Abstract
Proteins are not monolithic entities; rather, they can contain multiple domains that mediate distinct interactions, and their functionality can be regulated through post-translational modifications at multiple distinct sites. Traditionally, network biology has ignored such properties of proteins and has instead examined either the physical interactions of whole proteins or the consequences of removing entire genes. In this Review, we discuss experimental and computational methods to increase the resolution of protein– protein, genetic and drug–gene interaction studies to the domain and residue levels. Such work will be crucial for using interaction networks to connect sequence and structural information, and to understand the biological consequences of disease-associated mutations, which will hopefully lead to more effective therapeutic strategies.
A central challenge in biology is to understand how genotypes map to phenotypes. This mapping is complicated by the fact that genes, and their protein products, do not function independently of each other. Perturbations of multiple distinct genes can result in similar phenotypes (known as locus heterogeneity), whereas some phenotypes may only be observed in the presence of combinatorial perturbations (a type of epistasis1). Consequently, in order to understand the genotype-to-phenotype problem, it would be beneficial to study proteins and genes in a network context. Since the turn of the century, high-throughput interaction mapping has emerged as an extremely useful approach for providing this context. Large-scale networks have been generated in various model organisms, documenting which proteins physically interact, which gene pairs functionally interact and which genes functionally interact with specific drugs (TABLE 1). These networks have been enormously valuable both for understanding the function of individual genes2 and for elucidating the organizing principles of biological systems3 (FIG. 1). However, a necessary limitation of most large-scale network biology screens is that they treat proteins and genes as simple monolithic nodes in a network. In reality, most proteins are composed of multiple domains and peptide motifs that can bind to distinct partners4. Furthermore, their activity and cellular localization can be dynamically regulated by various post-translational modifications (PTMs)5. Such structural features of proteins are generally ignored by protein interaction screens, in which the typical reported result is of the form ‘protein A interacts with proteins B and C’. Similarly, genetic and drug–gene (chemogenetic) interaction screens typically report on the consequences of removing a gene altogether rather than on the effect of mutating specific residues. In isolation, these approaches can be used to assign function to whole genes or proteins but not to specific regions or residues. However, mapping at such a high resolution is necessary for understanding how protein structure relates to function. It is also necessary for elucidating how different mutations of the same gene may result in different phenotypic outcomes, which is particularly important in the context of understanding the consequences of genome sequence variation. Indeed, mutations that result in the complete loss of a gene or severe truncation of a protein are much rarer than those that alter a single nucleotide or residue6 (BOX 1).
Table 1.
Interaction networks in selected model organisms and in humans
| Species | Network type* | Details | Refs |
|---|---|---|---|
| Saccharomyces cerevisiae | Y2H | ~3,000 interactions; ~2,000 proteins | 14 |
| AP–MS | ~7,000 interactions; ~2,700 proteins | 16 | |
| AP–MS | ~500 complexes; ~2,700 proteins | 17 | |
| Drug–gene | ~6,000 genes; ~400 drugs or conditions | 152 | |
| Genetic | ~5.4 million measured interactions; ~4,500 genes | 40 | |
| Schizosaccharomyces pombe | Genetic | ~1.6 million measured interactions; ~2,400 genes | 38 |
| Drug–gene | ~440 genes; 21 drugs or conditions | 64 | |
| Drug–gene | ~2,500 genes; 6 drugs or conditions | 153 | |
| Caenorhabditis elegans | Genetic | ~65,000 measured interactions; ~162 genes | 50 |
| Y2H | ~3,800 interactions; ~2,600 proteins | 154 | |
| Drosophila melanogaster | AP–MS | ~550 complexes; ~5,000 proteins | 155 |
| Y2H | ~4,800 filtered interactions; ~4,700 proteins | 156 | |
| Genetic | ~30,000 measured interactions; 93 genes | 46 | |
| Genetic | ~17,000 measured interactions; ~500 genes | 157 | |
| Escherichia coli | AP–MS | ~6,000 interactions; ~1,800 proteins | 158 |
| Genetic | ~235,000 measured interactions; ~820 genes | 39 | |
| Drug–gene | ~4,000 genes; 324 drugs or conditions | 36 | |
| Homo sapiens | Fractionation–mass spectrometry | ~14,000 interactions; ~3,000 proteins | 159 |
| Y2H | ~3,200 interactions; ~1,700 proteins | 160 | |
| Y2H | ~2,800 interactions; ~1,500 proteins | 161 | |
| Drug–gene | 70 genes; 87 drugs | 145 | |
| Genetic | 878 validated interactions; 12 genes, each tested for interactions using genome-wide RNA interference | 47 | |
| Genetic | Pairwise genetic interactions among a set of 60 genes through double knockdown using RNA interference | 45 |
AP–MS, affinity purification–mass spectrometry; Y2H, yeast two-hybrid.
In cases in which multiple networks of the same type were available for a single species, details of the largest network are provided. For cases in which no network was clearly larger, both networks are included.
Figure 1. Interaction networks.
a | In protein–protein interactions, protein A interacts with proteins B, C, D and E, either directly (top panel) or within a complex (bottom panel). b | In genetic interactions, genes A and B operate in a parallel pathway to genes C and D, whereas genes E and F operate in a linear pathway or complex. c | In drug–gene (chemogenetic) interactions, genes A and B operate in parallel to a pathway (involving genes C and D) that is inhibited by drug G. Gene E works in a linear pathway with gene F that is inhibited by drug H. d | Profile similarity is shown. Rows represent genes and columns represent either genes (for genetic interaction screens) or drugs (for drug–gene interaction screens). Coloured squares display negative (blue), positive (yellow) or neutral (black) interaction scores. Genes A, B and C all have similar interaction profiles, which suggests that they function in the same pathway or complex. In an analogous manner, genes D and E have similar interaction profiles, which suggests that they function together. The tree on the right indicates a hierarchical clustering of the profiles.
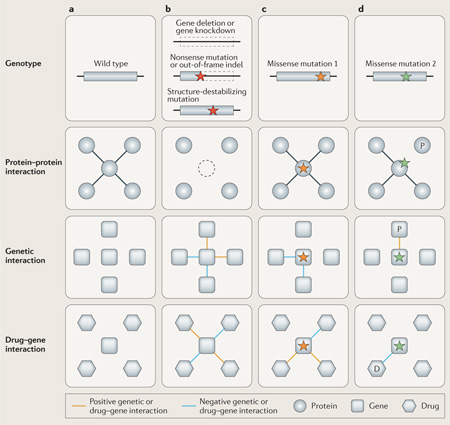
How sequence variation has an effect on proteins.
High-throughput sequencing has facilitated the rapid collection of genetic variation across human genomes. This includes both germline variation that is heritable and somatic mutations that occur in certain cell lineages (for example, as precursors to cancer126). Coupled with technologies that enrich DNA samples for protein-coding regions of the genome, exome sequencing has provided a wealth of information about genetic variants that potentially affect protein function. For somatic mutations in cancer, the range of effects is highly diverse and dependent on both cancer type and exposure to carcinogens (for example, tobacco smoke in lung cancers and ultraviolet radiation in skin cancers)126. For germline mutations, most functional coding variation is rare127 and is specific to single populations128. To illustrate the effect of sequence variation on proteins, we summarize the distribution of single-nucleotide variants across three possible categories: nonsense, missense and synonymous. Numerous computational techniques have recently been developed to predict the functional significance of amino acid substitutions; here, we use the PolyPhen-2 program to categorize missense variants as ‘benign’, ‘possibly damaging’ or ‘probably damaging’ (REF. 118).
Large-scale sequencing efforts, such as the 1,000 Genomes Project (TGP)129, have amassed a tremendous amount of data by sequencing thousands of individuals and have had an early emphasis on exome sequencing. If mutations were completely random, we would then expect nonsense and missense mutations to collectively make up ~72% of all coding variants observed, with a substantial fraction of these probably affecting protein function (see the figure, part a). This is nearly the case for the rarest of variants in the TGP (global frequency <0.1%), for which ~63% of variants are either nonsense or missense. However, purifying selection is an efficient evolutionary force that purges deleterious variation or that at least restricts them from reaching high frequency. Thus, almost all common amino acid variation is predicted to have no functional effect.
In addition to nucleotide substitutions, short insertions and deletions (indels) can also affect protein function. Frameshift indels (that is, indels with lengths that are not multiples of three) may be particularly deleterious, as they can have downstream effects during translation. The signature of purifying selection that operates against frameshift indels shows that the percentage of indels in the TGP129 that alter the reading frame of a protein decreases as the global allele frequency increases — from 66% for the rarest indels to 42% for the most common indels (see the figure, part b).
In this Review, we provide a description of three types of network biology screens: protein–protein, genetic and drug–gene interactions (FIG. 1). These three network types provide complementary views of the same cellular components. Protein–protein interactions are used to identify the pathways and complexes of the cell, and drug–gene interactions can be used to identify the activities that a pathway is involved in, whereas genetic interactions primarily report on the functional dependencies within and between pathways. We show how the approaches used to map these interactions are being extended to identify the parts of proteins that are responsible for specific interactions and to investigate how different mutations of the same protein can result in different functional consequences (BOX 2). Finally, we aim to place into context these high-resolution network biology approaches as a way to ultimately connect sequence with structure.
Understanding the consequences of mutations using network biology.
Traditionally, network biologists have used wild-type proteins to interrogate protein–protein interaction networks (see the figure, part a) and either complete gene deletion or gene knockdown to investigate genetic and drug–gene interaction networks (see the figure, part b). These approaches illuminate the global functions and interactions of proteins, but they do not provide much information about which parts of the protein are responsible for different interactions and how the function of a protein will be altered by different mutations. Some types of mutations, such as extreme truncations that result from nonsense mutations or complete structure disruption that results from frameshift insertions and deletions (indels), may be adequately modelled by the gene-knockout or gene-knockdown approach (see the figure, part b). However, others types of mutations, including missense mutations that do not destabilize the structure, require more detailed analyses (see the figure, parts c and d). The different screening approaches may offer different insights into the consequences of a single mutation; for example, missense mutation 1 results in no apparent change to the protein–protein interaction network but leads to unique genetic and drug–gene interactions. Combining the three approaches may offer a more direct insight into how genotype maps to phenotype; for example, missense mutation 2 results in the loss of a physical interaction with protein P, a positive genetic interaction with the gene coding for protein P and an increased sensitivity to drug D.
For brevity, we do not discuss gene regulatory networks, such as those derived from chromatin immunoprecipitation followed by sequencing experiments or from gene expression studies (reviewed in REFS 7,8), or the determination and modelling of dynamic signalling networks (reviewed in REFS 9–11). Moreover, we do not discuss the identification of enzyme– substrate relationships, such as those between kinases and their targets12. Similarly, we do not address approaches that seek to understand the functional changes that are caused by sequence variation in non-coding regions, as these have recently been reviewed elsewhere13.
Interaction network primer
Protein–protein interactions
Experimental methods for detecting protein–protein interactions can be generally grouped into two distinct categories: those that seek to identify direct ‘binary’ interactions, such as yeast two-hybrid (Y2H)14 and protein complementation methods15, and those that identify co-complex associations, such as the affinity purification–mass spectrometry (AP–MS) approach16,17 (FIG. 1a). All of the methods for detecting protein–protein interactions have different strengths and weaknesses, and it should be noted that no high-throughput approach has perfect specificity and sensitivity18. Initial reports suggested that the results of AP–MS experiments are of higher accuracy and higher reproducibility than Y2H methods19,20. However, subsequent analyses argue that this is a bias of the methods that were used to assess quality and that, in reality, both approaches favour the detection of different but complementary types of interactions14. For example, Y2H can identify transient and low-affinity interactions, whereas AP–MS can identify more stable indirect interactions, such as those that occur between proteins that belong to the same complex but that do not directly bind to each other14 (FIG. 1a). Large-scale interaction networks that use both approaches have been generated in various model organisms (TABLE 1). These networks are augmented by extensive literature curation efforts21,22, in which individual interactions from low-throughput experiments are manually identified from the literature. Interactions from both low- and high-throughput experiments are stored in databases23,24, which allow researchers to investigate many of the interactions that have been reported for a protein of interest. Computational methods have also been extensively used to predict protein–protein interactions using various sequence25, structure26 and genomic data27. Protein–protein interaction networks can be used to assign functionality to uncharacterized proteins through ‘guilt-by-association’, which essentially predicts the function of a protein on the basis of the function of its interacting partners (reviewed in REFS 2,28).
Genetic interactions
Genetic or epistatic interactions report on functional interactions between mutations and are identified when combinations of mutations produce a different phenotype than that expected from the phenotypes of individual mutations29. Although higher-order interactions have been measured30,31, for practical reasons experimental studies usually focus on interactions between pairs of mutations. Typically, genetic interactions are assigned a quantitative score on the basis of how the growth of a double mutant differs from that expected based on the growth of each of the two single mutants32. Negative interactions are identified when the growth is worse than expected and are typically interpreted as revealing redundant or parallel pathways. Positive interactions are detected when the growth is better than expected and often identify factors that function in linear pathways29 (FIG. 1b). The most extreme example of a negative genetic interaction is synthetic lethality, in which individually mutating two genes results in a viable organism, but mutating the two genes in combination results in cell death. Both in yeast and in bacteria, high-throughput genetic strategies have been developed to create strains that contain pairs of mutations33–37, and large-scale genetic interaction screens have been carried out using comprehensive gene deletion libraries38–41. Strategies have also been developed to disrupt either the expression or the stability of essential genes, in which a gene deletion would result in the loss of viability42–44. In metazoans, RNA interference (RNAi)-based approaches are more commonly used to simultaneously target two genes, and both cell growth45–49 and whole-organism growth50,51 have been used as phenotypes.
As with protein–protein interactions, high-throughput efforts to map genetic interactions are complemented by literature curation22, and the results are stored in centralized databases24. Compared with protein–protein interactions, efforts to predict genetic interactions computationally have been relatively limited — most methods focus on extending existing networks52,53 rather than on the de novo prediction of interactions. Nevertheless, there are a few examples of the de novo prediction approach54–56.
The set of partners that a gene interacts with — or in the case of quantitative screens, the set of scores for these interactions — is known as an interaction profile. The inhibition of genes that function in the same pathway or complex tends to result in similar genetic interaction profiles; that is, they interact with the same sets of genes in the same way (FIG. 1d). Although it is possible to predict the function of a gene from that of its genetic interaction partners, especially using positive genetic interactions, it is more common to predict gene function using genetic interaction profile similarity29.
Drug–gene interactions
Chemogenetic interactions report on the functional interactions between genes and drugs. They are conceptually similar to genetic interactions, and experimental screens for detecting such interactions are carried out in a similar manner. However, rather than simultaneously perturbing two genes, a single gene is perturbed in the presence of a compound. As with genetic interactions, they can be either negative, in which the combined effect of perturbing a gene in the presence of a drug is more severe than expected, or positive, in which the combined effect is less severe than expected. Similarly to genetic interactions, these can be interpreted as perturbing parallel or linear pathways respectively (FIG. 1c). However, owing to both off-target and nonspecific effects, such interpretations may be an oversimplification. It is important to note that a drug– gene interaction does not imply that the drug physically binds to the protein product of that gene. Rather, the phenotype that is associated with the perturbation of the gene is modified by the presence of the drug. This can be because the drug directly binds to the encoded protein, but it is more frequently because the drug induces a cellular state in which the requirement for the protein is altered. For example, many genes that are involved in DNA damage repair show negative interactions with the DNA-damaging agent methyl methanesulphonate (MMS)57, not because they directly bind to MMS but because their functionality becomes more important in the presence of the induced DNA damage. Gene function can be either directly inferred from interactions with a specific drug — for example, interaction with MMS could indicate that the gene functions in the DNA damage response — or indirectly inferred through profile similarity, as genes in the same pathway tend to interact with the same drugs (FIG. 1d).
Network integration
Each of the three interaction types discussed above provides mostly orthogonal information of the same cellular components. Consequently, by integrating multiple network types it is possible to obtain insights that are not obvious from analysing a single network in isolation. As these integrative approaches have been reviewed elsewhere58–60, we only mention a single example of the use of integrating each pair of networks here. Protein–protein and genetic interaction data have been integrated by various groups to identify functional modules; that is, sets of proteins that are physically connected and that show similar genetic interaction profiles. In addition to improving the identification of known complexes, this approach has revealed pairs of complexes that are linked by either all negative or all positive genetic interactions, which suggest parallel and linear dependencies, respectively58,61. Similarly, others have integrated protein complexes with chemogenetic interactions to identify conditionally essential complexes62 (the members of which all show negative interactions with a particular drug), which suggests that the function of the entire complex is required in the presence of that drug. Finally, genetic and chemogenetic interaction profiles have been successfully integrated to improve the identification of drug targets63,64.
High-resolution protein–protein interactions
Identifying which parts of a protein are responsible for different interactions is an important step towards predicting how its function will be affected by different mutations, as well as for understanding how a single protein can carry out multiple different functions.
Domain–domain interactions
One strategy to narrow down the region of a protein that is responsible for specific interactions is to screen multiple fragments of the same protein65–70. By comparing which fragments of a protein successfully interact with a given partner, it may be possible to determine which regions are responsible for that specific interaction (FIG. 2a). For example, a fragment-based Y2H approach was used to create a protein–protein interaction network for 749 factors that are involved in Caenorhabditis elegans early embryogenesis65, including members of the nuclear pore and the centrosome. An average of 40 different bait protein fragments for each of these 749 genes was screened against a library of full-length prey proteins. This approach was shown to be more sensitive than screening full-length proteins (that is, more interactions were identified) and was not associated with an obvious loss of specificity (that is, the interactions did not seem to be enriched for false positives). Owing to the high number of fragments screened for each protein, the authors were able to identify the minimal region of interaction — the smallest region shared by all fragments for which the interaction was observed — for many protein– protein interactions. Furthermore, they could identify multiple minimal regions of interaction on a single protein that corresponded to distinct interaction interfaces (FIG. 2a). Only a limited number of proteins were identified using exclusively full-length fragments, and these proteins were, on average, quite short in length — this led the authors to suggest that, in these cases, the entire protein consists of a single globular domain that cannot fold when truncated.
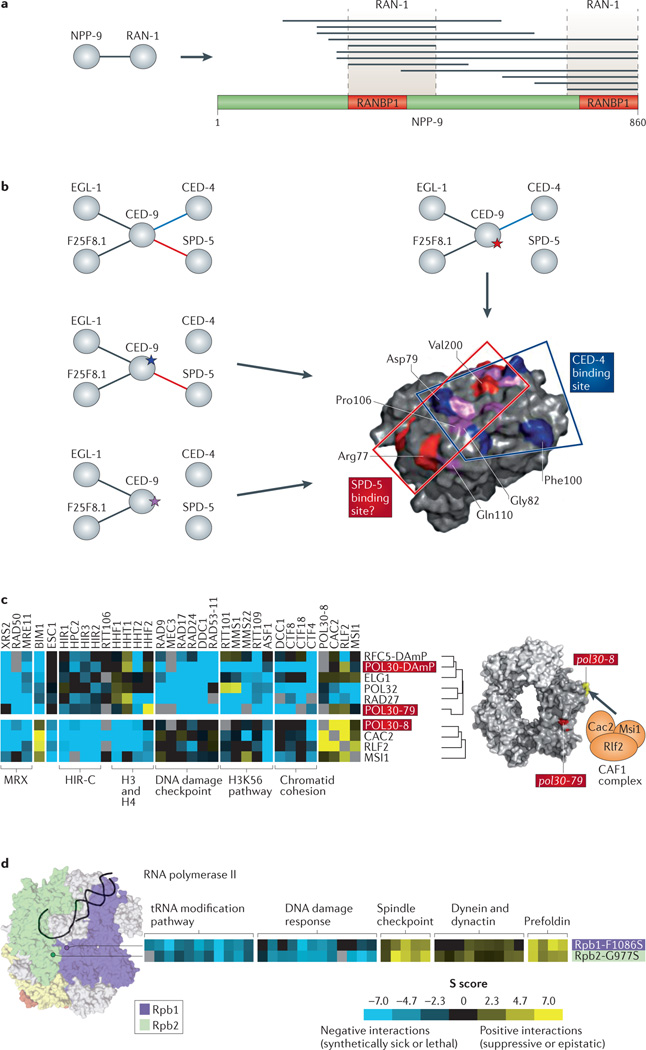
Figure 2. High-resolution physical and functional interactions.
a | In a Caenorhabditis elegans study, the interactions of NPP-9 with GTP-binding nuclear protein RAN-1 were studied to a resolution at the domain level by screening a high number of fragments for each protein using a yeast two-hybrid approach65. Shown on the right of the figure are minimal regions of interactions that have been identified on NPP-9. Each horizontal line corresponds to a fragment which interacts with RAN-1; the red RAN-binding protein 1 (RANBP1) boxes correspond to the locations of the known RAN-binding domains. b | ‘Edgetic’ interactions of apoptosis regulator CED-9 were identified in a C. elegans study that was based on a reverse yeast two-hybrid system78. Mutations that specifically perturb the interaction of CED-9 with CED-4 (blue star), with SPD-5 (red star) or with both (purple star) are shown. None of these mutations disrupt interactions of CED-9 with EGL-1 or F25F8.1. Mutated residues on the CED-9 structure are highlighted; the CED-4 binding site confirmed from this study and a potential binding site for SPD-5 are also shown. c | In a Saccharomyces cerevisiae study, different alleles of POL30 show different genetic interaction profiles; the tree to the right of the profile indicates a hierarchical clustering of the profiles. pol30-79 behaves in a similar way to pol30-DAmP — an allele with decreased mRNA expression — which suggests that it affects the core function of the protein. pol30-8 has similar behaviour to components of the chromatin assembly factor 1 (CAF1) complex, which suggests that both the specific interaction and the common function of Pol30 and this complex are perturbed. Shown on the right are the positions of the mutated residues of the Pol30 complex, as well as the subunits of the CAF1 complex. d | Mutations of residues that are on distinct subunits of RNA polymerase II but that are proximal in three-dimensional space show similar genetic interaction profiles in S. cerevisiae112. Highlighted on the Pol II structure are the locations of mutations to the Rpb1 subunit (purple) and the Rpb2 subunit (green). These mutations show negative genetic interactions with genes that are involved in the tRNA modification pathway and the DNA damage response, and positive genetic interactions with genes that are involved in the spindle checkpoint and the prefoldin complex. H3K56, histone H3 lysine 56; MRX, the S. cerevisiae homologue of the mammalian MRE11–RAD50–NBS1 (MRN) DNA damage repair complex. Part a is modified, with permission, from REF. 65 © (2008) Elsevier Science. Part b is modified, with permission, from REF. 78 © (2009) Macmillan Publishers Ltd. All rights reserved. Part c is modified, with permission, from REF. 29 © (2010) Elsevier Science.
The value of this approach in the context of human disease was demonstrated in a study of the Huntington’s disease-associated protein hungtingtin (HTT)67. A Y2H screen that was carried out with multiple fragments of the human HTT protein identified various novel interaction partners. Follow-on studies showed that one of these proteins, ARF GTPase-activating protein GIT1, influenced HTT aggregation — a phenotype that is linked to disease progression. Both GIT1 and HTT are large multidomain proteins, but by screening multiple fragments of each protein, the authors were able to narrow down the putative interacting regions to the amino terminus of HTT and the carboxyl terminus of GIT1.
Edgetic perturbations
An alternative approach to identify regions of proteins that are responsible for particular interactions is to identify ‘edgetic’ mutations that alter some, but not all, of the interactions of a protein71–78. Analogous to forward genetics and reverse genetics, this approach can be used in two different ways: ‘forward’ edgetics, the goal of which is to identify the interactions that are perturbed by a specific mutation of interest; and ‘reverse’ edgetics, in which mutations that perturb specific interactions are identified. Such approaches can be especially informative when integrated with structural models of the protein of interest — by mapping the mutated residues that perturb a specific interaction onto the three-dimensional structure of the protein, the regions that are important for the interacting interface can be inferred.
In a pioneering forward edgetics study, 35 different mutants of the Saccharomyces cerevisiae actin protein were screened using a Y2H approach71. This revealed three distinct types of mutations: those that disrupted all, none or specific interactions. The use of this approach to aid structural studies was shown by mapping onto the actin structure the locations of mutations that disrupted its interaction with a specific protein, which revealed a putative binding site. The use of forward edgetics in the context of human disease was highlighted in a systematic study of five proteins that are associated with distinct Mendelian diseases77. These proteins were selected because they each had multiple distinct disease-associated in-frame mutations and numerous known protein interactions. Twenty-nine alleles of these five proteins were screened using the Y2H method to identify any perturbed interactions. Only 5 of these alleles resulted in the loss of all interactions, whereas 16 of these resulted in the loss of specific interactions, which indicates that their associated disease phenotypes may be caused by a loss of specific protein–protein interactions rather than by a total loss of functionality.
A novel reverse edgetics experimental strategy78 based on the reverse Y2H system was used to identify edgetic mutations of CED-9, the C. elegans orthologue of the human oncoprotein B cell lymphoma-2 (BCL-2)78 (FIG. 2b). A library of full-length mutant alleles encoding CED-9 was used to identify mutants that perturbed interactions with any of four identified protein interaction partners. This approach identified 72 distinct alleles, 30 of which disrupted all interactions (that is, they were non-edgetic), and the rest perturbed a specific subset of interactions (that is, they were edgetic). By mapping onto the CED-9 structure the mutated residues resulting from these alleles, the authors found that edgetic residues are preferentially located in accessible regions, which suggests that they perturb specific interfaces, whereas non-edgetic residues were more likely to be found in the core, where they may destabilize the structure of the protein. Furthermore, in human cells edgetic mutants could be expressed at levels that are similar to those of wild-type CED-9, whereas the non-edgetic mutants were expressed at much lower levels, suggesting that they encode unstable proteins. As in the study of actin mutants in yeast, mapping the location of residues that are associated with edgetic perturbations onto the CED-9 structure suggested putative binding sites for specific interactions. For an interacting pair for which a co-crystal structure was available (CED-4–CED-9), this putative binding site agreed well with the known interaction interface (FIG. 2b).
By design, most studies of edgetic protein–protein interactions have focused on identifying mutations that lead to the loss of interactions. An initial set of proteins that interact with the wild-type protein of interest is often experimentally derived, and further analyses are then carried out to identify which of these interactions are lost by a specific mutation. However, it is likely that many mutations result in a gain of function, which leads to new interactions with previously unidentified partners. Investigating these mutations in the context of cancer is likely to be of particular interest, as many cancer-associated mutations are believed to confer gainof- function effects; for example, the tumour suppressor p53 is one of the most commonly mutated proteins in human cancer. Unusually for a tumour suppressor, most cancer-associated mutations of this protein are missense mutations rather than protein truncations or a complete gene loss. Several of these mutations result in both the loss of tumour suppression functionality and the gain of protein–protein interactions79. Specifically, mutant forms of p53 can interact with various transcription factors, which potentially results in substantially altered transcriptional regulation. There are other examples in the literature of a single disease-associated mutation causing both loss and gain of specific protein–protein interactions that result in changes in functionality80.
Computational approaches
Experimentally mapping interactions of multiple variants of each protein (for example, protein fragments or point mutants) is inherently more costly than screening a single variant. Consequently, an attractive alternative method for determining the domains and residues that are involved in specific interactions may be to compute a three-dimensional model of the corresponding macromolecular assembly by integrating existing protein–protein interaction networks with additional information81–88. Depending on the quantity and quality of the available information, such integrative modelling can map interactions at the resolution of subunits, domains or even individual residues (BOX 3). For example, both an interaction map and the localization of 456 constituent proteins in the yeast nuclear pore complex were determined by modelling that was based on low-resolution information from multiple sources, including affinity purification of protein subcomplexes, sedimentation analysis and electron microscopy89. Similarly, the 26S proteasome structure, which was determined from an electron microscopy map of the whole assembly, from proteomic information and from the subunit comparative models, revealed both the localization and interacting interfaces at the resolution of individual domains and even residues90. When atomic structures of individual constituent proteins are available, even sparse and low-resolution data on the quaternary structure can lead to an atomic model of interactions, as shown by the structure of the bacterial type II pilus system, the subunits of which were assembled from sparse NMR data91. Integrative structure determination makes it easy to take advantage of all data, which results in models that are generally more accurate, precise and complete than those that are based on any individual data set92.
Integrative structure determination of macromolecular assemblies.
The most detailed information about interactions between proteins is provided by three-dimensional structures of macromolecular assemblies. These structures generally contribute to our understanding about how the assemblies function and how they evolved, as well as how to control and possibly to modify their functions. Unfortunately, it is often difficult to solve these structures by traditional methods of structural biology, such as X-ray crystallography, NMR spectroscopy and electron microscopy. The reasons for this include the size, flexibility, transient nature, and compositional and structural heterogeneity of the assemblies, as well as the need for pure samples of sufficient quantity. To overcome these problems, integrative or hybrid approaches that combine data from multiple methods through computation were developed (reviewed in REFS 113,130,131). The resolution of the resulting hybrid structural models ranges from low, specifying only the positions of the protein subunits, to high, specifying the positions of each atom.
The integrative approach iterates through four stages: gathering structural information from as many sources as possible; defining how to represent and evaluate models on the basis of the available data; finding models that are consistent with the data; and analysing the input data as well as the output models. As integrative models are computed from all available data, they are often more accurate, precise and complete than those produced by traditional methods. Integrative modelling encourages the finding of all models that fit the data, not only one such model. At least in principle, it also facilitates the assessment of the data and the models. Finally, integrative modelling can provide feedback to guide future experiments, so that maximum model improvement is achieved for minimal effort.
Various structures have been solved by integrative approaches, including those of the bacterial type II pilus91, chromatin segments132 and the yeast nuclear pore complex89. We illustrate the integrative approach by its application to the regulatory particle of the 26S proteasome, which consists of 19 different protein subunits90 (see the figure). Structural information was first gathered, including atomic models of subunits or their domains that were either determined by X-ray crystallography or computed by comparative modelling based on known homologous structures; the shape of the regulatory particle that was defined by a cryo-electron microscopy map at 8.4 Å resolution; the positions of two subunits (Rpn10 and Rpn13) that were pinpointed in the cryo-electron microscopy electron-density maps of proteasomes without these two subunits; the proximities between pairs and larger subsets of subunits that were defined in publicly available protein–protein interaction data, including those from large-scale screens; and the proximities between specific residues across protein interfaces that were defined by residue-specific inter-subunit crosslinks. Next, all relative positions and orientations of subunits that minimally violated the data were found by a sophisticated structural sampling algorithm133. It turned out that a single cluster of solutions satisfied most of the data, thus providing a structural model of the 26S proteasome. This model was used to rationalize and to predict several aspects of the 26S proteasome function. In addition to the assessment of the model based on structural data that were not used in its calculation, the model was most convincingly validated by a completely independent structure determination based on cryo-electron microscopy maps for the entire regulatory particle and several of its subcomplexes113.
Figure is modified, with permission, from REF. 90 © (2012) US National Academy of Sciences.
Alternatively, more coarse-grained approaches may be used; for example, interactions from multiple databases were integrated with additional functional data to create a high-confidence protein–protein interaction network in S. cerevisiae83. The protein family interactions (iPfam) database93 — which contains interactions between pairs of Pfam94 domains that are supported by at least one representative structure in the Protein Data Bank95 — was used to identify a set of structurally characterized domain–domain interactions. The protein– protein interaction network was then filtered to include only interactions between proteins containing iPfam domains that were known to interact. This ‘structurally resolved’ interactome allowed the authors to distinguish between ‘simultaneously possible’ interactions, in which protein A interacts with proteins B and C through distinct domains, and ‘mutually exclusive’ interactions, in which protein A interacts with B and C using the same domain83. Notably, many of the previously observed relationships between network topology and other genomic features could be better explained using structural properties. For example, the authors elaborated on a previous observation that ‘hubs’ — proteins that are involved in many interactions — are more likely to be essential than random proteins, and they identified that hubs with multiple interaction interfaces are twice as likely to be essential as hubs with only a single interaction interface.
The same approach was recently used to create a structurally resolved human protein–protein interaction map onto which disease-associated mutations could be mapped87. Importantly, there was an enrichment of these mutations on protein–protein interaction interfaces, which again suggests that many diseases are the result of perturbed protein–protein interactions (that is, edgetic perturbations). Furthermore, in cases in which multiple mutations are found on the same protein, mutations of different interacting interfaces were significantly more likely to be associated with different diseases than mutations that affect the same interface. Finally, mutations that affect two distinct interacting proteins on their corresponding interacting domains are more likely to cause the same disease than mutations on domains that do not mediate their interaction87.
A disadvantage of these structurally resolved approaches is that they require a known three-dimensional structure onto which interacting domains can be mapped; such information is only available for a small proportion of domain–domain pairs. One alternative approach is to take an experimentally determined protein–protein interaction network and a list of the domains in each protein, and to use this information to predict the domain pairs that are most likely to be responsible for each interaction82,84–86. No structural information is used in this approach; instead, statistical r machine-learning methods are applied to compare the number of observed interactions between proteins that share a given domain pair with that expected from the overall frequency of each domain in the network. In principle, because of their greater coverage, these approaches could have an advantage over others that require three-dimensional structural information. However, their accuracy is difficult to assess, and there is a poor overlap between different prediction methods even when the same input data were provided96.
In addition to the identification of interactions between pairs of globular domains, the computational identification of interactions between globular peptide-recognition domains and short peptides is of great interest. Computational methods for predicting these interactions, which are of particular importance for cellular signalling4, have recently been reviewed elsewhere97.
An interesting approach is to invert the problem — rather than interpreting protein–protein interactions using structural information, it is possible to use the structural information to predict such interactions. A recent method showed that, by using a lenient threshold for structural similarity and by integrating orthogonal functional information about proteins, it is possible to computationally predict protein–protein interactions on a proteome-wide scale26. The predicted interactions were shown, at least for binary interactions in yeast, to be of similar accuracy to experimental methods. In addition to predicting protein–protein interactions for yeast and humans, this approach provides a crude model of the residues and domains that are involved in these interactions.
High-resolution (chemo)genetic interactions
Although the approaches detailed above may be used to identify the protein–protein interactions that are perturbed by a specific mutation, they cannot assess how these mutations affect specific cellular phenotypes of interest. Genetic and chemogenetic interaction screens may be used to close this gap between understanding the proximal mechanistic consequences of mutations and how the mutations ultimately affect specific phenotypes. Furthermore, in the case of a single protein that has several distinct functions in different pathways, genetic and chemogenetic interactions can be used to identify the specific parts of the protein that carry out these functions even when no change to the physical interaction network is detected. Currently, screens that analyse the genetic and chemogenetic interaction profiles of multiple alleles of the same gene have been limited and almost exclusively carried out in yeast; see BOX 4 for a discussion of some studies using human systems. Some yeast screens have used multiple mutant alleles of the same essential gene that either are temperature sensitive or have reduced mRNA expression40,43. Although such approaches are valuable for exploring the functionality of essential genes, they usually address the problem at the whole-protein level and do not identify the parts of proteins that are responsible for specific interactions or functions. However, a few studies have shown that genetic and chemogenetic interaction profiling can be extremely useful in this context98–100, particularly for investigating the functional consequences of PTMs98–102 that may not be detectable in the protein–protein interaction network. This can be achieved by mutating the specific residues that are subject to PTM, such that they mimic either their modified or unmodified state. By comparing the profiles of mutants with different modification status, it is possible to identify functional interactions that change owing to specific modifications.
Genetic and drug–gene interactions in human systems.
Genetic interactions and disease
A major challenge in cancer therapeutics is to kill tumour cells without harming other cells in the body. One means to achieve this is to exploit the genetic changes that distinguish cancer cells from normal cells and that may leave them vulnerable to targeted treatments134. In this context, the identification of drugs or genes that show a strong negative interaction (that is, synthetic lethality) with a specific oncogenic mutation has been a high priority135. Consequently, much of the early drug–gene (chemogenetic) and genetic interaction screens in mammalian cells were carried out not to dissect gene function but to identify potential targeted therapeutics.
To this end, various groups have used RNA interference (RNAi)-based approaches to identify genes that are only essential in specific cancer cell lines136, 137. By screening enough different cell lines, classical forward genetics approaches may be used to identify statistical associations between specific mutations and their sensitivity to the knockdown of specific genes. Such studies are promising but are complicated by the genetic heterogeneity of different cell lines. In the presence of multiple mutations, which may themselves genetically interact, a clear relationship between genotype and phenotype may be difficult to ascertain.
An alternative approach is to screen ‘isogenic’ cell lines that differ only by the mutation of a particular gene of interest138–140. The use of this approach was demonstrated in a study that identified genes which selectively inhibit growth in the presence of a specific activating point mutation (G13D) in the KRAS oncoprotein139. A genome-wide RNAi screen identified hundreds of candidate genetic interaction partners for the mutated KRAS, which were significantly enriched for components of the mitotic machinery. Subsequent analyses revealed that cells expressing KRAS-G13D showed increased sensitivity to an inhibitor of mitotic spindle function, which suggests that the KRAS oncogene causes increased mitotic stress. Furthermore, specific inhibition of PLK1, which is a mitotic kinase that was shown to genetically interact with KRAS-G13D, resulted in reduced tumour growth in a mouse model. This result highlights the value of investigating genetic interactions that are associated with specific cancer mutations, both for the identification of potential therapeutic targets and for an improved understanding of the oncogenic state. Similar RNAi-based approaches have recently been applied to the understanding of other diseases; for example, a recent RNAi screen identified genes that suppress the phenotype associated with cells that express a fragment of the mutant form of huntingtin (HTT). Interestingly, rather than identifying genes that inhibit cell growth, the authors identified genes that suppress caspase 3 activity, which is typically enhanced in cells that express the mutant HTT fragment141.
Drug–gene interactions and disease
As with genetic interactions, drug–gene interactions in cancer cell lines are of great interest owing to their potential therapeutic implications. Various large-scale studies have screened hundreds of cancer cell lines against drug libraries142–144. When combined with genotypic information, either standard forward genetics or more advanced machine-learning approaches can be used to try to associate specific mutations with sensitivity to specific drugs. As with genetic interaction screening, the genetic heterogeneity of different cancer cell lines can make the association of specific drugs with specific mutations difficult; for example, one recent study noted that “single gene–drug associations were only rarely able to explain the range of drug sensitivities observed across cell lines for any given drug” (REF. 143). Again, the alternative is to use isogenic cell lines to interrogate the drug sensitivity of a specific mutation of interest145,146. Such an approach was used to assess the ability of ~24,000 compounds to selectively kill 6 different tumorigenic cell lines but not their isogenic non-tumorigenic counterparts146. A drug–gene interaction screen was recently carried out to identify genes that, when inhibited, increase the sensitivity of KRAS-mutant cell lines to a specific drug (an inhibitor of MEK, which is a protein in the mitogen-activated protein kinase effector pathway of KRAS signalling)147. Such hybrid approaches may be more commonly used in the future to identify drug–gene interactions that are selectively lethal in the presence of specific cancer-associated mutations.
Recent developments and future directions
Experimental methods have recently been developed to improve both the throughput and the accuracy of genetic interaction mapping in mammalian cells45,47–49. In contrast to the studies of genetic interactions in cancer, in which a single query mutation is screened for interactions with a large RNAi library, these methods allow comprehensive analyses of all pairwise interactions between hundreds or thousands of genes. They have been used to carry out screens that are analogous to those using gene deletions in yeast, which were used to identify the dependencies between genes that are involved in chromatin regulation47–49 and ricin susceptibility45. Their reliance on RNAi knockdowns means that, at present, they will be primarily used to address interactions that are associated with whole genes, rather than with specific alleles of interest. However, recent improvements in genome editing and engineering, such as the transcription activator-like effector nucleases (TALEN) and the clustered regularly interspaced short palindromic repeat (CRISPR)–Cas9 systems148–151, allow the rapid introduction of mutations of interest into human cells. By combining this technology with RNAi-based genetic interaction screening and high-throughput chemogenetic interaction screening, it should be possible to create interaction profiles for many different mutations of the same gene, analogous to the studies that have been carried out in yeast. This will enable us to analyse structure–function relationships of human proteins at high resolution and also to investigate the functional consequences of different mutations to the same disease-associated protein (for example, KRAS).
This approach was used to identify the functional interactions that are mediated by the phosphorylation of Ies4, a subunit of the S. cerevisiae INO80 chromatin-remodelling complex100. Five serine residues that are differentially phosphorylated in response to MMS treatment were identified on this protein. Two mutant forms of the protein were created, such that all five serine residues mimicked either their phosphorylated or unphosphorylated state. No changes to the protein– protein interaction network could be detected as a result of these mutations, but a genetic interaction screen revealed subtle differences in the behaviour of these two mutants. Only the phosphomimetic mutant showed positive genetic interactions with genes that are involved in the DNA damage checkpoint, which suggests that the phosphorylation of Ies4 mediates its role in this process — a prediction that was ultimately confirmed100. A similar approach was used to investigate the role of acetylation in regulating the functions of the histone variant H2A.Z in S. cerevisiae101. The aminoterminal tail of this histone contains four lysine residues that can be acetylated, which potentially regulate its function. A series of mutants were created: mutants that were singly mutated (that is, only one of the four lysine residues was mutated), mutants that were singly acetylatable (that is, three of the four lysine residues were mutated), or mutants that were completely unacetylatable (that is, all four lysine residues were mutated). Genetic interaction screening revealed that mutants that were singly mutated or singly acetylatable showed few interactions and behaved in a similar way to wild-type controls. However, the completely unacetylatable mutant recapitulated a subset of the genetic interactions that are associated with a complete deletion of the H2A.Z gene, which suggests that the modifiable lysine residues are internally redundant and that the protein can correctly function if any one of these lysine residues is acetylated. These results indicate that genetic interaction screening can be used not only to identify the consequences of mutating specific residues but also to identify how combinatorial perturbations of the same gene can result in unique outcomes. Genetic interaction screening has also been used to study H2A.Z using truncations of varying lengths in S. cerevisiae102 or using targeted mutation in Schizosaccharomyces pombe103.
Edgetic interactions
A genetic interaction study104 that mirrors the earlier experiments using Y2H screening of actin point mutants71 identified gene deletion mutants that interact with haploinsufficient actin in S. cerevisiae. Six different point mutants of the actin gene were then tested for genetic interactions with these gene deletion mutants, which revealed that different point mutants interacted with different subsets of these partners. Furthermore, an analysis of the actin structure revealed that the mutations of two residues that were close together on the protein surface resulted in similar genetic interaction profiles, which suggests that genetic interaction profiles could be integrated with structural models to identify structure–function relationships.
In a striking example from a quantitative genetic interaction screen focused on factors that are involved in chromosome biology in S. cerevisiae105, different alleles of POL30 (the gene product of which is also known as Pcna) showed radically different genetic interaction profiles29 (FIG. 2c). POL30 is a multifunctional essential gene that is involved in both chromatin assembly, and DNA replication and repair. For example, the pol30-79 allele generated a similar interaction profile to pol30-DAmP (that is, a mutant of Pol30 with decreased abundance by mRNA perturbation), which suggests that the pol30-79 mutation has a general destabilizing effect on the protein. Another allele, pol30-8, elicited a profile that is similar to that observed for deletions of members of the chromatin assembly factor 1 (CAF1) complex. Furthermore, this allele showed strong positive genetic interactions with members of the CAF1 complex, which suggests that these mutations perturbed the same functional pathway. Indeed, Pol30 and CAF1 physically interact, and a pol30-8 mutant has previously been shown to severely weaken this interaction106 but not its interactions with other factors107. Thus, genetic interaction screening can be used to investigate allele-specific edgetic perturbations.
Comprehensive structure–function analyses
Although most studies have focused on a small number of alleles of a given gene, a few have highlighted the effectiveness of screening large numbers of alleles of a single gene, especially when the results are integrated with structural protein models. Histone proteins, in particular, have been a primary focus for such interaction screens. Indeed, various groups have created libraries of histone alleles, in which specific residues (for example, those on the protein surface and those that are subject to PTMs) have been systematically mutated, to facilitate the screening for drug sensitivity and other functional analyses108–110. In perhaps the most comprehensive functional analysis of individual proteins that has so far been carried out, a library of 486 alleles of the S. cerevisiae H3 and H4 histones was created108. Each residue of the two proteins was mutated one at a time: alanine residues were systematically mutated to serine, whereas all other residues were mutated to alanine. Remarkably, only ~10% of their point-mutant strains were completely inviable despite the very high interspecies sequence conservation. Each of the mutants was screened for growth defects in 14 different conditions, including 5 drug treatments, which allowed the fine-grained association of regions of the protein structure to different functions. For example, mutants that were sensitive to 6-azauracil, a compound that is associated with defects in transcriptional elongation, were over-represented on the lateral surface of the histones, which is the region that interacts with DNA. The HistoneHits database111 is an example of the integration of results from multiple mutant phenotyping screens of the same protein. This database provides a central repository of mutant–phenotype associations for histones. In addition to assessing the agreement of phenotype–residue associations across studies, it facilitates meta-analyses of these associations; for example, mutations of residues in the lateral surface of histones typically show more phenotypes than mutations elsewhere in the structure. Similarly, residues that are subject to PTMs have significantly more phenotypes than other residues. By contrast, mutations of residues in histone tails show significantly fewer phenotypes111.
A recent study genetically targeted a well-characterized, structurally defined protein machine — RNA polymerase II. To this end, 53 different point mutants in five evolutionarily conserved subunits of S. cerevisiae Pol II were identified and subjected to quantitative genetic interaction profiling112. The resulting point-mutant epistatic miniarray profile (pE-MAP) allowed the assignment of function to individual residues of this complex by comparing profiles that are associated with the point mutants of these residues with the existing profiles of gene deletion mutants. It uncovered a remarkable coordination of many processes that Pol II is involved in, including start-site selection, transcriptional elongation and mRNA splicing. Furthermore, it facilitated the discovery of new transcription factors and offered insights into how they function. Interestingly, there was a striking correlation between the similarity of genetic interaction profiles and the proximity of the corresponding residues in three-dimensional space, even when proximal residues were in different protein subunits (FIG. 2d). It will be of great interest to determine whether quantitative genetic data, which are often simply based on colony size, can be used for the structural modelling of macromolecular machinery89,113, especially those that are not biochemically tractable (such as membrane-associated complexes).
The creation of mutant libraries in which a single codon is edited is valuable but is also costly and time consuming. An alternative approach is to develop a strain in which the chromosomal copy of the gene to be mutated can be easily disabled, for example, by placing it under the control of a galactose-regulated promoter. Plasmids that contain mutated copies of the gene can then be introduced and their fitness assessed. Recently, by combining deep sequencing with a competitive growth assay114, such an approach was used to measure the fitness of every possible point mutant of ubiquitin in yeast115. By expanding this approach to measure fitness in the presence of different drugs or additional mutations, it should be possible to create high-resolution chemogenetic and genetic interaction profiles for all point mutants of any yeast gene.
Computational prediction of allele-specific interactions
Surprisingly, even in yeast there are few examples of computational methods to predict the phenotypic consequences (including increased sensitivity to drugs or to gene inhibition) of specific point mutations. Current approaches can predict, with reasonable accuracy, the phenotypes that are associated with the complete loss of gene function116,117. However, to our knowledge there are no computational tools that will either predict cases in which different mutations of the same gene result in different phenotypic consequences or predict which of the many phenotypes associated with a pleiotropic gene are likely to be altered by a specific mutation. There are various techniques that rely on either structural information or evolutionary conservation to predict whether a mutation will have a significant effect on fitness (such as the PolyPhen-2 program118,119 (BOX 1)). However, these approaches typically annotate mutations as either neutral or deleterious, but they do not state which phenotypes the mutation will alter. In light of the results presented above, the limitations of such a classification become apparent. Histone proteins and the components of RNAPII are among the most highly conserved of all eukaryotic proteins (the sequence identity of histones between humans and yeast ranges from 63% to 92%111), and consequently, one might reasonably expect most mutations in these proteins to be deleterious. However, only ~10% of the histone point mutants are completely inviable, whereas the remainder show severe growth defects only under specific conditions.
Conclusions
In this Review, we have highlighted computational and experimental approaches that seek to characterize the interactions and functions of genes and proteins at the resolution of domains or residues. For most species, even highly studied model organisms, the extant interaction networks are far from complete even at the protein– protein or genetic level. Consequently, it would seem to be overly ambitious to experimentally screen multiple variants of every gene or protein, at least using current approaches. Such high-resolution analyses are likely to be reserved for proteins that are of particular interest, either owing to their high conservation across species (such as histones and actin) or because of their association with a particular disease (such as KRAS (BOX 4) and hungtingtin). For other proteins, it is likely that single alleles will be screened and computational methods will be required to characterize the functions of their sequence and structure in greater detail.
The mapping of host–pathogen interaction networks is emerging as an important approach for understanding how pathogens hijack the machinery of the host cell120,121. So far, these studies have primarily focused on the interactions between the host and a specific viral or bacterial strain. However, given the rapidity with which viruses can evolve, it will be extremely informative to see how different alleles of the same viral proteins can result in changes to their interaction networks122.
As the available data on allele-specific interactions increase, it will be important to have centralized databases to store this information. Current interaction databases that integrate studies from multiple laboratories and from multiple organisms tend to document interactions between whole proteins or whole genes123, whereas databases that report allelic interactions tend to be associated with the screens from a specific laboratory105,124 or from a specific protein family111. A centralized database would facilitate the types of meta-analyses that have been carried out for standard interaction networks, and it would also provide training data for computational approaches to predict the network consequences of specific mutations.
The integrated analyses of protein–protein interaction networks with genetic interaction networks have revealed features that are not evident from studying either type of networks in isolation, and such analyses have offered an improved understanding of the relationship between these two networks58,61. One major goal is to extract from these networks mechanistic insights about the function of individual pathways, protein complexes, proteins and even individual domains or residues in these proteins125. As discussed above, structural information is often used to help to interpret such networks; however, in the future, it will be of great interest to determine how these types of data can ultimately be harnessed to inform structural studies, especially those involving protein machines that are mutated in different disease states, which have been uncovered through large-scale genomic studies. For these mutated machines, a deeper understanding at both the biophysical and structural levels may be needed to truly understand the underlying biology behind these detrimental effects. Ultimately, the information from an integrated pipeline — from sequence to systems to structure — will be crucial in helping to develop targeted therapeutic strategies that could be genetic, chemical or biological in nature.
Acknowledgements
The authors thank G. Cagney, D. Fitzpatrick and C. Maher for their comments and feedback. They also thank M. Shales and H. Braberg for suggestions and assistance with figures and K. Lasker for assistance with Box 3. C.J.R. is supported by ICON Plc and the University College Dublin Newman Fellowship Programme; P.C. is supported by a Howard Hughes Predoctoral Fellowship; A.S. is supported by the National Institutes of Health (R01 GM083960, U54 RR022220, U54 GM094662, P01 AI091575, and U01 GM098256); N.J.K. is supported by the US National Institutes of Health (P50 GM082250, R01 GM084448, P01 AI090935, P50G M081879, R01 GM098101, R01 GM084279 and P01 AI091575) and the Defense Advanced Research Projects Agency (DARPA-10-93-Prophecy-PA-008).
Glossary
- Epistasis
A phenomenon whereby the phenotype associated with a mutation is altered by the presence or absence of additional mutations.
- Domains
Distinct functional or structural regions of a protein, which can fold independently of the rest of the protein. A protein may contain several domains, and the same domain may be present in different proteins.
- Post-translational modifications
(PTMs). The chemical modifications of a protein after its translation, which can change the enzymatic activity, subcellular localization or interaction partners of the protein.
- Deletion libraries
Sets of mutant strains, each of which has a single gene removed. The removed gene is typically replaced with an antibiotic-resistant marker to allow easy selection in genetic experiments.
- Exome sequencing
The targeted sequencing of only known protein-coding regions.
- Nonsense
Pertaining to a mutation that changes an amino acid codon to a stop codon.
- Missense
Pertaining to a mutation that changes the encoded amino acid.
- Synonymous
Pertaining to a mutation that does not change the encoded amino acid.
- Forward genetics
The classical genetics approach, in which the genotypes that are associated with particular phenotypes are identified.
- Reverse genetics
The inverse approach to forward genetics, in which phenotypes that are associated with a particular genotype are identified. Such approaches are exemplified by studies of knockout mutants.
- Alleles
Multiple forms of a gene that occur at a specific locus.
- Reverse Y2H
(Reverse yeast two-hybrid). A genetic strategy to select against specific protein– protein interactions.
- Histone
A family of proteins that package DNA into nucleosomes. They consist of a globular domain and a tail that is subject to extensive post-translational modifications.
- Pleiotropic
Pertaining to a gene that is associated with multiple distinct phenotypes.
Footnotes
Competing interests statement
The authors declare no competing interests.
FURTHER INFORMATION
BioGRID: http://thebiogrid.org
DRYGIN: http://drygin.ccbr.utoronto.ca/
HistoneHits: http://histonehits.org
iPfam: http://ipfam.sanger.ac.uk/
Krogan lab interactome database: http://interactome-cmp.ucsf.edu/
Yeast fitness database: http://fitdb.stanford.edu/
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Phillips PC. Epistasis – the essential role of gene interactions in the structure and evolution of genetic systems. Nature Rev. Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol. Syst. Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barabasi AL. Scale-free networks: a decade and beyond. Science. 2009;325:412–413. doi: 10.1126/science.1173299. [DOI] [PubMed] [Google Scholar]
- 4.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 5.Beltrao P, et al. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–425. doi: 10.1016/j.cell.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furey TS. ChIP–seq and beyond: new and improved methodologies to detect and characterize protein–DNA interactions. Nature Rev. Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlebach G, Shamir R. Modelling and analysis of gene regulatory networks. Nature Rev. Mol. Cell Biol. 2008;9:770–780. doi: 10.1038/nrm2503. [DOI] [PubMed] [Google Scholar]
- 9.Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nature Rev. Mol. Cell Biol. 2010;11:414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nature Rev. Mol. Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 11.Ideker T, Krogan NJ. Differential network biology. Mol. Syst. Biol. 2012;8:565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linding R, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nature Biotech. 2012;30:1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarassov K, et al. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 16.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 17.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 18.Wodak SJ, Pu S, Vlasblom J, Seraphin B. Challenges and rewards of interaction proteomics. Mol. Cell Proteom. 2009;8:3–18. doi: 10.1074/mcp.R800014-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.von Mering C, et al. Comparative assessment of large-scale data sets of protein–protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 20.Bader GD, Hogue CW. Analyzing yeast protein–protein interaction data obtained from different sources. Nature Biotech. 2002;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- 21.Cusick ME, et al. Literature-curated protein interaction datasets. Nature Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolinski K, Chatr-Aryamontri A, Tyers M. Systematic curation of protein and genetic interaction data for computable biology. BMC Biol. 2013;11:43. doi: 10.1186/1741-7007-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licata L, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez SM, Noble WS, Rzhetsky A. Learning to predict protein–protein interactions from protein sequences. Bioinformatics. 2003;19:1875–1881. doi: 10.1093/bioinformatics/btg352. [DOI] [PubMed] [Google Scholar]
- 26.Zhang QC, et al. Structure-based prediction of protein–protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen R, et al. A Bayesian networks approach for predicting protein–protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 28.Wang PI, Marcotte EM. It’s the machine that matters: predicting gene function and phenotype from protein networks. J. Proteom. 2010;73:2277–2289. doi: 10.1016/j.jprot.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beltrao P, Cagney G, Krogan NJ. Quantitative genetic interactions reveal biological modularity. Cell. 2010;141:739–745. doi: 10.1016/j.cell.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 31.Haber JE, et al. Systematic triple-mutant analysis uncovers functional connectivity between pathways involved in chromosome regulation. Cell Rep. 2013;3:2168–2178. doi: 10.1016/j.celrep.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins SR, Roguev A, Krogan NJ. Quantitative genetic interaction mapping using the E-MAP approach. Methods Enzymol. 2010;470:205–231. doi: 10.1016/S0076-6879(10)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon SJ, et al. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proc. Natl Acad. Sci. USA. 2008;105:16653–16658. doi: 10.1073/pnas.0806261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butland G, et al. eSGA: E. coli synthetic genetic array analysis. Nature Methods. 2008;5:789–795. doi: 10.1038/nmeth.1239. [DOI] [PubMed] [Google Scholar]
- 35.Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nature Methods. 2007;4:861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- 36.Typas A, et al. High-throughput, quantitative analyses of genetic interactions in E. coli. Nature Methods. 2008;5:781–787. doi: 10.1038/nmeth.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 38.Ryan CJ, et al. Hierarchical modularity and the evolution of genetic interactomes across species. Mol. Cell. 2012;46:691–704. doi: 10.1016/j.molcel.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Babu M, et al. Genetic interaction maps in Escherichia coli reveal functional crosstalk among cell envelope biogenesis pathways. PLoS Genet. 2011;7:e1002377. doi: 10.1371/journal.pgen.1002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roguev A, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breslow DK, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nature Methods. 2008;5:711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davierwala AP, et al. The synthetic genetic interaction spectrum of essential genes. Nature Genet. 2005;37:1147–1152. doi: 10.1038/ng1640. [DOI] [PubMed] [Google Scholar]
- 44.Mnaimneh S, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 45.Bassik MC, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horn T, et al. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nature Methods. 2011;8:341–346. doi: 10.1038/nmeth.1581. [DOI] [PubMed] [Google Scholar]
- 47.Lin YY, et al. Functional dissection of lysine deacetylases reveals that HDAC1 and p300 regulate AMPK. Nature. 2012;482:251–255. doi: 10.1038/nature10804. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Roguev A, et al. Quantitative genetic-interaction mapping in mammalian cells. Nature Methods. 2013;10:432–437. doi: 10.1038/nmeth.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laufer C, Fischer B, Billmann M, Huber W, Boutros M. Mapping genetic interactions in human cancer cells with RNAi and multiparametric phenotyping. Nature Methods. 2013;10:427–431. doi: 10.1038/nmeth.2436. [DOI] [PubMed] [Google Scholar]
- 50.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nature Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 51.Byrne AB, et al. A global analysis of genetic interactions in Caenorhabditis elegans. J. Biol. 2007;6:8. doi: 10.1186/jbiol58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryan C, Greene D, Cagney G, Cunningham P. Missing value imputation for epistatic MAPs. BMC Bioinformatics. 2010;11:197. doi: 10.1186/1471-2105-11-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong SL, et al. Combining biological networks to predict genetic interactions. Proc. Natl Acad. Sci. USA. 2004;101:15682–15687. doi: 10.1073/pnas.0406614101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu X, Kensche PR, Huynen MA, Notebaart RA. Genome evolution predicts genetic interactions in protein complexes and reveals cancer drug targets. Nature Commun. 2013;4:2124. doi: 10.1038/ncomms3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pandey G, et al. An integrative multi-network and multi-classifier approach to predict genetic interactions. PLoS Comput. Biol. 2010;6:e1000928. doi: 10.1371/journal.pcbi.1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Folger O, et al. Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 2011;7:501. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl Acad. Sci. USA. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beyer A, Bandyopadhyay S, Ideker T. Integrating physical and genetic maps: from genomes to interaction networks. Nature Rev. Genet. 2007;8:699–710. doi: 10.1038/nrg2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharan R, Ideker T. Modeling cellular machinery through biological network comparison. Nature Biotech. 2006;24:427–433. doi: 10.1038/nbt1196. [DOI] [PubMed] [Google Scholar]
- 60.Mitra K, Carvunis AR, Ramesh SK, Ideker T. Integrative approaches for finding modular structure in biological networks. Nature Rev. Genet. 2013;14:719–732. doi: 10.1038/nrg3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bandyopadhyay S, Kelley R, Krogan NJ, Ideker T. Functional maps of protein complexes from quantitative genetic interaction data. PLoS Comput. Biol. 2008;4:e1000065. doi: 10.1371/journal.pcbi.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hillenmeyer ME, et al. Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. Genome Biol. 2010;11:R30. doi: 10.1186/gb-2010-11-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parsons AB, et al. Integration of chemical–genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nature Biotech. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 64.Kapitzky L, et al. Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol. Syst. Biol. 2010;6:451. doi: 10.1038/msb.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Boxem M, et al. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009.. This is a large-scale fragment-based protein–protein interaction screen that identifies the minimal regions of interaction for many interactions.
- 66.Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 67.Goehler H, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington’s disease. Mol. Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 68.Guglielmi B, et al. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.LaCount DJ, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103–107. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 70.Rain JC, et al. The protein–protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 71. Amberg DC, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. Nature Struct. Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28.. This is a pioneering study that highlights the use of integrating structural models with edgetic protein–protein interaction mapping.
- 72.Charloteaux B, et al. Protein–protein interactions and networks: forward and reverse edgetics. Methods Mol. Biol. 2011;759:197–213. doi: 10.1007/978-1-61779-173-4_12. [DOI] [PubMed] [Google Scholar]
- 73.Leanna CA, Hannink M. The reverse two-hybrid system: a genetic scheme for selection against specific protein/protein interactions. Nucleic Acids Res. 1996;24:3341–3347. doi: 10.1093/nar/24.17.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shih HM, et al. A positive genetic selection for disrupting protein–protein interactions: identification of CREB mutations that prevent association with the coactivator CBP. Proc. Natl Acad. Sci. USA. 1996;93:13896–13901. doi: 10.1073/pnas.93.24.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vidal M, Brachmann RK, Fattaey A, Harlow E, Boeke JD. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein–protein and DNA–protein interactions. Proc. Natl Acad. Sci. USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walhout AJM, et al. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 77.Zhong Q, et al. Edgetic perturbation models of human inherited disorders. Mol. Syst. Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dreze M, et al. ‘Edgetic’ perturbation of a C. elegans BCL2 ortholog. Nature Methods. 2009;6:843–849. doi: 10.1038/nmeth.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lim J, et al. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aloy P, et al. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- 82.Deng M, Mehta S, Sun F, Chen T. Inferring domain–domain interactions from protein–protein interactions. Genome Res. 2002;12:1540–1548. doi: 10.1101/gr.153002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 84.Prieto C, De Las Rivas J. Structural domain–domain interactions: assessment and comparison with protein–protein interaction data to improve the interactome. Proteins. 2010;78:109–117. doi: 10.1002/prot.22569. [DOI] [PubMed] [Google Scholar]
- 85.Riley R, Lee C, Sabatti C, Eisenberg D. Inferring protein domain interactions from databases of interacting proteins. Genome Biol. 2005;6:R89. doi: 10.1186/gb-2005-6-10-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H, et al. InSite: a computational method for identifying protein–protein interaction binding sites on a proteome-wide scale. Genome Biol. 2007;8:R192. doi: 10.1186/gb-2007-8-9-r192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang X, et al. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nature Biotech. 2012;30:159–164. doi: 10.1038/nbt.2106.. This study integrates high-throughput protein–protein interactions with three-dimensional structures of interacting interfaces to interpret human disease-associated mutations.
- 88.Schuster-Bockler B, Bateman A. Protein interactions in human genetic diseases. Genome Biol. 2008;9:R9. doi: 10.1186/gb-2008-9-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405.. This is a landmark example of the use of integrative approaches to determine the structure of a complex macromolecule — in this case, the nuclear pore complex that consists of 30 distinct proteins.
- 90.Lasker K, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl Acad. Sci. USA. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Campos M, Nilges M, Cisneros DA, Francetic O. Detailed structural and assembly model of the type II secretion pilus from sparse data. Proc. Natl Acad. Sci. USA. 2010;107:13081–13086. doi: 10.1073/pnas.1001703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ward AB, Sali A, Wilson IA. Biochemistry. Integrative structural biology. Science. 2013;339:913–915. doi: 10.1126/science.1228565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Finn RD, Marshall M, Bateman A. iPfam: visualization of protein–protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21:410–412. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- 94.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Berman HM, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yellaboina S, Tasneem A, Zaykin DV, Raghavachari B, Jothi R. DOMINE: a comprehensive collection of known and predicted domain–domain interactions. Nucleic Acids Res. 2011;39:D730–D735. doi: 10.1093/nar/gkq1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reimand J, Hui S, Jain S, Law B, Bader GD. Domain-mediated protein interaction prediction: from genome to network. FEBS Lett. 2012;586:2751–2763. doi: 10.1016/j.febslet.2012.04.027. [DOI] [PubMed] [Google Scholar]
- 98.Charles GM, et al. Site-specific acetylation mark on an essential chromatin-remodeling complex promotes resistance to replication stress. Proc. Natl Acad. Sci. USA. 2011;108:10620–10625. doi: 10.1073/pnas.1019735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fuchs SM, Kizer KO, Braberg H, Krogan NJ, Strahl BD. RNA polymerase II carboxyl-terminal domain phosphorylation regulates protein stability of the Set2 methyltransferase and histone H3 di- and trimethylation at lysine 36. J. Biol. Chem. 2012;287:3249–3256. doi: 10.1074/jbc.M111.273953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morrison AJ, et al. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell. 2007;130:499–511. doi: 10.1016/j.cell.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 101.Mehta M, et al. Individual lysine acetylations on the N terminus of Saccharomyces cerevisiae H2A.Z are highly but not differentially regulated. J. Biol. Chem. 2010;285:39855–39865. doi: 10.1074/jbc.M110.185967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang AY, Aristizabal MJ, Ryan C, Krogan NJ, Kobor MS. Key functional regions in the histone variant H2A.Z C-terminal docking domain. Mol. Cell. Biol. 2011;31:3871–3884. doi: 10.1128/MCB.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim HS, et al. An acetylated form of histone H2A.Z regulates chromosome architecture in Schizosaccharomyces pombe. Nature Struct. Mol. Biol. 2009;16:1286–1293. doi: 10.1038/nsmb.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haarer B, Viggiano S, Hibbs MA, Troyanskaya OG, Amberg DC. Modeling complex genetic interactions in a simple eukaryotic genome: actin displays a rich spectrum of complex haploinsufficiencies. Genes Dev. 2007;21:148–159. doi: 10.1101/gad.1477507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 106.Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 107.Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol. Cell. Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Dai J, et al. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019.. This paper describes both the systematic mutation of every individual residue of two histone proteins and the use of drug sensitivity screening to assess the functional effects of these mutations.
- 109.Matsubara K, Sano N, Umehara T, Horikoshi M. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells. 2007;12:13–33. doi: 10.1111/j.1365-2443.2007.01031.x. [DOI] [PubMed] [Google Scholar]
- 110.Nakanishi S, et al. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nature Struct. Mol. Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Huang HL, et al. HistoneHits: a database for histone mutations and their phenotypes. Genome Res. 2009;19:674–681. doi: 10.1101/gr.083402.108.. This paper reports a database that focuses on a specific protein family (histones) and that integrates the results of phenotyping screens of point mutants from several laboratories. It provides an interactive structure on which residues that are associated with specific phenotypes can be highlighted.
- 112. Braberg H, et al. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell. 2013;154:775–788. doi: 10.1016/j.cell.2013.07.033.. This study reports the functional dissection of RNA polymerase II by genetic interaction profiling of point mutants from multiple distinct subunits; it shows that the mutation of residues that are on distinct subunits but that are close together in the three-dimensional structure have similar genetic interaction profiles.
- 113.Alber F, Forster F, Korkin D, Topf M, Sali A. Integrating diverse data for structure determination of macromolecular assemblies. Annu. Rev. Biochem. 2008;77:443–477. doi: 10.1146/annurev.biochem.77.060407.135530. [DOI] [PubMed] [Google Scholar]
- 114.Hietpas R, Roscoe B, Jiang L, Bolon DN. Fitness analyses of all possible point mutations for regions of genes in yeast. Nature Protoc. 2012;7:1382–1396. doi: 10.1038/nprot.2012.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roscoe BP, Thayer KM, Zeldovich KB, Fushman D, Bolon DN. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J. Mol. Biol. 2013;425:363–1377. doi: 10.1016/j.jmb.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McGary KL, Lee I, Marcotte EM. Broad network-based predictability of Saccharomyces cerevisiae gene loss-of-function phenotypes. Genome Biol. 2007;8:R258. doi: 10.1186/gb-2007-8-12-r258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee I, et al. Predicting genetic modifier loci using functional gene networks. Genome Res. 2010;20:1143–1153. doi: 10.1101/gr.102749.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 120.Jager S, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shapira SD, et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Neveu G, et al. Comparative analysis of virus–host interactomes with a mammalian high-throughput protein complementation assay based on Gaussia princeps luciferase. Methods. 2012;58:349–359. doi: 10.1016/j.ymeth.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stark C, et al. The BioGRID interaction database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Koh JL, et al. DRYGIN: a database of quantitative genetic interaction networks in yeast. Nucleic Acids Res. 2010;38:D502–D507. doi: 10.1093/nar/gkp820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fraser JS, Gross JD, Krogan NJ. From systems to structure: bridging networks and mechanism. Mol. Cell. 2013;49:222–231. doi: 10.1016/j.molcel.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Maher MC, Uricchio LH, Torgerson DG, Hernandez RD. Population genetics of rare variants and complex diseases. Hum. Hered. 2012;74:118–128. doi: 10.1159/000346826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gravel S, et al. Demographic history and rare allele sharing among human populations. Proc. Natl Acad. Sci. USA. 2011;108:11983–11988. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lander GC, Saibil HR, Nogales E. Go hybrid: EM, crystallography, and beyond. Curr. Opin. Struct. Biol. 2012;22:627–635. doi: 10.1016/j.sbi.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Russel D, et al. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bau D, et al. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nature Struct. Mol. Biol. 2011;18:107–114. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lasker K, Topf M, Sali A, Wolfson HJ. Inferential optimization for simultaneous fitting of multiple components into a CryoEM map of their assembly. J. Mol. Biol. 2009;388:180–194. doi: 10.1016/j.jmb.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nature Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 135.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–38. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 136.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc. Natl Acad. Sci. USA. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Krastev DB, et al. A systematic RNAi synthetic interaction screen reveals a link between p53 and snoRNP assembly. Nature Cell Biol. 2011;13:809–818. doi: 10.1038/ncb2264. [DOI] [PubMed] [Google Scholar]
- 139. Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006.. This is a genome-wide RNAi screen of isogenic human cell lines to identify genes that are synthetically lethal with a specific oncogenic mutation.
- 140.Wang Y, et al. Critical role for transcriptional repressor Snail2 in transformation by oncogenic RAS in colorectal carcinoma cells. Oncogene. 2010;29:4658–4670. doi: 10.1038/onc.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Miller JP, et al. A genome-scale RNA-interference screen identifies RRAS signaling as a pathologic feature of Huntington’s disease. PLoS Genet. 2012;8:e1003042. doi: 10.1371/journal.pgen.1003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.McDermott U, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc. Natl Acad. Sci. USA. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Muellner MK, et al. A chemical–genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nature Chem. Biol. 2011;7:787–793. doi: 10.1038/nchembio.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 147.Corcoran RB, et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell. 2013;23:121–128. doi: 10.1016/j.ccr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ding Q, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nature Biotech. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Han TX, Xu XY, Zhang MJ, Peng X, Du LL. Global fitness profiling of fission yeast deletion strains by barcode sequencing. Genome Biol. 2010;11:R60. doi: 10.1186/gb-2010-11-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Simonis N, et al. Empirically controlled mapping of the Caenorhabditis elegans protein–protein interactome network. Nature Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Guruharsha KG, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 157.Bakal C, et al. Phosphorylation networks regulating JNK activity in diverse genetic backgrounds. Science. 2008;322:453–456. doi: 10.1126/science.1158739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hu P, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7:e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Havugimana PC, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Stelzl U, et al. A human protein–protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 161.Rual JF, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]