Abstract
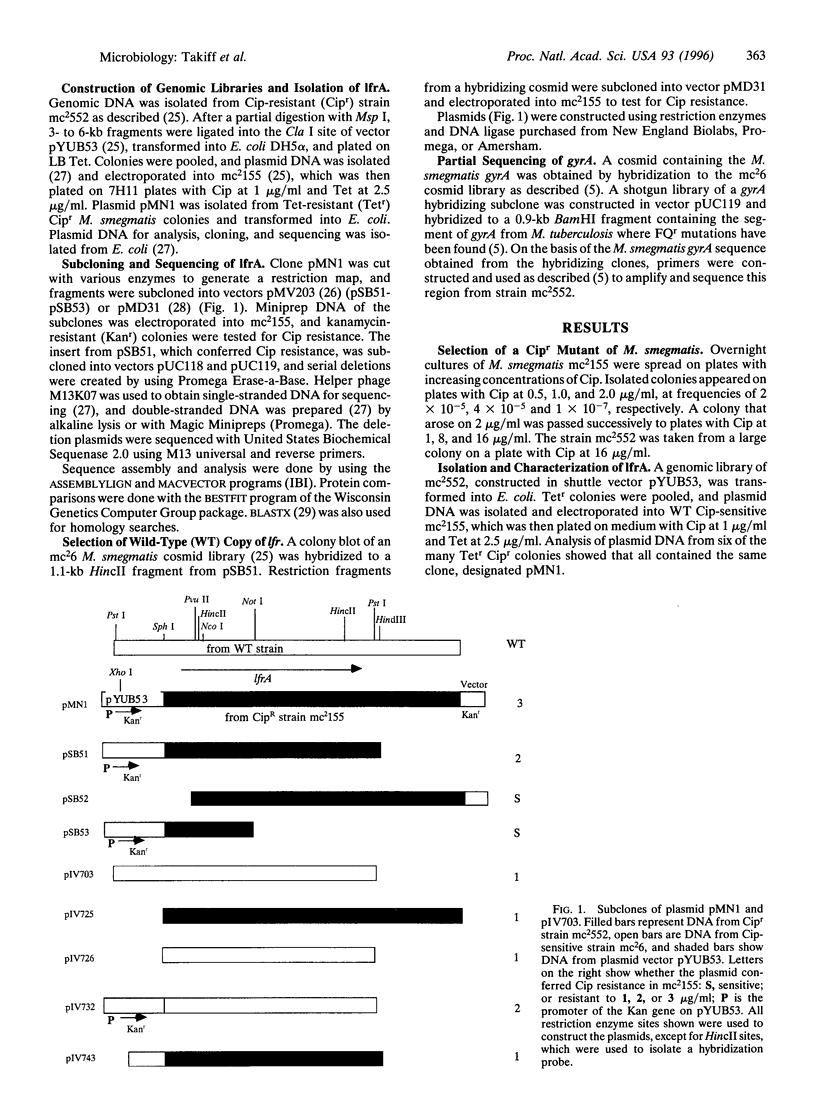
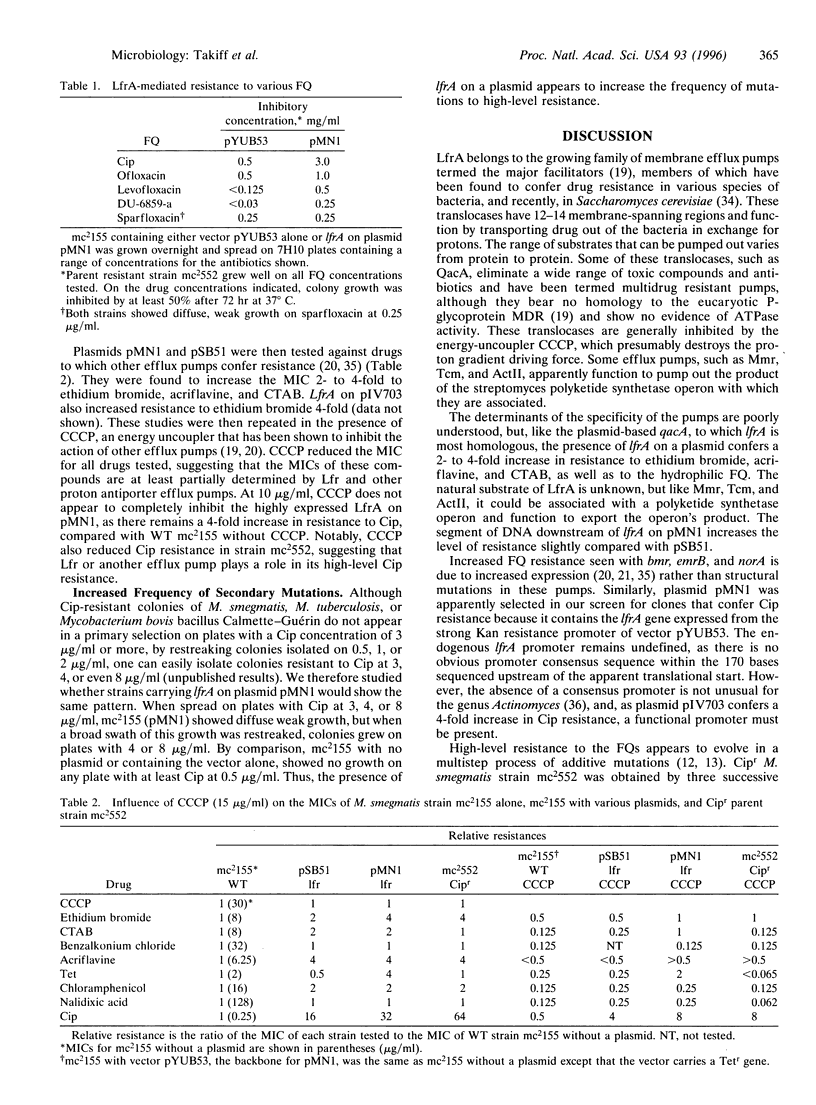
Due to the resurgence of tuberculosis and the emergence of multidrug-resistant strains, fluoroquinolones (FQ) are being used in selected tuberculosis patients, but FQ-resistant strains of Mycobacterium tuberculosis have rapidly begun to appear. The mechanisms involved in FQ resistance need to be elucidated if the effectiveness of this class of antibiotics is to be improved and prolonged. By using the rapid-growing Mycobacterium smegmatis as a model genetic system, a gene was selected that confers low-level FQ resistance when present on a multicopy plasmid. This gene, lfrA, encodes a putative membrane efflux pump of the major facilitator family, which appears to recognize the hydrophilic FQ, ethidium bromide, acridine, and some quaternary ammonium compounds. It is homologous to qacA from Staphylococcus aureus, tcmA, of Streptomyces glaucescens, and actII and mmr, both from Streptomyces coelicoler. Increased expression of lfrA augments the appearance of subsequent mutations to higher-level FQ resistance.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams D. E., Shekhtman E. M., Zechiedrich E. L., Schmid M. B., Cozzarelli N. R. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992 Oct 16;71(2):277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ball P. Emergent resistance to ciprofloxacin amongst Pseudomonas aeruginosa and Staphylococcus aureus: clinical significance and therapeutic approaches. J Antimicrob Chemother. 1990 Dec;26 (Suppl F):165–179. doi: 10.1093/jac/26.suppl_f.165. [DOI] [PubMed] [Google Scholar]
- Belland R. J., Morrison S. G., Ison C., Huang W. M. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol Microbiol. 1994 Oct;14(2):371–380. doi: 10.1111/j.1365-2958.1994.tb01297.x. [DOI] [PubMed] [Google Scholar]
- Cohen S. P., Hächler H., Levy S. B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993 Mar;175(5):1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker H., Motamedi H., Hutchinson C. R. Nucleotide sequences and heterologous expression of tcmG and tcmP, biosynthetic genes for tetracenomycin C in Streptomyces glaucescens. J Bacteriol. 1993 Jun;175(12):3876–3886. doi: 10.1128/jb.175.12.3876-3886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly-Wu M. K., Jacobs W. R., Jr, Hatfull G. F. Superinfection immunity of mycobacteriophage L5: applications for genetic transformation of mycobacteria. Mol Microbiol. 1993 Feb;7(3):407–417. doi: 10.1111/j.1365-2958.1993.tb01132.x. [DOI] [PubMed] [Google Scholar]
- Ehrenhofer-Murray A. E., Würgler F. E., Sengstag C. The Saccharomyces cerevisiae SGE1 gene product: a novel drug-resistance protein within the major facilitator superfamily. Mol Gen Genet. 1994 Aug 2;244(3):287–294. doi: 10.1007/BF00285456. [DOI] [PubMed] [Google Scholar]
- Fernández-Moreno M. A., Caballero J. L., Hopwood D. A., Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell. 1991 Aug 23;66(4):769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- Ferrero L., Cameron B., Manse B., Lagneaux D., Crouzet J., Famechon A., Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol Microbiol. 1994 Aug;13(4):641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- Fischl M. A., Daikos G. L., Uttamchandani R. B., Poblete R. B., Moreno J. N., Reyes R. R., Boota A. M., Thompson L. M., Cleary T. J., Oldham S. A. Clinical presentation and outcome of patients with HIV infection and tuberculosis caused by multiple-drug-resistant bacilli. Ann Intern Med. 1992 Aug 1;117(3):184–190. doi: 10.7326/0003-4819-117-3-184. [DOI] [PubMed] [Google Scholar]
- Gocke E. Mechanism of quinolone mutagenicity in bacteria. Mutat Res. 1991 May;248(1):135–143. doi: 10.1016/0027-5107(91)90095-6. [DOI] [PubMed] [Google Scholar]
- Heifets L. B. Antimycobacterial drugs. Semin Respir Infect. 1994 Jun;9(2):84–103. [PubMed] [Google Scholar]
- Heisig P., Tschorny R. Characterization of fluoroquinolone-resistant mutants of escherichia coli selected in vitro. Antimicrob Agents Chemother. 1994 Jun;38(6):1284–1291. doi: 10.1128/aac.38.6.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai K., Aoyama H., Suzue S., Irikura T., Iyobe S., Mitsuhashi S. Isolation and characterization of norfloxacin-resistant mutants of Escherichia coli K-12. Antimicrob Agents Chemother. 1986 Aug;30(2):248–253. doi: 10.1128/aac.30.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper D. C., Wolfson J. S. Fluoroquinolone antimicrobial agents. N Engl J Med. 1991 Feb 7;324(6):384–394. doi: 10.1056/NEJM199102073240606. [DOI] [PubMed] [Google Scholar]
- Hooper D. C., Wolfson J. S., Ng E. Y., Swartz M. N. Mechanisms of action of and resistance to ciprofloxacin. Am J Med. 1987 Apr 27;82(4A):12–20. [PubMed] [Google Scholar]
- Ishida H., Fuziwara H., Kaibori Y., Horiuchi T., Sato K., Osada Y. Cloning of multidrug resistance gene pqrA from Proteus vulgaris. Antimicrob Agents Chemother. 1995 Feb;39(2):453–457. doi: 10.1128/aac.39.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs W. R., Jr, Kalpana G. V., Cirillo J. D., Pascopella L., Snapper S. B., Udani R. A., Jones W., Barletta R. G., Bloom B. R. Genetic systems for mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- Jarlier V., Nikaido H. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol Lett. 1994 Oct 15;123(1-2):11–18. doi: 10.1111/j.1574-6968.1994.tb07194.x. [DOI] [PubMed] [Google Scholar]
- Kato J., Suzuki H., Ikeda H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J Biol Chem. 1992 Dec 25;267(36):25676–25684. [PubMed] [Google Scholar]
- Levy S. B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992 Apr;36(4):695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z., Livermore D. M., Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob Agents Chemother. 1994 Aug;38(8):1732–1741. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya O., Lewis K. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Cook D. N., Hearst J. E., Nikaido H. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 1994 Dec;2(12):489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- Miller P. F., Gambino L. F., Sulavik M. C., Gracheck S. J. Genetic relationship between soxRS and mar loci in promoting multiple antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother. 1994 Aug;38(8):1773–1779. doi: 10.1128/aac.38.8.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal R. J., Chater K. F. Nucleotide sequence analysis reveals similarities between proteins determining methylenomycin A resistance in Streptomyces and tetracycline resistance in eubacteria. Gene. 1987;58(2-3):229–241. doi: 10.1016/0378-1119(87)90378-7. [DOI] [PubMed] [Google Scholar]
- Neyfakh A. A., Bidnenko V. E., Chen L. B. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng E. Y., Trucksis M., Hooper D. C. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob Agents Chemother. 1994 Jun;38(6):1345–1355. doi: 10.1128/aac.38.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994 Apr 15;264(5157):382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- Rouch D. A., Cram D. S., DiBerardino D., Littlejohn T. G., Skurray R. A. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol Microbiol. 1990 Dec;4(12):2051–2062. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- Silver L. L., Bostian K. A. Discovery and development of new antibiotics: the problem of antibiotic resistance. Antimicrob Agents Chemother. 1993 Mar;37(3):377–383. doi: 10.1128/aac.37.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover C. K., de la Cruz V. F., Fuerst T. R., Burlein J. E., Benson L. A., Bennett L. T., Bansal G. P., Young J. F., Lee M. H., Hatfull G. F. New use of BCG for recombinant vaccines. Nature. 1991 Jun 6;351(6326):456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- Strohl W. R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992 Mar 11;20(5):961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan E. A., Kreiswirth B. N., Palumbo L., Kapur V., Musser J. M., Ebrahimzadeh A., Frieden T. R. Emergence of fluoroquinolone-resistant tuberculosis in New York City. Lancet. 1995 May 6;345(8958):1148–1150. doi: 10.1016/s0140-6736(95)90980-x. [DOI] [PubMed] [Google Scholar]
- Takiff H. E., Salazar L., Guerrero C., Philipp W., Huang W. M., Kreiswirth B., Cole S. T., Jacobs W. R., Jr, Telenti A. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994 Apr;38(4):773–780. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucksis M., Hooper D. C., Wolfson J. S. Emerging resistance to fluoroquinolones in staphylococci: an alert. Ann Intern Med. 1991 Mar 1;114(5):424–426. doi: 10.7326/0003-4819-114-5-424. [DOI] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Bogaki M., Nakamura M., Yamanaka L. M., Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother. 1991 Aug;35(8):1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Bogaki M., Nakamura S., Ubukata K., Konno M. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol. 1990 Dec;172(12):6942–6949. doi: 10.1128/jb.172.12.6942-6949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]




