Abstract
Cellular reprogramming holds tremendous potential for cell therapy and regenerative medicine. Recently, fibroblasts have been directly converted into induced neurons (iNs) by overexpression of the neuronal transcription factors Ascl1, Brn2 and Myt1L. Hypothesizing that cell-topography interactions could influence the fibroblast-to-neuron reprogramming process, we investigated the effects of various topographies on iNs produced by direct reprogramming. Final iN purity and conversion efficiency were increased on micrograting substrates. Neurite branching was increased on microposts and decreased on microgratings, with a simplified dendritic arbor characterized by the reduction of MAP2+ neurites. Neurite outgrowth increased significantly on various topographies. DNA microarray analysis detected 20 differentially expressed genes in iNs reprogrammed on smooth versus microgratings, and quantitative PCR (qPCR) confirmed the upregulation of Vip and downregulation of Thy1 and Bmp5 on microgratings. Electrophysiology and calcium imaging verified the functionality of these iNs. This study demonstrates the potential of applying topographical cues to optimize cellular reprogramming.
1. Introduction
Neurodegenerative disorders caused by the loss of specific cells in the nervous system are on the rise in the aging population – presenting an increasingly urgent challenge for the society. Wernig et al. in 2010 directly reprogrammed mouse fibroblasts into functional neurons by lentiviral delivery of the transcription factors (TFs) Brn2, Ascl1, and Myt1l - referred to as induced neuronal (iN) cells [1]. Further studies confirmed that iNs could be obtained from adult human cells [2–4]. Continuing progress saw the generation of induced functional dopaminergic neurons (iDAs) [5–8], cholinergic motor neurons (iMNs) [9] and induced oligodendroglial cells (iOPCs) [10] from various cocktails of transcription factors. Neuronal reprogramming of fibroblasts from patients with familial Alzheimer’s disease (fAD) could generate a model that recapitulated hallmarks of the disease [2]. These advances of generating a new source of functional neurons render cell replacement therapy a promising approach to combat neurodegenerative diseases.
We [11] and Maucksch et al. [12] have respectively demonstrated that iNs and neural precursor-like cells can be produced without viral vectors, paving the way to a clinically viable cell source for translation. Another issue is the continuing need to optimize the reprogramming process to yield a therapeutic quantity of iNs. To date, all neuronal reprogramming has been conducted on conventional tissue culture dishes. During reprogramming a drastic change in cell morphology and gene expression occurs. Considering the recent findings that cell-topography interactions modulated many cell phenotypes [13, 14] and improved induced pluripotent stem cell (iPSC) generation [15], we hypothesized that topographical cues can influence the neuronal conversion process and the subsequent phenotype of the induced neurons.
In vivo, all adherent cells encounter topographical features from neighboring cells and the extracellular matrix. The function of cells in general – and of neurons in particular – depends on their shape and polarity. Neurons explore their immediate extracellular environment to make the appropriate connections to build functional neural circuitry. For instance, neurons and PC12 cells polarized along gratings [16] and this polarity is controlled by the confinement of focal adhesions on the gratings [17, 18]. Focal adhesions have been shown to play an important role in topography sensing [19]. In this study we investigated if topographical features imprinted on polystyrene would affect the direct reprogramming of fibroblasts to neurons by means of lentiviral vectors. We chose posts and grating topographies because these have been shown to have an effect on neuronal cells [20]. Here we report the resultant changes compared to conventional planar tissue-culture polystyrene (TCPS), in terms of efficiency, morphology, and gene expression, suggesting a role of cell-topography interactions in shaping iNs.
2. Materials and Methods
2.1. Substrate Preparation
2 × 2 cm2 metal masters of pits of 1 μm diameter spaced by 4 μm (F (1.4)) and pits of 4 μm diameter spaced by 1 μm (F (4.1)) were produced by a lithographic process reference in Suppl. Text. A master for the 5 μm grating features was produced by UV photolithography. PDMS substrates were fabricated from the masters and used for imprinting onto polystyrene (PS) for cell culture.
2.2. Cell culture
Primary mouse embryonic fibroblasts (PMEFs) were isolated as previously described [1] or purchased (ATCC, Manassas, VA). PMEF (Passage 2–4) at 15,000 cells cm−2 were seeded on PS substrates at 37 °C and 5% CO2 in complete PMEF medium as previously described [1]. For neurite tracing experiments a seeding density of 7,500 cells cm−2 was used to enable the attribution of the neurites to their cell body. 24 hours after seeding, PMEFs were infected in PMEF medium containing 8 mg mL−1 sequabrene (Sigma, St. Louis, MO) with TFs in lentiviral vectors Ascl1 (Addgene, Cambridge, MA, plasmid # 27150), Brn2 (Addgene plasmid # 27151), Myt1L (Addgene plasmid # 27152) [1], and M2rttA (Addgene plasmid # 20342) [21]. 24 hours after infection, PMEF medium was replaced with PMEF medium containing 2 mg mL−1 doxycycline (Sigma). 72 hours after infection, PMEF medium containing doxycycline was replaced with neurogenic N3 medium containing: DMEM/F-12 (Life Technologies/Invitrogen), 25 μg mL−1 bovine insulin (Gemini Bio-Products, West Sacramento, CA), 50 μg mL−1 human apo-transferrin, 30 nM sodium selenite, 20 nM progesterone, 100 μM putrescine (Sigma), 10 ng mL−1 human bFGF2 (Stemgent, Cambridge, MA), 25 μg mL−1 gentamicin (Life Technologies/Invitrogen) and 2 mg mL−1 doxycycline (Sigma). After 48 hrs in N3 medium with doxycycline, the medium was exchanged every 48 hrs with N3 medium without doxycycline. To identify iNs from non-converted fibroblasts, the cells were infected with a synapsin promoter-driven RFP expression plasmid (Addgene, plasmid # 22909) [22] at day 10 in electrophysiology, microarray and flowcytometry experiments. In some experiments, 25 μM blebbistatin (Millipore, Billerica, MA) – an inhibitor of non-muscle myosin II ATPase – was added to the culture medium throughout the entire cell culture. Three independent experiments with triplicate substrates were performed for each analysis.
2.3. Immunocytochemistry and Antibodies
Cells were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature, permeabilized, and stained in a blocking solution containing the antibodies, 0.03 g/ml bovine serum albumin (BSA, Sigma), 10% goat serum (Sigma), and 0.3% Triton X-100 (Sigma) in PBS for 2 hr. The samples were washed before incubation with secondary antibodies for 1h at RT. The samples were then mounted in Fluoro-Gel (Electron Microscopy Sciences, Hatfield, PA, USA) for fluorescent imaging.
Antibodies against the following proteins were applied for immunofluorescence (IF). Monoclonal antibodies: Neuronal class III β-tubulin (Tuj1) 1:500 (Covance, Princeton, NJ), MAP2 1:500 (BD Biosciences, Franklin Lakes, New Jersey). Rabbit polyclonal antibodies: Neuronal class III β-tubulin (Tuj1) 1:500 (Covance). Secondary antibodies used for IF were Alexa Fluor 594 goat anti-rabbit secondary antibody 1:200 (Life Technologies/Invitrogen), or Alexa Fluor 488 goat anti-mouse secondary antibody 1:200 (Life Technologies/Invitrogen) and cell nuclei were stained with 4,6-Diamidino-2-phenylindole (DAPI) 1:5000 (Life Technologies/Invitrogen).
2.4. Image Analysis
Wide field images were collected for measurement of Tuj1+ cells using a Nikon Eclipse TE2000-U fluorescence inverted microscope and a 4x objective. Large areas corresponding to 6 single images were scanned with an automated stage and the images stitched together from each of three replicate substrates: planar PS, 5 μm gratings PS, F (1.4) PS and F (4.1) PS. At least 705 cells from three independent experiments with at least 235 cells per substrate were analyzed for each condition using a FIJI macro [23], implemented by a tester blinded to the identity of each condition. Briefly, Bernsen’s thresholding method was used to define regions of high local contrast within each image, corresponding to the cell bodies. Using FIJI’s “Analyze Particles” command, ellipses were fitted to these outlines, and an angle of deviation from the horizontal axis was measured for each, with 0 ° (90 °) denoting a cell parallel (perpendicular) to the grating direction or line of posts. If an angle was greater than 90 ° then the angle was subtracted from 180 °, so all angles fell between 0 ° and 90 °. Very small and large (non-cell) debris particles were excluded with a size threshold. For Figure 1M all measured angles were averaged. An average angle of deviation of 45 ° represents a random orientation with respect to the horizontal axis. For Figure 1N the distribution of the measured angles from 0 ° to 90 ° is displayed. To quantify cell attachment, cells were fixed and DAPI stained 24h after seeding. Images of DAPI-stained cell nuclei were acquired by scanning each of the three replicate substrates. Bernsen’s thresholding method was used to define regions of high local contrast within each image corresponding to the cell’s nucleus, and the “Analyze Particles” command was used to count the number of nuclei for each substrate. For neurite tracing, an initial fibroblast seeding density of 7500 cells cm−2 was used and the iNs produced were stained for Tuj1 expression. Higher magnification images of iNs were collected by wide field microscopy by an experimenter blinded to the experimental conditions. A blinded experimenter traced neurites using the “Simple Neurite Tracer” plugin in FIJI. The cell soma was chosen as starting point and the longest neurite was designated as the primary neurite. The number of neurites per soma was determined by the sum of primary and secondary neurites extending from the cell body. The primary neurite length was determined by comparison of the average length of the primary neurite per iN for each substrate topography. The length of the neurite arbor was determined by comparison of the length of all neurites per iN for each substrate topography.
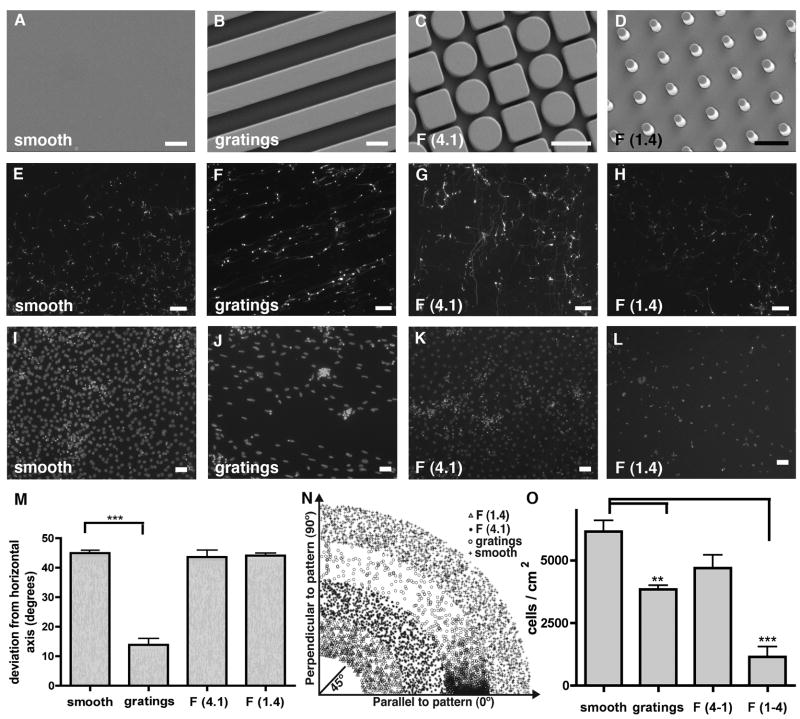
Figure 1. Tuj1+ induced neurons cultured on different topographies.
(A–D) Images of substrate topographies with micron-sized features fabricated by micro-imprinting. Scale bar 5 μm. (E) Neurites of tuj1+ iNs on smooth substrates displayed random orientation. Scale bar 100 μm. (F) Neurites of tuj1+ iNs on 5 μm gratings showed alignment with the underlying substrate features. Scale bar 100 μm. (G, H) Neurites of tuj1+ iNs on F (1.4) and F (4.1) substrates displayed random orientation, similar to smooth substrates. Scale bar 100 μm. (I–L) DAPI stain of cell nuclei attached to the different topographies. Scale bar 50 μm. (M) Quantification of iN’s directionality; 45° represents completely random orientation. A global one-way ANOVA of cell alignment revealed a significant effect of substrate gratings on iN alignment (p < 0.0001). (N) Visual representation of the iN’s angle in respect to the pattern axis. X-axis represents the axis parallel to the pattern, whereas y-axis represents iN alignment perpendicular to the pattern. (O) Quantification of cell attachment to the different topographies. Variation of cell attachment to the different substrates was observed by a global one-way ANOVA with a significant reduction of cell attachment to F (1.4) and grating substrates (p < 0.001).
2.5. Electrophysiology
iNs for patch clamp analysis were identified by synapsin promoter-driven RFP expression after 12 days of culture in N3 medium. Micropipettes with resistances between 3–7 MΩ were filled with internal solution containing 130 mM KMeSO3, 10 mM HEPES, 10 mM sodium phosphocreatine, 4 mM MgCl2, 4 mM Na2ATP, 0.4 mM Na2GTP, 3 mM sodium L-ascorbic acid, with pH 7.24 and an osmolarity of 290 mM. Giga-ohm membrane seals were formed under voltage-clamp conditions. Action potentials were then recorded using an Axon Multiclamp 700B Microelectrode Amplifier (Molecular Devices, Sunnyvale, CA) by stepwise whole-cell current clamp injections, and analyzed with MATLAB programs developed in-house. The cells were perfused with artificial cerebral spinal fluid (ACSF) saturated with 5% O2 and 95% CO2 and containing: 130 mM NaCl, 2.5 mM KCl, 2 mM NaHCO3, 1.25 mM NaH2PO4, 25 mM glucose, 2 mM CaCl2, and 2 mM MgCl2.
2.6. Calcium Imaging
Cells were infected with a GCaMP5 construct under control of a MAP2 promoter at day 9 after infection with the Ascl1, Myt1L, and Brn2. Imaging experiments were performed 4 – 5 days thereafter in imaging solution (NaCl 140 mM, KCl 5 mM, 0.8 mM MgCl2, Hepes 10 mM, CaCl 2 mM, glucose 10 mM). Calcium imaging was performed using a Nikon Eclipse TE2000-U fluorescence inverted microscope and a 20x objective. The time-series images were acquired at a rate of 5 fps. 50 mM KCl, 50 mM NaCl and 1 mM glutamate in imaging solution was directly pipetted onto the iN during perfusion. Calcium imaging was performed at room temperature. To produce ΔF/F traces, ROIs were drawn with FIJI, containing either the iN cell body or the background, and the average fluorescent intensity was measured for each time point.
2.7. Flow Cytometry
Cells were washed briefly with phosphate-buffered saline (PBS) (Mediatech, Washington, DC), and released from the substrates with 0.25% Trypsin-EDTA (Invitrogen/Life Technologies). Trypsin was inactivated with serum-containing media and the cells were centrifuged at 4 °C, and resuspended in ice-cold N3 medium if they were to be sorted for RNA extraction, or fixed in ice-cold PBS containing 1% paraformaldehyde (PFA) for 10 min for flow cytometric analysis. Cells were analyzed for tau promoter-driven expression of EGFP, or synapsin promoter-driven expression of RFP with FACS Canto II (Becton Dickinson, Franklin Lakes, NJ), or sorted and analyzed with FACSVantage SE (Becton Dickinson, Franklin Lakes, NJ), and collected for RNA extraction. For each analysis we conducted three independent experiments with triplicate substrates and at least 930 cells were analyzed.
2.8. Microarray
Total RNA samples were assessed for quality with an Agilent 2100 Bioanalyzer G2939A (Agilent Technologies, Santa Clara, CA) and Nanodrop 8000 spectrophotometer (Thermo Scientific/Nanodrop, Wilmington, DE). Hybridization targets were prepared with MessageAmp™ Premier RNA Amplification Kit (Applied Biosystems/Ambion, Austin, TX) from total RNA, hybridized to GeneChip® Mouse 430A 2.0 arrays in an Affymetrix GeneChip® hybridization oven 645, washed in an Affymetrix GeneChip® Fluidics Station 450, and scanned with an Affymetrix GeneChip® Scanner 7G according to standard Affymetrix GeneChip® Hybridization, Wash, and Stain protocols (Affymetrix, Santa Clara, CA). Data processing and statistical analysis: Partek Genomics Suite 6.5 (Partek Inc., St. Louis, MO) was used to perform data analysis. Robust multi-chip analysis (RMA) normalization was performed on the entire data set. Multi-way ANOVA and fold-change were performed to select target genes that were differentially expressed between previously sorted synapsin-RFP− (immature or non-converted cells) and synapsin-RFP+ iNs, and between synapsin-RFP+ iNs on smooth substrates and synapsin-RFP+ iNs on 5 μm grating topography. Top differentially expressed genes were selected with a p value cutoff of 0.05 based on ANOVA test, and a fold-change cutoff of ±1.5. Hierarchical clustering was performed on differentially expressed genes based on average linkage with Pearson’s dissimilarity.
2.9. Real-time qPCR
Total RNA samples were assessed for quality with a Nanodrop 8000 spectrophotometer (Thermo Scientific/Nanodrop, Wilmington, DE). Comparative CT real-time reverse transcription-PCR was performed in 20 μl reactions using the QuantiTect SYBR Green RT-PCR Kit (QIAGEN, Valencia, CA) with 10 ng of starting mRNA isolated with RNeasy and QIAshredder kits (QIAGEN). PCR proceeded for 40 cycles in an ABI 7900HT Real-Time PCR System (Applied Biosystems, Carlsbad, CA). Target gene expression levels were normalized to endogenous GAPDH references, and presented as a fold-change relative to expression levels from synapsin-RFP− PMEFs.
2.10. Statistical Analysis
One-way ANOVA with Bonferroni-corrected post-hoc testing was used to compare cell attachment and alignment. Student’s t-tests were used to compare conversion efficiency for flow cytometry and for qPCR analysis. Global one-way ANOVA with Dunn’s posthoc testing was used to compare the length of primary neurites and neurite arbor on different substrates.
3. Results
3.1. Tuj1+ induced neurons on substrate topographies
The embossed polystyrene (PS) substrates used in this study consisted of smooth controls (Fig. 1A), microgratings of 5 μm width with a 1:1:1 aspect ratio (Fig. 1B), posts of 1 μm diameter spaced by 4 μm (F (1.4)) with a depth of 5 μm (Fig. 1C) and posts of 4 μm diameter spaced by 1 μm (F (4.1)) with a depth of 5 μm (Fig. 1D). PMEFs were seeded at 1.5 x104 cells/cm2, infected with Ascl1, Myt1L, and Brn2 (BAM factors), and cultured on the different substrates (Fig. S1). Cell attachment was assessed 24 hours after seeding. On smooth substrates cells adhered well with 6.1 ± 0.6 x104 cells/cm2 (Fig. 1I), on grating substrates the cell attachment was significantly reduced to 3.8 ± 0.2 x 104 cells/cm2 (Fig. 1J), and the cell nuclei displayed a more elliptic shape. On the F (4.1) topography, which consists of pits of 4 μm diameter spaced by 1 μm, the cells adhered well with 4.7 ± 0.7 x 104 cells/cm2 (Fig. 1K). On the F (1.4) topography, where 1 μm diameter pits are spaced by 4 μm, sparse DAPI staining depicts significantly diminished cell attachment with 1.2 ± 0.6 x 104 cells/cm2 (Fig. 1L). To assess iN orientation and alignment the cells were stained with neuron-specific beta-III tubulin (Tuj1), a pan-neuronal marker. On smooth control substrates the converted iNs displayed typical neuronal morphologies and neurites extended in random directions (Fig. 1E). On gratings substrates iNs adhered primarily on top of the grating ridges and extended their neurites in the grating direction (Fig. 1F). On the F (4.1) topography and the F (1.4) topography the neurites extended randomly along the posts (Fig. 1G, Fig. 1H) Fig. 1O shows the quantification of the difference in cell attachment to the different substrates. Fig. 1M indicates the average angle measured of the neurites with respect to the grating direction or the line of posts. On smooth substrates neurite extensions from the cell body displayed an average angle of 44.9° (randomly aligned), compared with an angle of 13.9° (parallel to the grating axis) for cells on the grating substrates (p < 0.0001). No significant differences were observed among the smooth, F (4.1) and F (1.4) substrates (Fig. 1M). For a more visual representation, angles of each individual iN were plotted in relation to the pattern, with X = 0° for angles parallel to the pattern direction and Y = 0° for angles perpendicular to the pattern direction (Fig. 1N).
3.2. iN reprogramming efficiency on topography
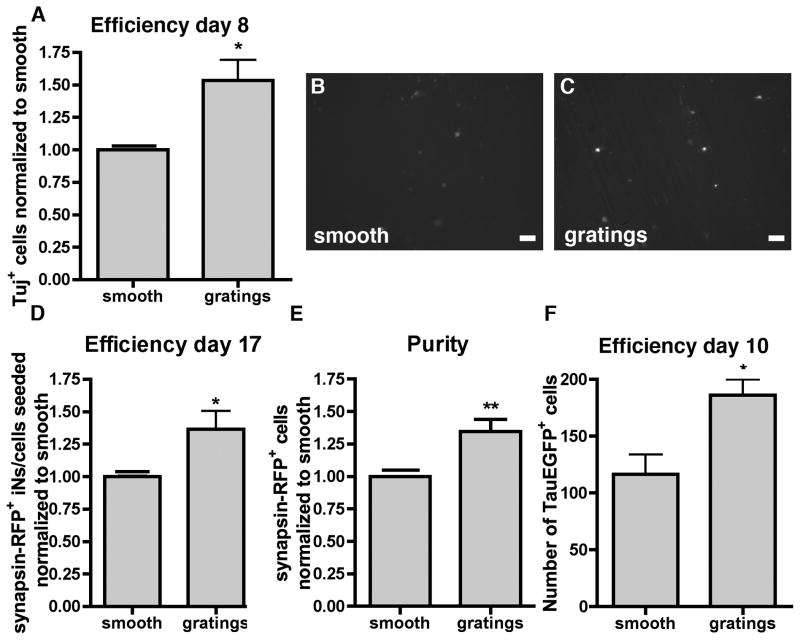
Micrograting topography polarized the cells effectively along its axis, influenced the phenotype of iNs dramatically and still allowed the recovery of enough cells for large scale analysis. Additionally, when we quantified the number of Tuj+ cells, we observed a significant increase (1.53 ± 0.27 fold) in the number of Tuj1+ cells on gratings compared to smooth substrates (Fig. 2A). Tuj1 however is occasionally expressed in cells with fibroblastic morphology that fail to fully convert to iNs. To assess if grating topography had an effect on the generation of functional iNs, we chose a reporter that expresses RFP under control of the neuronal synapsin promoter. RFP+ iNs were more fully converted and capable of generating action potentials [11]. We infected the cells with the synapsin-RFP reporter plasmid (Fig. 2B–C) and subjected the cells to flow cytometry. The normalized efficiency defined as the ratio of RFP+ cells at the end of the experiment to the number of cells seeded was significantly higher (1.36 ± 0.14 fold) for gratings compared with smooth substrates (Fig. 2D), corresponding to 11.6% and 16.1% respecitvely. The normalized purity of iNs, defined as the percentage of synapsin-RFP+ cells to the total number of cells sorted, was also significantly higher (1.35 ± 0.09 fold) on gratings substrates compared to smooth (Fig. 2E). To address if grating topography had an effect on the kinetics of iN reprogramming, we used fibroblasts from mice expressing EGFP under a neuron specific Tau promoter [1]. When infected with the BAM factors, cells expressing Tau-EGFP were considered iNs. Tau-EGFP could be detected at day 10 post infection, and at this time point we could observe an increase in Tau-EGFP+ iNs on grating topography (186 ± 13.8) compared to smooth surface (116.3 ± 17.6) (Fig. 2F).
Figure 2. Increase of grating topography on iN generation.
(A) Quantification of the number of Tuj+ cells on grating substrates normalized to smooth substrates. Images of synapsin-RFP+ iNs on (B) smooth and on (C) gratings substrates. Scale bar 50 μm. (D) Efficiency of iN defined as the ratio of synapsin-RFP+ iNs to cells seeded and normalized to smooth substrates. (E) Quantification of iN purity defined as the ratio of synapsin-RFP+ cells to the total number of cells sorted normalized to the percentage of synapsin-RFP+ cells on smooth substrates. (F) Number of Tau-EGFP+ iNs at day 10. All data displayed as mean ± SEM.
3.3. Neurite branching and outgrowth on topographies
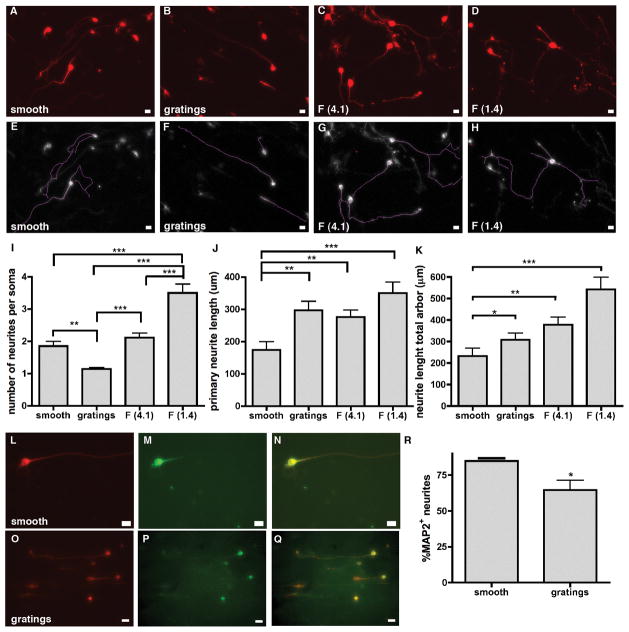
Next we analyzed neurite branching and neurite outgrowth, as they are critical for the development of functional neuronal circuitry. Higher magnification images and lower seeding density cultures (7500 cells cm−2) of Tuj1+ iNs on smooth, 5 μm gratings, F (4.1) and F (1.4) topographies (Fig. 3A–D) were used to trace the neurites (Fig. 3E–F). The number of neurites per cell soma and the neurite outgrowth was influenced by the underlying substrate topography. On smooth substrates the iNs extended an average of 1.85 ± 0.15 neurites per soma, which was similar to the 2.1 ± 0.14 neurites per iN on the F (4.1) topography. The number of neurites per soma on the 5 μm gratings topography was significantly reduced to 1.14 ± 0.05. On the F (1.4) topography however, the number of neurites per iN was significantly increased compared to smooth, gratings, and F (4.1) topographies, with 3.5 ± 0.27 neurites per iN (Fig. 3I). To investigate if the underlying topography influences neurite outgrowth, we averaged the length of the longest (primary) neurite traced (Fig. 3J) as well as the total length of all neurites per cell (Fig. 3K) for each substrate. On smooth substrates, iNs developed the shortest neurites, with an average length of 174 ± 15.9 μm for the primary neurite and 237.8 ± 37.6 μm for the total length of all neurites per cell. iNs on the 5 μm grating substrates formed primary neurites of comparable length to the F (4.1) topography, with average lengths of 296.6 ± 2.18 μm and 275.8 ± 13.4 μm, respectively, and the total length of all neurites combined were 308.9 ± 30.3 μm and 378.6 ± 29.9 μm, respectively. iNs on the F (1.4) topography developed the longest neurites, with an average length of the primary neurite of 349.8 ± 14.4 μm and a total length for all neurites combined as 551.4 ± 57.7 μm.
Figure 3. Influence of substrate topographies on neurite branching and outgrowth of Tuj1+ iNs.
(A–D) typical images of iNs on different topographies with neurites stained by Tuj1. Scale bar 10 μm. (E–H) Traces of neurites from widefield microscopy images obtained using Fiji. (I) Quantification of the number of neurites per cell soma. iNs on grating topography displayed significantly less neurite extensions per cell soma compared to smooth, F (4.1) and F (1.4) topographies, whereas neurites of iNs on F (1.4) were significantly more branched compared to neurites of iNs on smooth, gratings or F (4.1) topography. Graph shows analysis by a global one-way ANOVA with Dunn’s posthoc testing (** p < 0.01; *** p < 0.0001). (J) neurite outgrowth determined by the length of the primary path and (K) total neurite arbor was significantly enhanced on gratings, F (4.1) and F (1.4) topographies compared to neurites of iNs on smooth control substrates. Graph shows analysis by a global one-way ANOVA with Dunn’s posthoc testing (* p < 0.05; ** p < 0.01; *** p < 0.001). (L,O) Tuj staining on smooth and gratings substrates; (M,P) MAP2 staining on smooth and gratings substrates; scale bar 10 μm (N,Q) overlay of Tuj and MAP2 staining; (R) % of MAP2 expressing neurites. All data displayed as mean ± SEM.
Since on gratings we observed dramatically reduced neurite branching we stained for the dendritic marker MAP2 in addition to Tuj1 (Fig. 3L–Q). We observed 84.8 % ± 0.23 of neurites expressing MAP2 on smooth substrates, compared to only 64.6% ± 6.7 of neurites expressing MAP2 on grating substrates (Fig. 3R).
3.4. iN gene expression in response to topography
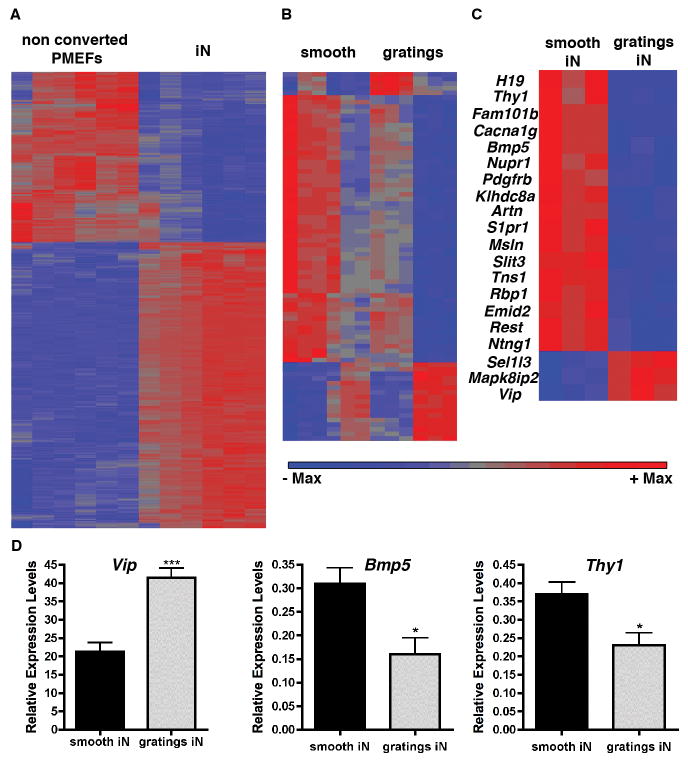
To understand the topography-mediated changes of iN phenotype at the gene expression level, we conducted a microarray experiment on cells reprogrammed on smooth surface with those on 5 μm gratings, the two substrates generating iNs with the greatest morphological differences. Synapsin-RFP+ iNs from smooth surface or gratings were sorted by flow cytometry and compared with Synapsin-RFP− non-converted PMEFs. As expected, a large number of genes (9308) were differentially expressed between the iNs and the non-converted PMEFs (Fig. 4A). When comparing gene expression of all cells (synapsin-RFP+ and synapsin-RFP−) on smooth substrates to 5 μm gratings we detected 81 differentially expressed genes (Fig. 4B). When we grouped these genes into functional annotation clusters using DAVID functional annotation bioinformatics microarray analysis, we observed enrichment in genes for the following clusters: extracellular matrix, glycoproteins, signaling and secreted proteins, which comprised 34 different genes; TGFβ pathway with artemin, Bmp5, inhibinβA, and C-type-lectin domain family 11; genes encoding for membrane associated proteins, like ephrinA2, growth arrest specific 1, netrin G1, and Thy1; and finally genes encoding proteins implicated in neuronal differentiation and cell projection organization like artemin, netrin, slit3, Thy1, growth arrest specific protein 1, and sphingosine-1-phosphate receptor 1. We then compared only synapsin-RFP+ iNs produced on smooth versus 5 μm gratings topographies, and discovered 20 differentially expressed genes (Fig. 4C). The functional annotation classification using DAVID grouped the genes into the following categories: neuron projection morphogenesis, axonogenesis, neuron projection development, containing the genes artemin, netrin G1 and slit homologue 3; genes associated with the cell membrane structure, including genes for voltage-gated calcium channel T type, alpha 1G subunit, mesothelin, netrin G1, platelet derived growth factor receptor beta polypeptide (PDGFβ), tensin1, thymus cell antigen 1 theta (Thy1), sphingosine-1-phosphate receptor 1; genes involved in neuronal differentiation comprising Thy1, slit homologue 3, netrin, artemin and sphingosine-1-phosphate receptor 1; and genes associated with signal transduction and secretion such as mitogen-activated protein kinase 8 interacting protein 2 (Mapk8ip2) vasointestinal polypeptide (Vip), bone morphogenic protein 5 (Bmp5), mesothelin, Pdgf, Thy1, EMI domain containing 2, slit 3, artemin and sphingosine-1-phosphate receptor 1. To validate these results we performed real-time qPCR analysis for Vip, Thy1 and Bmp5, which confirmed the results obtained by the microarray experiment.
Figure 4. Hierarchical clustering of gene transcripts.
(A) Hierarchical clustering of transcripts from non-converted PMEFs not expressing the synapsin-RFP reporter (left) and converted synapsin-RFP+ iNs (right). (B) Hierarchical clustering of expressed transcripts differentially regulated on smooth versus on 5 μm gratings topography. (C) Hierarchical clustering of expressed transcripts differentially regulated in iNs on smooth substrates (left) and on 5 μm gratings substrates (right). 20 genes expressed in iNs were found to be differentially regulated in response to the grating substrates compared to the smooth controls. (D) Quantitative, real time qPCR for Vip, Thy1 and Bmp5.
3.5. Functional Assessment of iNs on topography
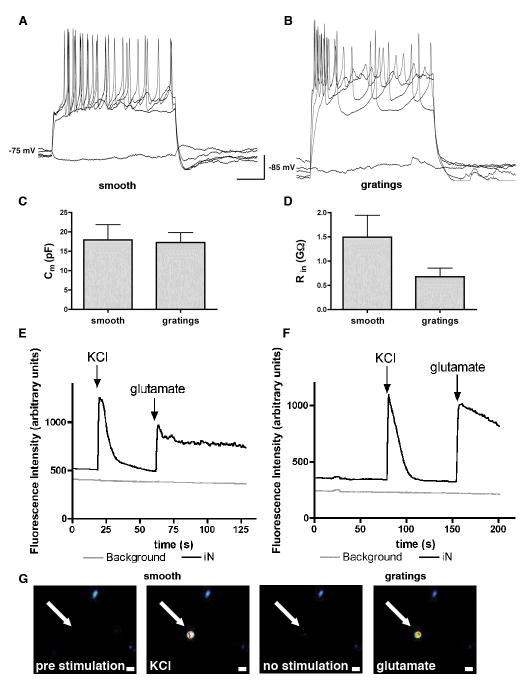
To evaluate if the iNs generated on smooth surface versus gratings showed any difference in electrophysiological characteristics, synapsin-RFP+ iNs were subjected to whole-cell patch clamp at day 16 post infection. iNs on both substrates were capable of firing action potentials: 10 out of 11 synapsin-RFP+ cells on smooth substrates were capable of firing at least one action potential in response to step depolarizing current injection under the current-clamp mode (500 ms). A representative trace of a train of action potentials is shown in Figure 5A. Similarly, 8 out of 9 synapsin-RFP+ cells on gratings fired action potentials in response to depolarizing current injection (Fig. 5B). The average membrane potential of iNs on smooth substrates (−76 mV) was similar that on gratings (−82 mV). No difference was observed in membrane capacitance, with an average of 17.94 pF ± 3.9 (SEM, n=10) on smooth substrate versus 17.27 pF ± 2.5 (SEM, n=8) on gratings (Fig. 5C). The input resistance of iNs on gratings was 0.68 GΩ ± 0.18 (SEM, n=8), slightly but not significantly lower compared to the 1.6 GΩ ± 0.48 (SEM, n=10) measured for iNs on smooth substrates (Fig. 5D). Additionally, iNs expressing GCaMP5 under control of a MAP2 promoter were depolarized with 50 mM of KCl and stimulated with 50 mM NaCl and 1 mM glutamate. An increase in fluorescence intensity represents the opening of Ca2+ channels and an increase in intracellular Ca2+ in response to the stimulation. Figure 5E depicts a typical trace in response to KCl and glutamate on smooth substrates and Figure 5F shows the response on gratings. On smooth substrates 20/24 (83.3%) cells responded to KCl stimulation compared to 13/22 (59.1%) cells on gratings, and 14/24 (58.3%) cells responded to stimulation with glutamate compared to 13/22 (59.1%) cells on gratings. In addition, 10/24 (41.7%) cells and 10/22 (45.5%) cells displayed spontaneous Ca2+ oscillations on smooth and grating substrates, respectively.
Figure 5. Action potentials of iNs generated on smooth substrates and 5 μm gratings.
(A) A representative trace of a train of action potentials in response to step depolarizing current injection of a synapsin-RFP+ iN on smooth substrate. (B) A train of action potentials in synapsin-RFP+ iN on 5 μm gratings in response to depolarizing current injection. (C) No difference was observed in membrane capacitance between iNs on smooth or grating substrates. (D) The input resistance of iNs on 5 μm gratings was slightly lower compared to iNs on smooth substrates. (E) A representative trace of fluorescent intensity measurements in iN on smooth and (F) on grating substrates in response of KCl and glutamate stimulation. (G) Image sequence of a representative iN (arrow) before stimulation, stimulated with KCl, between stimulations and stimulated with glutamate. Warm colors white, yellow and red represent high fluorescent intensities. Scale bar 10 μm.
3.6. Contribution of Myosin II in topography-mediated changes of iNs
We speculated that the underlying mechanism of topography-altered neuronal reprogramming has to do with the fibroblasts and/or neurons mechanically sensing the microenvironment. Non-muscle myosin II has been implicated in topography-sensing of fibroblasts [24]. This prompted us to ask if the reduction in branching of iNs due to underlying grating topography could be mediated by non-muscle myosin II. Therefore, we treated PMEFs with blebbistatin (a non-muscle myosin II ATPase inhibitor) before and during the direct reprogramming protocol, fixed and stained the cells with Tuj1 after 12 days in N3 medium (Fig. S2C,D), and compared with controls without the blebbistatin treatment (Fig. S2A,B). The number of neurite branches of iNs treated with blebbistatin was significantly increased in iNs on grating topography. We also observed a slight but not significant increase in branching of iNs cultured on smooth substrates (Fig. S2E).
4. Discussion
The current study illustrates that underlying substrate topography can influence the direct reprogramming of fibroblasts to iNs. Grating topography yielded iNs of higher purity and at a higher efficiency. Interestingly, the microarray and subsequent qPCR analysis revealed a reduced Bmp5 expression in iN generated on grating topography. Blocking the Bmp pathway with small molecules is known to increase neuronal differentiation from human embryonic stem cells and induced pluripotent stem cells [25], and improve the efficiency of iN conversion from human fibroblasts [26]. It is possible that grating topography similarly inhibits Bmp signaling to produce more iNs. Alternatively, or in addition, grating topography could promote iN adhesion and survival. iN morphology is clearly altered by substrate topography, a phenomenon observed for a number of other cell types on a variety of topographies [20, 27, 28]. The polarity of iNs in the grating direction and the guidance of their neurites is especially pronounced along the 5 μm grating topography. This phenomenon has been previously reported for hippocampal neurons and PC12 cells [29, 30] on micron-sized gratings. Another striking morphological difference of iNs on the different topographies is their branching pattern. Neuronal dendrites and axons are usually branched. Their branching can range from simple daughter branches from a common neurite to elaborated arbors. These branching patterns are formed during development and are tailored to the functional needs of the neural network they assemble [31]. The iNs on grating topography were mostly monopolar, with very limited branching, whereas the iNs on the F (1.4) topography showed significantly increased branching. Additional staining for MAP2, a marker for dendrites, on smooth and grating substrates demonstrated that iNs generated on grating substrates displayed a reduced dendritic arbor. The reduced branching on grating substrates is consistent with the microarray finding that the expression of Slit3, artemin and Netrin G genes associated with neurite branching, is downregulated. Slit/Robo signaling has been shown to regulate arborization of sensory neurons in vivo. Experiments in Slit 1 and Slit 2/Slit 3 mutant mice showed that both the arbor size and the branch point numbers are sensitive to the loss of Slits [31]. Artemin belongs to the subfamily of glial derived neurotrophic factor (GDNF) [32] and plays a role in neurite outgrowth and actin polymerization in DRG neurons [33]. Bmp5 has also been reported to play a role in dendritic arbor formation with its suppression resulting in reduced dendritic arbor [34], consistent with our observation of reduced MAP2+ neurites and reduced Bmp5 expression in iNs on grating topography. Mechanical cues such as substrate stiffness have been shown to impact neurite branching. Neurons grown on softer hydrogels formed up to 3 times more branches compared to neurons cultured on stiffer gels [35]. Taken together, the results support our hypothesis that physical cues from substrate topography can influence the morphology and branching pattern of iNs derived from the reprogramming of fibroblasts. Moreover, Thy1 expression was reduced in iNs on grating topography. Thy1 encodes a surface protein highly expressed in fibroblasts but is also found to a lesser extent in glutamatergic neurons. The reduced expression of Thy1 could indicate a more pronounced silencing of fibroblastic genes in iNs generated on grating substrates. The iNs on gratings showed increased Vip expression and lower branching, consistent with the literature report that neurons expressing Vip are characterized by a slender morphology with limited branching [36]. Despite the simplified morphology and reduction in dendritic arbor of iNs generated on gratings, our electrophysiological experiments showed that they are capable of generating action potentials, and calcium imaging showed that these iNs responded to KCl and glutamate just like iNs generated on smooth substrates.
During the reprogramming of fibroblast to iN important cytoskeleton rearrangements take place and actin-myosin contractility is important in this process. Non-muscle myosin II has been implicated in the cytoskeleton rearrangements in response to substrate topography and substrate stiffness [24]. Interestingly, the reduction of neurite branching on 5 μm grating is reversed when non muscle myosin II is inhibited, indicating that topography acts via non-muscle myosin II as a mechanical cue to impact the fibroblast-to-neuron reprogramming process.
5. Conclusion
Transcription factors would be expected to be the most powerful effectors of the reprogramming process. However, we have demonstrated in this study that cell-topography interactions can shape the iNs in terms of gene expression, neurite branching, outgrowth and iN reprogramming efficiency. Presentation of topographical cues suggests a new strategy to augment or tune the production of specialized neuronal cell types via direct reprogramming.
Supplementary Material
PMEFs were seeded on topography on day 0, infected with the doxycycline inducible TFs Ascl1, Brn2, Myt1L on day 1 and doxycycline was added on day 2 for 5 days and the medium was replaced by neurogenic medium on day 4 and exchanged every 48h. The culture was infected with the synapsin-promotor driven RFP+ construct on day 10.
Tuj1+ iNs cultured on (A) smooth substrates and (C) on 5 μm grating substrates with traces of neurites from widefield microscopy images obtained using Fiji. Tuj1+ iNs cultured in presence of blebbistatin on (B) smooth substrates and (D) on 5 μm grating substrates with traces of neurites from widefield microscopy images obtained using Fiji. (E) Quantification of the number of neurites per cell soma. A significant increase in the number of neurites/soma was observed for iNs cultured on 5 μm grating substrates in presence of blebbistatin. Analysis by global one-way ANOVA with Dunn’s posthoc testing (** p < 0.01). Scale bar 20 μm. All data displayed as mean ± SEM.
Acknowledgments
We acknowledge the help of Dr. Michael Cook and Lynn Martinek with the flow cytometry, Dr. Morton Foss for the metal masters of the F (1.4) and F (4.1) topographies. We thank Dr. Marius Wernig, Dr. Russell Addis, Dr. Didier Trono, Dr. Rudolf Jaenisch and Dr. Edward Callaway for the plasmids used in this study. We thank Dr. Marius Wernig for fibroblasts from Tau-EGFP mice. We thank the Duke Microarray Core facility (a Duke National Cancer Institute and a Duke Institute for Genome Sciences and Policy shared resource facility), especially Laura-Leigh Rowlette and Zhengzheng Wei, for their technical support, microarray data management and feedback on the generation of the microarray data reported in this manuscript. We also acknowledge Dr. Anne West, Dr. Blanche Capel, Michael Czerwinski and Dr. Nicolas Christoforou for helpful discussions. This work was funded by the NIH grant EB 015300 and the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qiang L, Fujita R, Yamashita T, Angulo S, Rhinn H, Rhee D, et al. Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell. 2011;146(3):359–71. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476(7359):228–31. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476(7359):220–3. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caiazzo M, Dell’anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476(7359):224–7. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 6.Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011;108(25):10343–8. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, Su SC, Wang H, Cheng AW, Cassady JP, Lodato MA, et al. Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell. 2011;9(5):413–9. doi: 10.1016/j.stem.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Li F, Stubblefield EA, Blanchard B, Richards TL, Larson GA, et al. Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 2012;22(2):321–32. doi: 10.1038/cr.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9(3):205–18. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang N, Zuchero JB, Ahlenius H, Marro S, Ng YH, Vierbuchen T, et al. Generation of oligodendroglial cells by direct lineage conversion. Nat Biotechnol. 2013;31(5):434–9. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adler AF, Grigsby CL, Kulangara K, Wang H, Yasuda R, Leong KW. Nonviral direct conversion of primary mouse embryonic fibroblasts to neuronal cells. Mol Ther Nucleic Acids. 2012;1(e32) doi: 10.1038/mtna.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maucksch C, Firmin E, Butler-Munro C, Montgomery JM, Dottori M, Connor B. Non-viral generation of neural precursor-like cells from adult human fibroblasts. JSRM. 2012;8(3):162–170. doi: 10.46582/jsrm.0803009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yim EK, Darling EM, Kulangara K, Guilak F, Leong KW. Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials. 2009;31(6):1299–306. doi: 10.1016/j.biomaterials.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMurray RJ, Gadegaard N, Tsimbouri PM, Burgess KV, McNamara LE, Tare R, et al. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat Mater. 2011;10(8):637–44. doi: 10.1038/nmat3058. [DOI] [PubMed] [Google Scholar]
- 15.Downing TL, Soto J, Morez C, Houssin T, Fritz A, Yuan F, et al. Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater. 2013;12(12):1154–62. doi: 10.1038/nmat3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrari A, Cecchini M, Dhawan A, Micera S, Tonazzini I, Stabile R, et al. Nanotopographic control of neuronal polarity. Nano Lett. 2011;11(2):505–11. doi: 10.1021/nl103349s. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari A, Cecchini M, Serresi M, Faraci P, Pisignano D, Beltram F. Neuronal polarity selection by topography-induced focal adhesion control. Biomaterials. 2010;31(17):4682–94. doi: 10.1016/j.biomaterials.2010.02.032. [DOI] [PubMed] [Google Scholar]
- 18.Becker T, McLane MA, Becker CG. Integrin antagonists affect growth and pathfinding of ventral motor nerves in the trunk of embryonic zebrafish. Mol Cell Neurosci. 2003;23(1):54–68. doi: 10.1016/s1044-7431(03)00018-6. [DOI] [PubMed] [Google Scholar]
- 19.Kulangara K, Leong KW. Substrate topography shapes function. Soft Matter [Highlight] 2009;5(21):4072–6. [Google Scholar]
- 20.Hanson JN, Motala MJ, Heien ML, Gillette M, Sweedler J, Nuzzo RG. Textural guidance cues for controlling process outgrowth of mammalian neurons. Lab Chip. 2009;9(1):122–31. doi: 10.1039/b803595d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3(3):346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nathanson JL, Yanagawa Y, Obata K, Callaway EM. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience. 2009;161(2):441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frey MT, Tsai IY, Russell TP, Hanks SK, Wang YL. Cellular responses to substrate topography: role of myosin II and focal adhesion kinase. Biophys J. 2006;90(10):3774–82. doi: 10.1529/biophysj.105.074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27(3):275–80. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ladewig J, Mertens J, Kesavan J, Doerr J, Poppe D, Glaue F, et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods. 2012;9(6):575–8. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- 27.Micholt L, Gartner A, Prodanov D, Braeken D, Dotti CG, Bartic C. Substrate topography determines neuronal polarization and growth in vitro. PLoS One. 2013;8(6):e66170. doi: 10.1371/journal.pone.0066170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berry CC, Campbell G, Spadiccino A, Robertson M, Curtis AS. The influence of microscale topography on fibroblast attachment and motility. Biomaterials. 2004;25(26):5781–8. doi: 10.1016/j.biomaterials.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 29.Dowell-Mesfin NM, Abdul-Karim MA, Turner AM, Schanz S, Craighead HG, Roysam B, et al. Topographically modified surfaces affect orientation and growth of hippocampal neurons. J Neural Eng. 2004;1(2):78–90. doi: 10.1088/1741-2560/1/2/003. [DOI] [PubMed] [Google Scholar]
- 30.Ferrari A, Faraci P, Cecchini M, Beltram F. The effect of alternative neuronal differentiation pathways on PC12 cell adhesion and neurite alignment to nanogratings. Biomaterials. 2010;31(9):2565–73. doi: 10.1016/j.biomaterials.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 31.Ma L, Tessier-Lavigne M. Dual branch-promoting and branch-repelling actions of Slit/Robo signaling on peripheral and central branches of developing sensory axons. J Neurosci. 2007;27(25):6843–51. doi: 10.1523/JNEUROSCI.1479-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenblad C, Gronborg M, Hansen C, Blom N, Meyer M, Johansen J, et al. In vivo protection of nigral dopamine neurons by lentiviral gene transfer of the novel GDNF-family member neublastin/artemin. Mol Cell Neurosci. 2000;15(2):199–214. doi: 10.1006/mcne.1999.0817. [DOI] [PubMed] [Google Scholar]
- 33.Park S, Hong YW. Transcriptional regulation of artemin is related to neurite outgrowth and actin polymerization in mature DRG neurons. Neurosci Lett. 2006;404(1–2):61–6. doi: 10.1016/j.neulet.2006.05.041. [DOI] [PubMed] [Google Scholar]
- 34.Beck HN, Drahushuk K, Jacoby DB, Higgins D, Lein PJ. Bone morphogenetic protein-5 (BMP-5) promotes dendritic growth in cultured sympathetic neurons. BMC Neurosci. 2001;2:12. doi: 10.1186/1471-2202-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flanagan LA, Ju YE, Marg B, Osterfield M, Janmey PA. Neurite branching on deformable substrates. Neuroreport. 2002;13(18):2411–5. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magistretti PJ. VIP neurons in the cerebral cortex. Trends Pharmacol Sci. 1990;11(6):250–4. doi: 10.1016/0165-6147(90)90253-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PMEFs were seeded on topography on day 0, infected with the doxycycline inducible TFs Ascl1, Brn2, Myt1L on day 1 and doxycycline was added on day 2 for 5 days and the medium was replaced by neurogenic medium on day 4 and exchanged every 48h. The culture was infected with the synapsin-promotor driven RFP+ construct on day 10.
Tuj1+ iNs cultured on (A) smooth substrates and (C) on 5 μm grating substrates with traces of neurites from widefield microscopy images obtained using Fiji. Tuj1+ iNs cultured in presence of blebbistatin on (B) smooth substrates and (D) on 5 μm grating substrates with traces of neurites from widefield microscopy images obtained using Fiji. (E) Quantification of the number of neurites per cell soma. A significant increase in the number of neurites/soma was observed for iNs cultured on 5 μm grating substrates in presence of blebbistatin. Analysis by global one-way ANOVA with Dunn’s posthoc testing (** p < 0.01). Scale bar 20 μm. All data displayed as mean ± SEM.