Abstract
Objective
X-linked dominant hypophosphatemia (XLH) is the most prevalent form of inherited rickets/osteomalacia in humans. The aim of this study was to identify PHEX gene mutations and describe the clinical features observed in 6 unrelated Chinese families and 3 sporadic patients with hypophosphatemic rickets/osteomalacia.
Methods
For this study, 45 individuals from 9 unrelated families of Chinese Han ethnicity (including 16 patients and 29 normal phenotype subjects), and 250 healthy donors were recruited. All 22 exons and exon-intron boundaries of the PHEX gene were amplified by polymerase chain reaction (PCR) and directly sequenced.
Results
The PHEX mutations were detected in 6 familial and 3 sporadic hypophosphatemic rickets/osteomalacia. Altogether, 2 novel mutations were detected: 1 missense mutation c.1183G>C in exon 11, resulting in p.Gly395Arg and 1 missense mutation c.1751A>C in exon 17, resulting in p.His584Pro. No mutations were found in the 250 healthy controls.
Conclusions
Our study increases knowledge of the PHEX gene mutation types and clinical phenotypes found in Chinese patients with XLH, which is important for understanding the genetic basis of XLH. The molecular diagnosis of a PHEX genetic mutation is of great importance for confirming the clinical diagnosis of XLH, conducting genetic counseling, and facilitating prenatal intervention, especially in the case of sporadic patients.
Introduction
Hypophosphatemic rickets/osteomalacia is characterized by defective renal phosphate re-absorption and abnormal bone mineralization [1]–[4]. Clinical features of the disease include short stature, bone and/or joint pain, lower extremity deformities, calcification of the ligaments, and dental abnormalities. X-linked dominant hypophosphatemia (XLH), autosomal dominant hypophosphatemic rickets (ADHR), and autosomal recessive hypophosphatemic rickets (ARHR) are the three main types of hypophosphatemic rickets and are associated with mutations in the phosphate-regulating endopeptidase gene (PHEX), the fibroblast growth factor 23 gene (FGF23), and the dentin matrix acidic phosphoprotein1 gene (DMP1), respectively. XLH (MIM 307800), which was first reported in 1939 [5], is the most common genetic form of hypophosphatemic rickets/osteomalacia and has an incidence rate of 3.9–5.0 per 100,000 [6]–[8]. In familial hypophosphatemic rickets (FHR), hypophosphatemic rickets/osteomalacia can be inherited in either an X-linked autosomal dominant or autosomal recessive manner. The most common disease-causing genetic mutations in these cases occur in the PHEX gene and cause 87% of familial XLH and 72% of the sporadic cases [9]. XLH is characterized by renal phosphate wasting, which causes hypophosphatemia and normal to low 1,25-dihydroxy-vitamin D3 serum levels [10]; together, these indicate defects in phosphate and vitamin D metabolism.
The PHEX gene is located on chromosome Xp22.1, consists of 22 exons, spanning 220 kb with 6172 bp of transcript length, and encodes a membrane-bound metalloprotease composed of 749 amino acids [10], [11]. The PHEX protein shares a common overall structure with other members of the neutral endopeptidase family, including neprilysin, two endothelin-converting enzymes (ECE-1 and -2), the KELL antigen, and the damage-induced neuronal endopeptidase/X-converting enzyme [3], [10]. The structure consists of a short N-terminal intracellular region, a single N-terminal hydrophobic region that corresponds with the transmembrane domain, a highly conserved zinc-binding domain in exons 17 and 19, and a large extracellular C-terminal domain [3], [10]. The PHEX protein is predominantly expressed in cartilage, osteoblasts, and odontoblasts but not in the kidney [10], [12], [13]. Although the exact mechanism of how PHEX mutations cause rickets/osteomalacia remains unknown, some studies have shown that PHEX may inactivate bone mineralization inhibitors and that one of the extraosseous consequences of PHEX inactivation includes an increase in the level of FGF-23 [14].
Currently, 329 mutations in the PHEX gene have been reported in the PHEX mutation database (http://www.phexdb.mcgill.ca; Mar 17 12:35∶21 2014), which mostly occur in European (Northern European), North American, and Far Eastern populations. According to the PHEX mutation database, the frequencies of the different types of mutations include the following: 27% frameshifts, 19.8% abnormal splicing, 19.4% missense, 18% nonsense, 28% deletions, and 2.4% polymorphisms. However, only 14 mutations (2 deletion mutation, 3 nonsense mutations, 1 frameshift mutation, 3 donor splice site mutations, and 5 missense mutations) in the PHEX gene have been reported in Chinese patients with familial XLH [15]–[20]. In this study, we identified PHEX gene mutations in Chinese patients (familial and sporadic) with hypophosphatemic rickets/osteomalacia in order to elucidate the PHEX gene mutation types and clinical features observed in Chinese patients.
Materials and Methods
Study Subjects
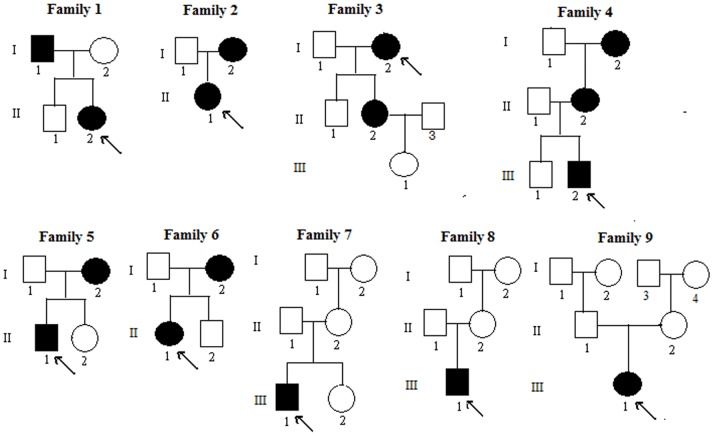
The Department of Osteoporosis and Bone Diseases recruited all of the subjects involved in the study over a 6-year period. All of the subjects were of Chinese Han ethnicity and had non-consanguineous parents. Diagnosis of XLH was based on clinical manifestations, radiology results, skeletal deformities, growth impairment, and laboratory results that indicated the occurrence of hypophosphatemia and renal phosphate wasting. Altogether, 45 individuals including 16 patients from 9 unrelated Chinese families were investigated in our study. Three patients were from family 4. Family 7, 8 and 9 had only 1 patient each. The other families (Family 1, 2, 3, 5 and 6) had 2 patients each. The pedigrees of X-linked hypophosphatemic rickets are shown in Figure 1. The fasting blood was collected from each of the subjects during a clinic visit and was analyzed in the central laboratory of Shanghai JiaoTong University Affiliated Sixth People’s Hospital. Serum samples were used to measure calcium (Ca), phosphate (P), parathyroid hormone (PTH) concentrations, alkaline phosphatase (ALP) activity, and the Serum 25-hydroxyvitamin D [25(OH)D] level. Serum concentrations of Ca, P and ALP were measured by using Hitachi 7600 automatic biochemical analyzer. Serum 25(OH)D and PTH were measured by ECLIA Elecsys autoanalyzer (E170; Roche Diagnostic GmbH, Mannheim, Germany). The intra-assay coefficients of variation (CVs) for 25(OH)D were 5.7% at a level of 25.2 ng/mL, 5.7% at a level of 39.9 ng/mL and 5.4% at a level of 65.6 ng/mL, respectively. The inter-assay CVs for 25(OH)D were 9.9% at a level of 25.2 ng/mL, 7.3% at a level of 39.9 ng/mL and 6.9% at a level of 65.6 ng/mL, respectively. The intra-assay and inter-assay CVs were 1.4% and 2.9%, respectively, for PTH. All of the laboratory data were collected prior to treating the patients with surgery or medicine.
Figure 1. Pedigree of the familial patients with XLH.
Black symbols represent the affected individuals, and open symbols represent the unaffected individuals. Circles and squares represent the females and males, respectively. The arrows identify the probands in the families.
Ethics Statement
The Ethics Committee of the Shanghai JiaoTong University Affiliated Sixth People’s Hospital approved this study and the project was conducted following the terms of “Declaration of Helsinki”. Signatures confirming informed consent were obtained from the participating subjects before starting the project. In addition, we obtained written informed consent from the parents on the behalf of the minor/children participants involved in our study.
Mutation Analysis
Genomic DNA was isolated from peripheral blood leukocytes using the conventional phenol-chloroform extraction method. We screened the PHEX gene completely for mutations in 16 patients, the other phenotype normal family members, and 250 healthy ethnically matched controls (125 males and 125 females, Chinese Han ethnicity). The DNA sequence for the PHEX gene was obtained from available online database (NCBI Reference Sequence Accession Number: NG_007563.2; the mRNA Accession Number: NM_000444.5.). The PCR and sequencing primers were the same as we used in our previous study [18]. All 22 exons and exon-intron boundaries in the PHEX gene were amplified by polymerase chain reaction (PCR). Hot Start PCR reaction was performed in our study, and HotStar Taq DNA polymerase (Qiagen Inc.) was used for highly specific amplification in hot-start PCR reaction. The cycling program of amplification was 95°C for 15 minutes; 11 cycles of 94°C for 15 seconds, 62°C per cycle for 40 seconds, 72°C for 1 minutes; 24 cycles of 94°C for 15 seconds, 57°C for 30 seconds, 72°C for 1 minute, 72°C for 2 minutes. Direct sequencing was performed using the BigDye Terminator Cycle Sequencing Ready Reaction Kit, version 3.1 (Applied Biosystems, Foster, CA, USA), and the cycling program of sequencing was 96°C for 1 minute; 28 cycles of 96°C for 10 seconds, 50°C for 5 seconds, 60°C for 4 minutes. The resulting PCR products were directly sequenced using an automated ABI PRISM 3130 sequencer (Foster, CA). Meanwhile, once a mutation was detected, we performed PCR amplification in the same DNA sample again by using HotStar HiFidelity polymerase (Qiagen Inc.) for highly sensitive and reliable high-fidelity hot-start PCR. Then, the purified PCR product was sequenced from the other strand to further verify the mutation. Single-nucleotide polymorphisms (SNPs) were identified using Polyphred (http://droog.mbt.washington.edu/poly_get.html). Novel mutations were identified using HGMD (http://www.hgmd.cf.ac.uk/). Mutations were confirmed using Mutalyzer 2.0 (http://mutalyzer.nl/check). The DNA sequences obtained were aligned with homologous sequences that had been deposited into GenBank using the CluxtalX 1.83 algorithm [16].
Mutation Prediction
Polyphen-2 [19] and Sorting Intolerant from Tolerant (SIFT) were used to determine the functional effects of all the missense mutations in the PHEX gene [8]. Polyphen-2 (http://genetics.bwh.Harvard.edu/pph2/) and SIFT (http://sift.jcvi.Org/www/SIFT_enst_submit) are tools that predict the possible impacts of an amino acid substitution on the structure and function of a human protein using a straightforward physical comparative analysis [21]–[24]. For Polyphen-2, the following three empirically derived outcomes were used: most likely damaging (with a high confidence level, the substitution will affect protein function or structure), possibly damaging (the substitution is supposed to affect protein function or structure), and benign (the substitution most likely lacks any phenotypic effect). The SIFT score [8], [25] represents the normalized probability that the amino acid change is tolerated. The SIFT score <0.05 are predicted to be deleterious.
Results
Clinical Features of the Subjects
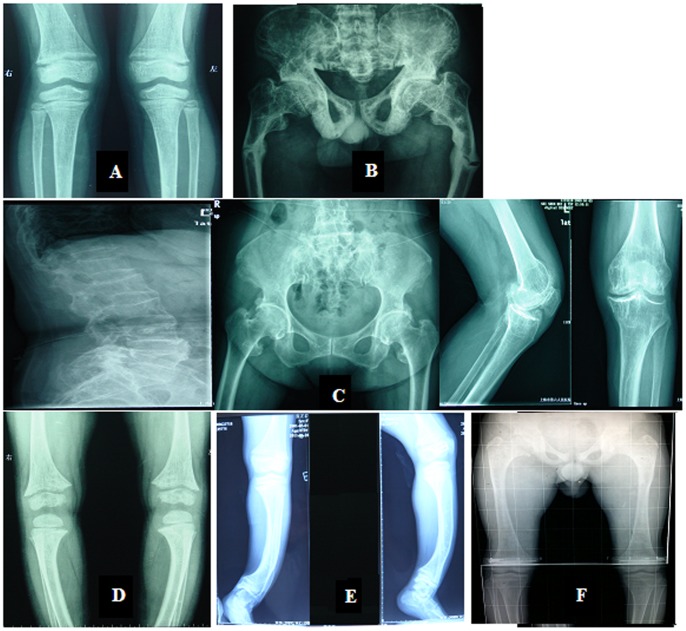
The general features and laboratory results of patients are shown in Table 1. The radiology results of the patients are shown in Figure 2. The phenotypes and laboratory results for the family members except patients were all normal (data not shown).
Table 1. General features and laboratory results for the XLH patients studied.
| Family No. | Patient No./Gender | Age (year) | Height (cm) | Weight (kg) | BMI (kg/m2) | Height percentile | Ca (mmol/l) | P (mmol/l) | ALP (u/l) | PTH (u/l) | 25(OH)D (ng/ml) |
| 1 | II2/F | 5 | 92 | 13.5 | 15.95 | <1st | 2.35 | 0.73 | 475 | 78.56 | 20.20 |
| 1 | I1/M | 34 | 147 | 54 | 24.99 | <5th | 2.30 | 0.68 | 168 | 49.80 | 15.30 |
| 2 | II1/F | 4 | 95 | 15 | 16.62 | <3rd | 2.43 | 0.98 | 752 | 83.04 | 6.78 |
| 2 | I2/F | 27 | 139 | 45 | 23.29 | <5th | 2.28 | 0.54 | 45 | 67.08 | 5.63 |
| 3 | II2/F | 43 | 150 | 51 | 22.67 | <25th | 2.67 | 0.63 | 121 | 55.14 | 19.70 |
| 3 | I2/F | 73 | 145 | 53 | 25.21 | <25th | 2.36 | 0.63 | 157 | 132.10 | <4 |
| 4 | III2/M | 4 | 99 | 17 | 17.35 | <15th | 2.38 | 0.74 | 507 | 50.62 | 17.99 |
| 4 | II2/F | 33 | 141 | 41 | 20.62 | <5th | 2.17 | 0.83 | 66 | 71.21 | 9.66 |
| 4 | I2/F | 58 | 127 | 48 | 29.76 | <5th | 2.26 | 0.91 | 168 | 67.52 | 18.77 |
| 5 | II1/M | 14 | 137 | 31.5 | 16.78 | <1st | 2.32 | 0.71 | 409 | 61.25 | <4 |
| 5 | I2/F | 37 | 137.5 | 47 | 24.86 | <5th | 2.18 | 0.70 | 79 | 77.62 | <4 |
| 6 | II1/F | 13 | 142 | 45 | 22.32 | <3rd | 2.34 | 0.67 | 122 | 84.52 | 13.08 |
| 6 | I2/F | 37 | 138 | 41 | 21.53 | <5th | 2.19 | 0.70 | 109 | 101.80 | 17.48 |
| 7 | III1/M | 16 | 145 | 40.5 | 19.26 | <1st | 2.25 | 0.85 | 534 | 50.12 | 21.23 |
| 8 | III1/M | 3 | 82 | 12 | 17.85 | <1st | 2.32 | 0.89 | 414 | 87.41 | 27.36 |
| 9 | III1/F | 11 | 98 | 21 | 21.87 | <1st | 2.22 | 0.65 | 359 | 107.63 | 5.38 |
Footnotes: Abnormal data are bolded. The normal range for phosphate is 0.8–1.6 mmol/l; for calcium is 2.08–2.60 mmol/l; for alkaline phosphatase (ALP) is 15–112 u/l; for parathyroid hormone (PTH) is 15–65 µ/l; and for 25-OH vitamin D [25(OH)D] is 20–35 ng/ml. F: female, M: male. BMI (Body Mass Index) is defined as the individual’s body mass divided by the square of their height. BMI normal range is 18.5–25 Kg/m2. The data of height percentile referenced the standard provided by the World Health Organization.
Figure 2. Radiology results for the patients.
(A): A general decrease in bone density, widened metaphyses, bilateral femoral distal metaphysis epiphyseal line blurred, and similar features in the lower tibia are shown. (B): A high bone mineral density, genu varum deformities in the lower limbs, and a Looser Zone in the bilateral femoral shaft are shown. (C): A general decrease in the bone density of the vertebrae and pelvis, bilateral hip and knee joint degeneration, and multiple vertebral wedge changes are shown. (D): Widened metaphyses is shown. (E): A high bone mineral density and anterior bowing of the lower limbs are shown. (F): A general decrease in the bone density of the vertebrae and pelvis, an enlargement of the epiphyseal and metaphyseal portions of the under section of the femoral, a short and wide femoral neck, and a large capital femoral epiphyses are shown.
In family 1, the proband (II2, 5 years) had a short stature (92 cm tall) with an abnormal gait and retarded dentition, whose imaging results are shown in Figure 2A. While her father (I1, 34 years) even had more severe phenotypes, besides a short stature (147 cm height) and an abnormal gait. His onset age was 1.5 years and had genu varum with an “O” appearance. In addition, he suffered a bilateral femoral fracture at 7 years of age and the imaging results are shown in Figure 2B. In family 2, the proband (II1, 4 years) was characterized by delayed dentition and genu varum. While her mother (I2) had more severe phenotypes with earlier onset age (1.5 years), consisted of an abnormal gait, delayed dentition, odontodysplasia, and genu varum with an “O” appearance. The symptoms were even worse as aging. Her teeth started falling out at 22 years of age, and only 5 teeth remained currently. In family 3, the proband’s (I2, 73 years) clinical features consisted of an abnormal gait, kyphosis, and hip and knee joint pain. The X-ray results are shown in Figure 2C. However, her daughter (II2, 43 years) only had mild genu varum and bone pain. In family 4, the proband (III2, 4 years) had short stature and mild genu varum. The X-ray results are shown in Figure 2D. His mother (II2, 33 years) and grandmother (I2, 58 years) had the same symptoms as he. In family 5, the proband (II1, 14 years) was delivered by Caesarean section and had a low birth weight of 2.45 kg. The proband had obvious growth retardation and stopped growing after 9 years of age. He also suffered from hip and knee joint pain. His mother’s (I2, 37 years) first clinical abnormalities were detected at 4 years of age and consisted of an abnormal gait and growth retardation. Genu varum with an “O” appearance developed as aging, and her height stopped growing at 16 years of age after the onset of her menstrual cycle. Her teeth began to fall out at 17 years of age, and only 1 tooth remained at present. In family 6, the proband (II1, 13 years) was characterized by goyectyposis. The X-ray results are shown in Figure 2E. Her mother (I2, 37 years) had the similar symptoms as her daughter. The first sporadic patient (III1, 16 years) was from family 7. When he was 9 months old, he experienced swelling of his hip joints and was diagnosed with rickets when he was 11 months old. He was characterized by genu varum, hip pain and growth retardation. Imaging results are shown in Figure 2F. The second sporadic patient (III1, 3 years) was from family 8. When he began to walk at 1.5 years of age, genu varum was detected. The X-ray results revealed cephalus quadratus, bilateral femur bending, and mild bilateral genu varum in the knees (data not shown). The third sporadic patient (III1, 11 years) was from family 9. Her first clinical abnormality was detected when she was 16 months old and characterized by an abnormal gait (genu varum with “O” appearance).
Mutation Analysis of the PHEX Gene
The mutational analysis of the PHEX gene in our patients revealed the following 2 novel mutations and they both were detected in family 2∶1 missense mutation, c.1183G>C, in exon 11, resulting in p.Gly395Arg and 1 missense mutation, c.1751A>C in exon 17, resulting in p.H584P. The main clinical results from the genetic testing of the patients at the time of this study are summarized in Table 2.
Table 2. Summary of the patient clinical data.
| Family No. | Patient | Gender/Age of onset (year) | Clinical findings | Mutation site | PHEX mutation | Inheritance |
| 1 | (II-2) daughter | F/3 | Retarded dentition | Exon 20 | Trp660X (c.1980G>A) | Familial |
| 1 | (I-1) father | M/1.5 | Genu varum | Exon 20 | Trp660X (c.1980G>A) | Familial |
| 2 | (II-1) daughter | F/2 | Genu varum; retarded dentition | Exon 17 | His584Pro (c.1751A>C) | Familial |
| 2 | (I-2) mother | F/1.5 | Genu varum; retarded dentition; odontodysplasia; teeth falling out | Exon 17 Exon 11 | His584Pro (c.1751A>C) Gly395Arg (c.1183G>C) | Familial |
| 3 | (II-2) daughter | F/4 | Genu varum and bone pain | Exon 12 | Trp444X (c.1332 G>A) | Familial |
| 3 | (I-2) mother | F/5 | Hip and knee joint pain; kyphosis; bone pain | Exon 12 | Trp444X (c.1332 G>A) | Familial |
| 4 | (III2) son | M/1.5 | Genu varum | Intron 15 | c.1646-2A>T | Familial |
| 4 | (II2) mother | F/2 | Genu varum | Intron 15 | c.1646-2A>T | Familial |
| 4 | (I2) grandmother | F/5 | Genu varum | Intron 15 | c.1646-2A>T | Familial |
| 5 | (II1) son | M/5 | Genu varum (mild); bone pain; growth retardation | Intron 10 | c.1174-1G>A | Familial |
| 5 | (I2) mother | F/4 | Genu varum; odontodysplasia; teeth falling out; growth retardation | Intron 10 | c.1174-1G>A | Familial |
| 6 | (II1) daughter | F/2 | Bowing of legs | Exon 16 | Tyr565Phefsx5 (c.1694delA) | Familial |
| 6 | (I2) mother | F/2 | Bowing of legs | Exon 16 | Tyr565Phefsx5 (c.1694delA) | Familial |
| 7 | (III1) son | M/0.75 | Genu varum; hip pain; growth retardation | Intron 17 | c.1768+2T>G | Sporadic |
| 8 | (III1) son | M/1.5 | Genu varum; cephalus quadratus | Exon 21 | Trp692IlefsX2 (c.2077_*4delinsA) | Sporadic |
| 9 | (III1) daughter | F/1.3 | Genu varum | Exon 15 | Arg549X (c.1645C>T) | Sporadic |
Footnotes: Novel mutations are bolded.
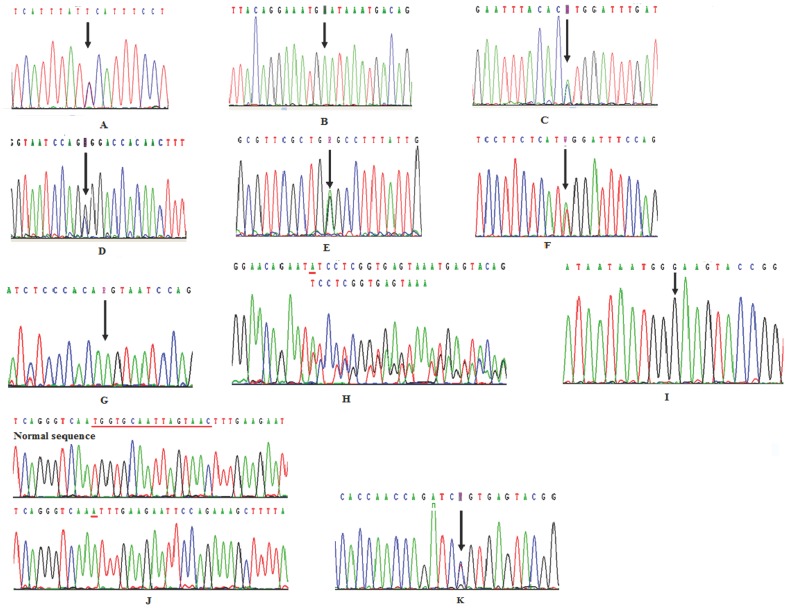
In family 1, the clinical diagnosis of XLH in the proband (II2) and her father (I1) was confirmed by the detection of a nonsense mutation in codon 660 in exon 20 of the PHEX gene (c.1980G>A), which results in the replacement of a tryptophan residue with a premature stop codon (p.Trp660X). (The sequences of II2 and I1 were shown in figure 3A and 3B, respectively). In family 2, a novel missense mutation was detected in the proband (II1) and her mother (I2) in which a proline is substituted for a histidine at position 584 as a result of a mutation in exon 17 of the PHEX gene (c.1751A>C; p.His584Pro; Figure 3C). Interestingly, the mother was harbouring another novel missense mutation in codon 395 in exon 11 of the PHEX gene (c.1183G>C), which results in an arginine replacing a glycine (p.Gly395Arg; Figure 3D). In family 3, sequence analysis of the proband (I2) and her daughter (II2) revealed a nonsense mutation in codon 444 in exon 12 of the PHEX gene (c.1332G>A), which results in a premature stop codon replacing a tryptophan residue (p.Trp444X; Figure 3E). In family 4, the proband (III2), his mother (II2), and his grandmother (I2) carried a putative aberrant splicing mutation c.1646-2A>T in intron 15 at splicing acceptor sites (Figure 3F). In family 5, the proband (II1) and his mother (I2) carried a putative aberrant splicing mutation c.1174-1G>A in intron 10 at splicing acceptor sites (Figure 3G). In family 6, the proband (II1) and her mother (I2) carried a heterozygous deletion of one nucleotide (A) in codon 565 (c.1694delA), which causes a phenylalanine to be substituted for a tyrosine at position 565 and replaces the next 5 amino acids with a stop codon (exon 16 of the PHEX gene; c.1694delA; p.Tyr565Phefsx5; Figure 3H).
Figure 3. PHEX gene mutation sequencing diagram.
(A): A nonsense mutation c.1980G>A in exon 20 was detected in Proband (II-2) from family 1. (B): Proband’s father (I-1) was hemizygous for a nonsense mutation, c.1980G>A in exon 20 from family 1. (C): A novel missense mutation, c.1751A>C in exon 17, was identified in proband (IV-1) and her mother (III-2) from family 2. (D): A novel missense mutation, c.1183G>C in exon 11, was identified in III-2 from family 2. (E): A nonsense mutation, c.1332G>A in exon 12, was identified in proband (I-2) and her daughter (II-2) from family 3. (F): A putative aberrant splicing mutation, c.1646-2A>T in intron 15, was identified in proband (III 2), his mother (II2), and his grandmother (I2) from family 4. (G): A putative aberrant splicing mutation, c.1174-1G>A in intron 10, was identified in proband (II1) and his mother (I2) from family 5. (H): A deletion mutation, c.1694delA in exon 16, was identified in proband (II1) and her mother (I2) from family 6. (I): A splicing mutation, c.1768+2T>G in intron 17, was identified in proband (III1) from family 7. (J): A del-insertion mutation, c.2077_*4delinsAin exon 21, was identified in proband (III1) from family 8. (K): A nonsense mutation, c.1645C>T in exon 15, was identified in proband (III1) from family 9.
In the 3 sporadic cases (family 7, 8, and 9), the proband (III1) from family 7 carried a splicing mutation c.1768+2T>G in intron 17 (Figure 3I); the proband (III1) from family 8 carried a deletion mutation c.2077_*4delinsA in exon 21 that results in p.Trp692IlefsX2 (Figure 3J); and the proband (III1) from family 9 carried a nonsense mutation c.1645C>T in exon 15 that results in p.Arg549X (Figure 3K).
No mutation was detected in the phenotype normal family members and 250 ethnically matched control individuals.
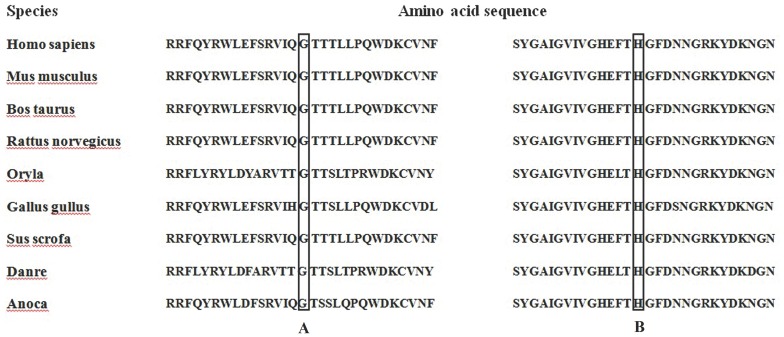
To evaluate the consequence of the p.Gly395Arg and p.His584Pro mutations, PolyPhen-2 and SIFT analyses of the mutations were performed. Both mutations were predicted to be most likely damaging (PolyPhen-2 score of 0.98 and 0.99, respectively. Both SIFT score <0.05). Meanwhile, the aminoacid residues at p.395 and p.584 are highly conserved across 9 different biological species (Figure 4A and 4B).
Figure 4. Partial amino acid sequences of the PHEX gene from 9 species.
The amino acids at p.395 in exon 11 (A) and p.584 in exon 17 (B) are highly conserved in 9 different species.
Discussion
In this study, we identified 10 different PHEX mutations (including 2 novel mutations) in 16 patients from 9 unrelated families (probands from family 7, 8, and 9 are sporadic patients) with XLH and reported the different clinical features observed in these Chinese patients.
The nonsense mutations p.Trp660X in exon 20, p.Trp444X in exon 12, and p.Arg549X in exon 15 may result in the translation of truncated proteins that lack exons 20 to 22, exons 12 to 22, and exons 15 to 22, respectively. Four cysteine residues are located within this C-terminal region and are highly conserved in the PHEX protein [3], [10]. These 4 cysteine residues are most likely involved in disulfide bond formation, and losing them could result in a defective secondary protein structure that could greatly inhibit the enzymatic activity of the protein. Therefore, of all of the mutations detected in this study, these 3 mutations are the most likely to affect the function of the PHEX protein. It is known that approximately 27% of the mutations in the PHEX gene are nonsense mutations [26]. After searching the PHEX mutation database, 15 mutations were detected in exon 20 [18], indicating that exon 20 may be a mutational hotspot.
Two novel missense mutations were detected in family 2: p.His584Pro in exon 17 and p.Gly395Arg in exon 11. The PHEX gene contains 10 highly conserved cysteine residues, all of which are located in a very large extracellular domain. These cysteine residues may be involved in disulfide bond formation and protein folding [24]. The p.His584Pro and p.Gly395Arg mutations affect 2 of these cysteine residues. Mutations at both sites would most likely result in changes to the protein structure and would result in the loss of protein function. Additionally, glycine (G) and proline (P) are non-polar hydrophobic amino acids, and arginine (R) and histidine (H) are polar alkaline hydrophilic amino acids. Therefore, it is predicted that substituting G with R and H with P will alter the biochemical properties at these positions. Furthermore, p.His584Pro and p.Gly395Arg are non-conservative, affect evolutionarily highly conserved amino acids from nine different species (Fig. 4) and were predicted in silico by all bioinformatic tools used to be of pathogenic relevance (PolyPhen-2 score of 0.98 and 0.99, respectively. Both SIFT score <0.05). The proband’s mother (I2) in family 2 carried both mutations (p.His584Pro and p.Gly395Arg), therefore has more severe phenotypes than her daughter. These phenotypic symptoms included a lower blood phosphorus level, an earlier onset age, odontodysplasia, delayed dentition, and teeth falling out at the age of 22 years.
Two deletion mutations were identified in our study. One deletion mutation was p.Tyr565Phefsx5 in exon 16. This mutation consists of a heterozygous deletion of one nucleotide (A) in codon 565 (c.1694delA), which leads to a phenylalanine substitution for a tyrosine at P.565 and a subsequent premature truncation at p.570, which results in a nonfunctioning PHEX product. The other mutation is an insertion-deletion mutation, c.2077_*4delinsA. In this mutation, 16 nucleotides (TGGTGCAATTAGTAAC) from codon 718 to codon 723 are deleted and 1 nucleotide (A) is inserted at p.N718, which results in 6 amino acids missing from p.N718 to p.N723 and a lysine insertion (at codon 718). These changes most likely alter the biochemical properties at these positions and affect the PHEX protein function.
Three splice site mutations were identified in our study. Splice site mutations at introns 10, 15, and 17 (IVS10-1G>A, IVS15-2A>T, and IVS17+2T>G, respectively) were predicted to cause exons 11, 16, and 18 to be skipped, respectively. These changes would result in a reading frame shift and truncated proteins.
The PHEX gene mutations in the 3 sporadic patients (probands from family 7, 8, and 9) were not inherited from their parents and are most likely de novo mutations. These types of mutations are caused by a mutation in the germ cell or germ cell mosaicism in the gonads of one of the parents or by a mutation in the fertilized egg itself. Studies have shown that male mutation bias frequently occurs among higher organisms, and in humans, the male to female bias ratio is approximately 6 to 1 because of differences in male and female gamete formation [24], [27]. Furthermore, the male germline accumulates more DNA replication errors because of the higher number of germline cell divisions in males than females [28]. Therefore, PHEX mutagenesis in paternal germ cells is likely more frequent in sporadic patients and would only affect the female offspring [24], [29], which is accordance with our finding from family 9 (proband III1 is female). Interestingly, however, that the 2 sporadic patients (probands from family 7 and 8) in our study are males, which differs from the demographics in previous studies. This finding indicates that the mutated PHEX alleles in sporadic male patients probably resulted from the mutagenesis in the X chromosome of the maternal germ cell.
From our study, there are no significant differences of gene mutation types and mutation locations in the PHEX gene in Chinese XLH patients compare to non-Chinese patients. However, the same mutations in different races can cause different clinical features. For example, p.Trp444X was firstly reported by Beck-Nielsen SS, et al. [30] in a sporadic patient, a Danish male, with a normal height, mild skeletal and endodontic phenotype. Whereas, in our study, the mutation was found in familial patients with abnormal gait, kyphosis, and hip and knee joint pain. In addition, we identified that the proband and her daughter carried the non-sense mutation (p.Trp444X) which consisted of a heterozygous G to A transition at c.1332 in exon 12, while, The mutation reported by Beck-Nielsen SS, et al. is c.1331G>A affecting one nucleotide upstream the one described in our manuscript (c.1332G>A). Although, the result is the same at the protein level with a tryptophan at position 444 being substituted by a stop codon and truncation at p. 444, the clinical features are quite different in Chinese patients compare to non-Chinese patients.
Although, no evident genotype phenotype correlation could be established in our study, 2 novel mutations were detected and different clinical features were described. Therefore, Functional studies investigating the PHEX gene mutation should be performed to elucidate the complex relationship between genotype and phenotype.
Acknowledgments
The authors give thanks to all patients, their families and ethnicity-matched healthy controls for their excellent collaboration. We thank our colleagues working in the Department of Osteoporosis for recruiting all the subjects and we are grateful for the help of our colleagues working in the central laboratory of Shanghai JiaoTong University Affiliated Sixth People’s Hospital for serum analysis and help us complete the study. Greatly appreciated for the efforts to improve the quality of our study made by all the Reviewers and the Academic Editors.
Funding Statement
The study was supported by grants from the National Natural ScienceFoundation of China (No. 81000360, 81170803, 81070692, and 81370978), Shanghai Rising-Star Program (No. 11QA1404900), and Academic Leaders in Health Sciences in Shanghai (No XBR2011014). The founder (Prof. Zhen-lin Zhang( with the National Natural Science Foundation of China (81170803 and 81070692) and Academic Leaders in Health Sciences in Shanghai (No XBR2011014) played a role in conceiving, designing, revising the study and data collection as well. The founder (Dr. Hua Yue) with the National Natural Science Foundation of China (No. 81000360) and Shanghai Rising-Star Program (No. 11QA1404900) was responsible for data analysis, writing, and preparation of the manuscript. The founder with the National Natural Science Foundation of China (No. 81370978) had a role in data collection.
References
- 1. Rowe PS (1994) Molecular biology of hypophosphataemic rickets and oncogenic osteomalacia. Hum Genet 94: 457–467. [DOI] [PubMed] [Google Scholar]
- 2. Rowe PS, Oudet CL, Francis F, Sinding C, Pannetier S, et al. (1997) Distribution of mutations in the PHEX gene in families with X-linked hypophosphataemic rickets (HYP). Hum Mol Genet 6: 539–549. [DOI] [PubMed] [Google Scholar]
- 3. Rowe PS (1998) The role of the PHEX gene (PEX) in families with X-linked hypophosphataemic rickets. Curr Opin Nephrol Hypertens 7: 367–376. [DOI] [PubMed] [Google Scholar]
- 4. Quarles LD, Drezner MK (2001) Pathophysiology of X-linked hypophosphatemia, tumor-induced osteomalacia, and autosomal dominant hypophosphatemia: a perPHEXing problem. J Clin Endocrinol Metab 86: 494–496. [DOI] [PubMed] [Google Scholar]
- 5. Albright F, Butler A, Bloomberg E (1939) Rickets resistant to vitamin D therapy. American Journal of Disease of Children 54: 529–547. [Google Scholar]
- 6. Davies M, Stanbury SW (1981) The rheumatic manifestations of metabolic bone disease. Clinics in Rheumatic Disease 7: 595–646. [Google Scholar]
- 7. Beck-Nielsen SS, Brock-jacobsen B, Gram J, Brixen K, Jensen TK (2009) Incidence and prevalence of nutritional and hereditary rickets in southern Denmark. Eur J Endocrinol 160: 491–497. [DOI] [PubMed] [Google Scholar]
- 8. Ruppe MD, Brosnan PG, Au KS, Tran PX, Dominguez BW, et al. (2011) Mutational analysis of PHEX, FGF23 and DMP1 in a cohort of patients with hypophosphatemic rickets. Clin Endocrinol (Oxf) 74: 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Quinlan C, Guegan K, Offiah A, Neill RO, Hiorns MP, et al. (2012) Growth in PHEX-associated X-linked hypophosphatemic rickets: the importance of early treatment. Pediatr Nephro 27: 581–588. [DOI] [PubMed] [Google Scholar]
- 10. Clausmeyer S, Hesse V, Clemens PC, Engelbach M, Kreuzer M, et al. (2009) Mutational analysis of the PHEX gene: novel point mutations and detection of large deletions by MLPA in patients with X-linked hypophosphatemic rickets. Calcif Tissue Int 85: 211–220. [DOI] [PubMed] [Google Scholar]
- 11. Francis F, Strom TM, Hennig S, Boddrich A, Lorenz B, et al. (1997) Genomic organization of the human PEX gene mutated in X-linked dominant hypophosphatemic rickets. Genome Res 7: 573–585. [DOI] [PubMed] [Google Scholar]
- 12. Du L, Desbarats M, Viel J, Glorieux FH, Cawthorn C, et al. (1996) cDNA cloning of the murine Pex gene implicated in X-linked hypophosphatemia and evidence for expression in bone. Genomics 36: 22–28. [DOI] [PubMed] [Google Scholar]
- 13. Thompson DL, Sabbagh Y, Tenenhouse HS, Roche PC, Drezner MK, et al. (2002) Ontogeny of Phex/PHEX protein expression in mouse embryo and subcellular localization in osteoblasts. J Bone Miner Res 17: 311–320. [DOI] [PubMed] [Google Scholar]
- 14. Addison WN, Nakano Y, Loisel T, Crine P, McKee MD (2008) MEPE-ASARM peptides control extracellular matrix mineralization by binding to hydroxyapatite: an inhibition regulated by PHEX cleavage of ASARM. J Bone Miner Res 23: 1638–1649. [DOI] [PubMed] [Google Scholar]
- 15. Yang L, Yang J, Huang X (2013) PHEX gene mutation in a Chinese family with six cases of X-linked hypophosphatemic rickets. J Pediatr Endocrinol Metab 26: 1179–1183. [DOI] [PubMed] [Google Scholar]
- 16. Xia W, Meng X, Jiang Y, Li M, Xing X, et al. (2007) Three Novel Mutations of the PHEX Gene in Three Chinese Families with X-linked Dominant Hypophosphatemic Rickets. Calcif Tissue Int 81: 415–420. [DOI] [PubMed] [Google Scholar]
- 17. Lo FS, Kuo MT, Wang CJ, Chang CH, Lee ZL, et al. (2006) Two novel PHEX mutations in Taiwanese patients with X-linked hypophosphatemic rickets. Nephron Physiol 103: 157–163. [DOI] [PubMed] [Google Scholar]
- 18. Kang QL, Xu J, Zhang Z, He JW, Lu LS, et al. (2012) Three novel PHEX gene mutations in four Chinese families with X-linked dominant hypophosphatemic rickets. Biochem Biophys Res Commun 423: 793–798. [DOI] [PubMed] [Google Scholar]
- 19. Jap TS, Chiu CY, Niu DM, Levine MA (2011) Three novel mutations in the PHEX gene in Chinese subjects with hypophosphatemic rickets extends genotypic variability. Calcif Tissue Int 88: 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qiu G, Liu C, Zhou J, Liu P, Wang J, et al. (2010) Prenatal diagnosis for a novel splice mutation of PHEX gene in a large Han Chinese family affected with X-linked hypophosphatemic rickets. Genet Test Mol Biomarkers 14: 385–91. [DOI] [PubMed] [Google Scholar]
- 21. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sunyaev S, Ramensky V, Bork P (2000) Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet 16: 198–200. [DOI] [PubMed] [Google Scholar]
- 23. Doyle AJ, Doyle JJ, Bessling SL, Maragh S, Lindsay ME, et al. (2012) Mutations in the TGF-β repressor SKI cause Shprintzen-Goldberg syndrome with aortic aneurysm. Nat Genet 44: 1249–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Durmaz E, Zou M, Al-Rijjal RA, Baitei EY, Hammami S, et al. (2012) Novel and de novo PHEX mutations in patients with hypophosphatemic rickets. Bone 52: 286–291. [DOI] [PubMed] [Google Scholar]
- 25. Ng PC, Henikoff S (2003) SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Filisetti D, Ostermann G, von Bredow M, Strom T, Filler G, et al. (1999) Non-random distribution of mutations in the PHEX gene, and under-detected missense mutations at non-conserved residues. Eur J Hum Genet 7: 615–619. [DOI] [PubMed] [Google Scholar]
- 27. Makova KD, Li WH (2002) Strong male-driven evolution of DNA sequences in humans and apes. Nature 416: 624–626. [DOI] [PubMed] [Google Scholar]
- 28. Goetting-Minesky MP, Makova KD (2006) Mammalian male mutation bias: impacts of generation time and regional variation in substitution rates. J Mol Evol 63: 537–544. [DOI] [PubMed] [Google Scholar]
- 29. Zhu X, Li M, Pan H, Bao X, Zhang J, et al. (2010) Analysis of the parental origin of de novo MECP2 mutatiopns and X chromosome inactivation in 24 sporadic patients with Rett syndrome in China. J Child Neurol 25: 842–848. [DOI] [PubMed] [Google Scholar]
- 30. Beck-Nielsen SS1, Brixen K, Gram J, Brusgaard K (2012) Mutational analysis of PHEX, FGF23, DMP1, SLC34A3 and CLCN5 in patients with hypophosphatemic rickets. J Hum Genet. 57: 453–458. [DOI] [PubMed] [Google Scholar]