCGATGTCGACT16-3′) and a specific primer. The 5′ end resulted in the A being located 230 bp upstream of the translational start site of the BCAL2822 gene (Fig. 2A).
To further confirm that RND-4 was responsible for resistance to 11026103, a wild-type copy of BCAL2823 was cloned into pAP20 constitutive expression vector (kindly provided by Miguel Valvano). The B. cenocepacia CU4 mutant was transformed with the recombinant vector by triparental mating (13), and the MIC of 11026103 was determined for both the CU4 strain transformed with the empty vector and for the same strain carrying the BCAL2823/pAP20 vector. The MIC of the mutant transformed with the wild-type copy of BCAL2823 was the same as that of the wild-type B. cenocepacia J2315, indicating that complementation occurred. This was further confirmed by a quantitative RT-PCR experiment, which was conducted also on the complemented strain, and the value achieved was the same as for the wild type.
In conclusion, a new compound, which has been shown to be effective against M. tuberculosis, has been demonstrated to be promising also for the treatment of B. cenocepacia infections. This is very important for CF patients, as the current therapy is not effective. Moreover, the RND-4 transporter was confirmed to have a prominent role as an efflux transporter in this bacterium, and in the future, it will be important to find drugs that are able to block this pump and not be extruded by this protein.
ACKNOWLEDGMENT
This work was supported by the Italian Cystic Fibrosis Research Foundation (FFC project no. 10/2012 to G. Riccardi, adopted by the Associazione Trentina FC onlus in ricordo di Zaira Tutino, Gruppo di Sostegno FFC di Palermo-in ricordo di Elisa Pepe, Delegazione FFC di Imola).
Footnotes
Published ahead of print 6 January 2014
REFERENCES
- 1.LiPuma JJ. 2010. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 23:299–323. 10.1128/CMR.00068-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones AM, Dodd ME, Govan JRW, Barcus V, Doherty CJ, Morris J, Webb AK. 2004. Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax 59:948–951. 10.1136/thx.2003.017210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson RL, Burns JL, Ramsey BW. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918–951. 10.1164/rccm.200304-505SO [DOI] [PubMed] [Google Scholar]
- 4.Waters V. 2012. New treatments for emerging cystic fibrosis pathogens other than Pseudomonas. Curr. Pharm. Des. 18:696–725. 10.2174/138161212799315939 [DOI] [PubMed] [Google Scholar]
- 5.Aaron SD, Ferris W, Henry DA, Speert DP, Macdonald NE. 2000. Multiple combination bactericidal antibiotic testing for patients with cystic fibrosis infected with Burkholderia cepacia. Am. J. Respir. Crit. Care Med. 161:1206–1212. 10.1164/ajrccm.161.4.9907147 [DOI] [PubMed] [Google Scholar]
- 6.Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren MS, Boyer E, Chamberland S, Lee VJ. 1999. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:1340–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan YY, Tan TM, Ong YM, Chua KL. 2004. BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 48:1128–1135. 10.1128/AAC.48.4.1128-1135.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buroni S, Pasca MR, Flannagan RS, Bazzini S, Milano A, Bertani I, Venturi V, Valvano MA, Riccardi G. 2009. Assessment of three resistance-nodulation-cell division drug efflux transporters of Burkholderia cenocepacia in intrinsic antibiotic resistance. BMC Microbiol. 9:200. 10.1186/1471-2180-9-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bazzini S, Udine C, Sass A, Pasca MR, Longo F, Emiliani G, Fondi M, Perrin E, Decorosi F, Viti C, Giovannetti L, Leoni L, Fani R, Riccardi G, Mahenthiralingam E, Buroni S. 2011. Deciphering the role of RND efflux transporters in Burkholderia cenocepacia. PLoS One 6:e18902. 10.1371/journal.pone.0018902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salina E, Ryabova O, Kaprelyants A, Makarov V. 2013. New 2-thiopyridines as potential candidates for killing both actively growing and dormant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58:55–60. 10.1128/AAC.01308-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikaido H, Pagès JM. 2012. Broad specificity efflux pumps and their role in multidrug resistance of Gram negative bacteria. FEMS Microbiol. Rev. 36:340–363. 10.1111/j.1574-6976.2011.00290.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234. 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 13.Craig FF, Coote JG, Parton R, Freer JH, Gilmour NJ. 1989. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 135:2885–2890 [DOI] [PubMed] [Google Scholar]