Abstract
Eukaryotes employ combinatorial strategies to generate a variety of expression patterns from a relatively small set of regulatory DNA elements. As in any other language, deciphering the mapping between DNA and expression requires an understanding of the set of rules that govern basic principles in transcriptional regulation, the functional elements involved, and the ways in which they combine to orchestrate a transcriptional output. Here, we review current understanding of various grammatical rules, including the effect on expression of the number of transcription factor binding-sites, their location, orientation, affinity and activity; co-association with different factors; and intrinsic nucleosome organization. We review different methods that are used to study the grammar of transcription regulation, highlight gaps in current understanding, and discuss how recent technological advances may be utilized to bridge them.
Keywords: Transcriptional regulation, gene expression, transcription factor, binding site, nucleosome
Introduction
Proper control of mRNA levels is critical in nearly all biological processes. Since much of this control is encoded within non-coding regulatory regions, deciphering the mapping between DNA sequence and expression levels is key for understanding transcriptional control. Eukaryotes are known to employ combinatorial strategies to generate a variety of expression patterns from a relatively small set of regulatory motifs1 and exploit motif geometry as another dimension of combinatorial power for regulating transcription. Understanding the fundamental principles governing transcriptional regulation could allow us to predict expression from DNA sequence, with far reaching implications. Most notably, in many human diseases, genetic changes occur in non-coding regions such as gene promoters and enhancers. However, without understanding the grammar of transcriptional regulation we cannot tell which sequence changes affect expression and how. For example, even for a single binding site, we do not know the quantitative effects on expression of its location, orientation, and affinity; whether these effects are general, factor-specific, and/or promoter-dependent; and how they depend on the intrinsic nucleosome organization. Similarly, we do not know which properties determine whether multiple sites contribute additively or cooperatively to expression, what types of cooperativity functions can be achieved, how they depend on the affinity of the sites and identity of the factors, and whether their mechanistic basis involves protein-protein interactions and/or nucleosome eviction. Unraveling this transcriptional grammar will allow us to understand, predict and design expression patterns from regulatory sequences (Figure 1).
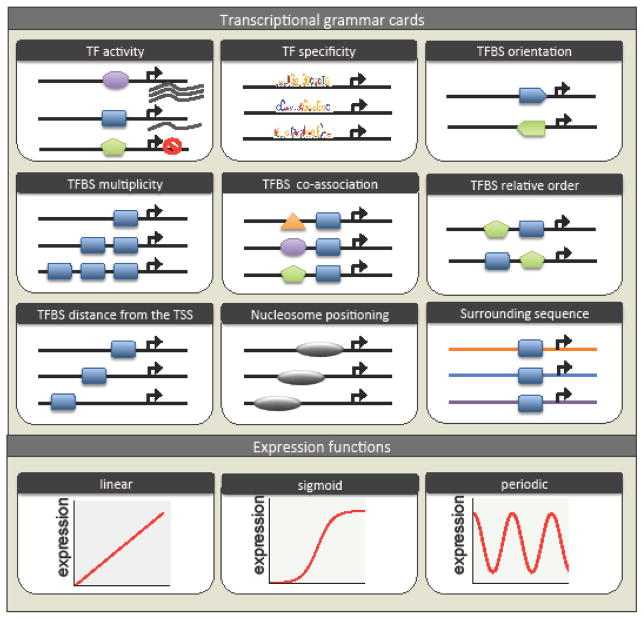
Figure 1. Illustration of grammatical rules and their effect on gene expression.
Transcriptional grammar. The rules of transcription regulation are illustrated as “grammar cards”. Each card represents a different rule. For example, the number of binding sites for the same TF, the distance of one TF from the TSS, the orientation of the binding-site relative to the TSS, etc.
Expression function. The function by which each rule affects expression. For example: cooperative binding results in a sigmoidal curve since one binding event increases the probability of another binding to occur, resulting in higher expression. The x-axis changes according to the rule identity. For example, for the rule of binding-site location, the x-axis represents distance from the TSS (in bp).
Addressing this challenge requires knowledge of both the functional elements and the ways in which such elements combine to orchestrate a transcriptional output. Testing the effect of designed DNA mutations has been successfully employed for several decades in the research of transcriptional control, but on the scale of a handful of sequences per study. A major hindrance to progress is the limited ability to measure the transcriptional effect of a large number of designed DNA sequences in which specific regulatory elements are systematically varied. Recently developed technologies increases the throughput of these experiments by ~1000-fold, allowing us to gain considerably more insight into how information is encoded in the language of DNA. In this review, we discuss several examples of grammatical rules in transcription, highlight the main gaps, and discuss how these may be bridged using recent technological advances.
Methods to decipher the grammar of transcription
A broad range of methods exist for annotating and testing functional regulatory elements in non-coding DNA sequences in order to decipher the principles governing transcription regulation. These include comparative computational models2–4, high-throughput assays to map functional elements in the genome such as TF binding sites and nucleosomes5–9, and classical genetic techniques including reporter assays for quantitative activity measurements10–12. Accumulation of genome-wide data on gene expression (RNA-seq)5, TF binding landscape (Chip-seq)6, chromatin state (DNase-seq7 and FAIRE-seq8), and physical DNA interactions (5C)9 led to the identification of potential promoter and enhancer regions, the TFs bound to these regions, and the chromatin architecture13. However, although revealing an unprecedented number of regulatory elements in the genome, these studies do not assay the mechanism and functional activity of these elements. For example, we cannot tell which of the binding sites of a TF affect transcription and in which manner. Genome-wide quantitative measurements of native enhancers were facilitated by recent developed methods such as self-transcribing active regulatory region sequencing (STARR-Seq)14. Yet, native enhancers differ in many sequence elements making it hard to attribute the measured expression differences to any single sequence change. Thus, it is difficult to infer systematic rules of transcriptional grammar solely by quantitatively measuring native sequences. Another approach uses computational models for learning the complex combinatorial code underlying gene expression2–4. These studies utilize mRNA expression data and DNA-sequence elements in the promoters of the corresponding genes to decipher the effect of motif strength, orientation, and relative position on gene expression. However, although computational studies generate a large number of mechanistic hypotheses, experimental validation is still required.
One direct and quantitative way to measure the activity of regulatory element is to fuse a DNA sequence to a reporter gene and measure its expression with biochemical assays such as luciferase assay. Researches have utilized this approach successfully to determine the activity of promoters15, enhancers16 and insulators17,18. However, the construction of these reporters by traditional cloning techniques is slow and labor-intensive, limiting throughput to at most dozens of regulatory elements per experiment. Several medium-scale19–21 and large-scale22–25 libraries were created in bacteria, yeast and mammalian cells, in which regulatory elements were randomly ligated, mutagenized or synthesized in tandem and the expression of the resulting promoters was measured. These studies provided much insight, but their random nature imposes limitations on the repertoire of promoters constructed and thus they cannot systematically dissect basic principles of transcriptional grammar. For example, studying the effect of binding site location on expression requires measurements of promoters that differ only in the location of the site and sampling many such locations. Such collection of promoters could not be generated by random ligation of regulatory elements. Systematic manipulation of some specific promoters10–12 led to profound insights, but since the variants were constructed one by one, time and cost considerations have limited the scale of previous studies, such that to date, only a modest number of elements have been characterized at high resolution.
Recent advances in the fields of DNA synthesis and deep sequencing provided a fertile ground for the development of new high-throughput approaches that address this technological barrier. These approaches provide the ability to accurately measure the activity of tens of thousands different designed regulatory elements within a single experiment. Although they differ in the way in which they measure expression, these methods utilize the power of massive synthesis of DNA oligonucleotides on a microarray platform, harvest these oligos as pool and proceed with this complex library as input for a single experiment26–30 (Figure 2). The first approach26–28,30 is applicable both in-vitro and in-vivo and measures expression by the counts of mRNA molecules transcribed from each tested sequence using RNA-seq. To evaluate transcriptional activity in-vitro26, each oligonucleotide in the library is designed to include a unique barcode sequence downstream to the TSS. The oligos are transcribed in vitro, and the resulting transcripts are then sequenced. The relative abundance of each barcode provides a digital readout of the transcriptional efficiency of its cis-linked designed sequence. This protocol was recently adapted to measure transcription in-vivo in mammalian cells27,28,30. To that end, the oligonucleotides pool is inserted into plasmids, where each plasmid contains one of the designed sequences upstream to an arbitrary open reading frame (ORF) followed by a unique barcode sequence. Plasmids are co-transfected into cells, where each designed sequence drives the transcription of mRNAs containing the barcode in their 3′ UTRs. To estimate their relative activities, the barcodes in the reporter mRNAs and the plasmids pools are then sequenced and the ratios of these counts are calculated.
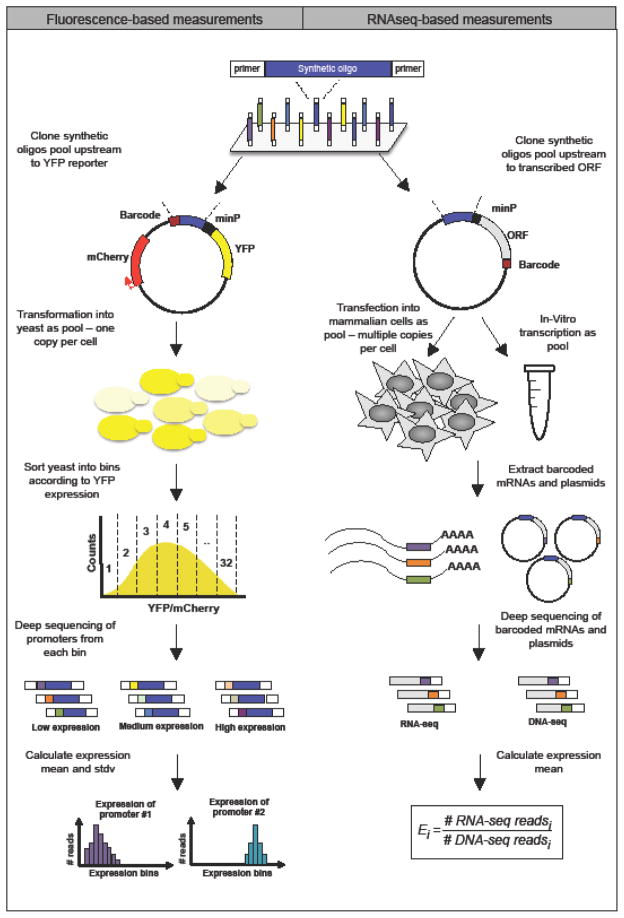
Figure 2. High-throughput measurements of thousands of systematically designed regulatory sequences.
Fluorescence-based measurements. ssDNA oligos are synthesized on an array and harvested as a single pool. The entire library is cloned into a plasmid upstream of a fluorescent reporter (YFP). A plasmid pool is transformed into yeast such that each cell expresses a single plasmid. Cells are sorted into bins according to their YFP/mCherry ratio using fluorescence activated cell sorter (FACS). Promoters from each bin are then amplified and sent for deep sequencing. In the final step, the activity of each sequence in the library is computed by calculating reads distribution among expression bins.
RNA-seq based measurements. ssDNA oligos are synthesized on an array and harvested as a single pool. The entire library is cloned into a plasmid upstream of an open reading frame. Each plasmid carries a unique barcode sequence downstream to the ORF. Plasmids can be in-vitro transcribed or transfected into mammalian cells for in-vivo studies. Next, plasmids and mRNAs are extracted from cells, barcodes are amplified and sent for deep sequencing. In the final step, the activity of each sequence in the library is computed by calculating the ratio between RNAseq and DNAseq reads number (Ei).
Another approach uses fluorescence to measure expression in vivo29. Although it was developed in yeast, it can in principle be adapted to mammalian cells. In this approach, each oligonucleotide in the library, which includes a unique barcode at the 5′ end, is ligated into a plasmid upstream of a yellow fluorescent reporter (YFP). The pool of plasmids is then transformed into yeast such that every transformed strain carries one plasmid expressing YFP from one promoter in the library. Using fluorescence activated cell sorter (FACS), the resulting library of yeast cells are then sorted into ~32 bins according to YFP expression. Next, the promoters of each bin are sequenced such that for every barcode (representing one promoter), the total number of reads is counted. Finally, by examining the distribution of reads of every promoter across the expression bins, both its mean and standard deviation of expression can be calculated.
Although currently limited by the length of the synthesis DNA oligo (200bp), these technologies provide ~1,000-fold increase in throughput over previous methods, thereby providing new methodologies for accelerating research in the field of transcriptional regulation.
Grammatical rules of transcription
TF specificity
The first step in understanding the grammar of a language is to dissect its most basic building blocks. In transcriptional regulation, one of the most important building blocks are binding sites for transcription factors (TFBS). For each of the ~1,500 TFs in the human genome, we would like to know the length and composition of its sequence preferences, which nucleotides are essential for its binding, and how sensitive is the TF to sequence alterations in its binding site. For TFs that form homo/hetero-dimers, we should determine the spacing between its half-sites and any other constraint that may apply to its sequence. In addition, we should analyze the effect of other elements, which are not directly bound by the TF, on its specificity.
In-vitro and in-vivo binding profiles generated by protein-binding microarrays31,32, high-throughput SELEX and ChIP-seq33 determined the binding specificities of hundreds TFs. However, despite this growing information about TF binding specificity, much less is known about the effect of TF site affinity on expression. A recent study utilized high-throughput measurements technology to compare the expression of 2,104 enhancers in which the binding sites for 7 TFs were varied34. As expected, this study found that changes in expression correlate with the change in motif match score, indicating that gene expression is correlated with binding affinity when all else is maintained. However, sites with different affinities for the same TF are rarely placed in the exact same context, making it hard to study the relationship between binding site strength and expression. Indeed, examining binding sites for the transcription factor p53 in the genome reveals that many sites with predicted high-sequence specificity are not bound by p5335. These sites were able to activate transcription when cloned in a plasmid in front of a luciferase gene, indicating that they can interact productively with p53 when removed from their native context and chromatin environment.
Despite these advances, the recognition sites of many TFs are still missing and for the known ones we still do not understand the effect of context, site accessibility, chromatin, and TF concentration on expression. Some parameters such as chromatin and TF concentration vary between conditions and are expected to affect binding in a condition-specific manner. To date, we cannot tell how many sequence changes in its recognition site can a TF tolerate before it will affect its activity, whether this number is uniform for most TFs or changes for different TFs families. Answering such questions requires a systematic analysis measuring the effect of TF site affinity on expression for a large number of TFs.
TF activity
Once a TF is bound to the DNA it can enhance or repress transcription by promoting or blocking the recruitment of RNA-polymerase. However, we cannot predict whether the TF functions as an activator or as a repressor from the sequence of its binding-site. In addition, the expression levels, localization and activity of TFs vary between cell types, different phases of cell cycle, and in response to stress conditions. Therefore, we cannot tell in advance which of TF site will affect expression and to what extent. Even the most comprehensive mapping of TF binding-sites cannot provide information about the activity of the TF in each of these sites. To achieve that, direct measurements of expression are needed in various cell types and conditions.
Large scale data from the ENCODE project yielded genome-wide binding profiles for ~100 TFs in various cell types, providing insight on the actual occupancy of binding-sites in the human genome36. However, the functionality of these binding events was not determined. A recent study37 combined information gained from these binding profiles with computational methods to identify TFBS and to measure their activity within a functional assay. The authors predicted and mutagenized 455 binding sites for six TFs in human promoters and measured expression using luciferase assay in four cell lines, and found that 30% of TFBSs were not functional in any cell line and only 14% of the sites were verified in all four cell-lines. These results emphasize that the presence of a TFBS by itself is not sufficient to determine whether it will be active in vivo and highlights the importance of in vivo functional measurements of TFBSs. Despite being more comprehensive than previous studies, this study still examined a total of only six TFs, and due to the native nature of the tested sequences, the promoter background was not kept constant. Thus, differences in activity levels cannot be attributed solely to the binding site, as they may also represent the effect of the context, including binding sites for co-activators, GC content, and nucleosome positioning signals. A more direct assay for comparing activity measurements of different TFs involves planting sites for different TFs at the exact same location and within the same background sequence. Utilizing a new approach for measuring the expression of thousands of designed regulatory sequences within one experiment, a recent study compared the activity of 75 different yeast TFs to each other, by separately planting each TF site within the same promoter sequence29.
Despite these advances above, to date we do not know the actual fraction of functional TFBSs in a given cell, which of these sites act as an activator or a repressor (or both), and how the activity of different TFs compares to one another. One way to answer such questions is to perform a full survey of all known TF binding-site in various contexts and conditions through a synthetic high-throughput approach similar to that recently done in yeast29.
The effect of TFBS orientation
Unlike regulatory elements playing a role at the mRNA level, which are restricted to the coding strand of the DNA, regulatory elements in promoters can appear on both the coding and non-coding strands. This mechanism has some advantages such as doubling the probability to find TFBS in a specific location and preventing steric interference between two neighboring TFs if they bind the DNA surface from opposite sides. On the other hand, since transcription is directional, some of these elements should be orientation-sensitive in order to place the RNA-polymerase in the right direction. In order to recognize the functional TFBSs in a given promoter sequence we should know for each TF whether it can activate transcription from one of the DNA strands or from both.
One way to achieve orientation-insensitive regulation is by using palindromic sequences as binding-sites. Indeed, palindromes are predominant in regulatory elements38. A palindromic design has two main advantages. First, palindromes can be bound on either strand of DNA, thus doubling the local concentration of the TF binding-site and increasing productive encounters between the TF and the DNA. Second, palindromes can be bound by TFs that work as dimers. However, palindromic binding-sites are flanked by non-palindromic sequences, which in some cases contribute to TF binding and are therefore sensitive to the orientation2. There are also examples for non-palindromic binding-sites that can equally activate transcription when placed in the forward or the reverse orientation39. Thus, we cannot predict orientation-sensitivity just by the appearance of palindromic sequence in the TF binding-site. A genome-wide computational study investigated the directionality of promoter motifs by comparing conservation on the forward and the reverse strands in four mammalian genomes40. This study found that conservation of promoter motifs is largely symmetric on both strands of the DNA in contrast to 3′-UTR motifs that have a strand preference, suggesting that most TFs are not affected by the orientation of their binding site. Consistent with this finding, a functional experiment that systematically compared the activity of 75 yeast TF binding-sites in two orientations found strong effects on expression for only 8% of the binding-sites29.
Taken together, we do not yet know the orientation-sensitivity of the ~1,500 TFs in human, providing another hindrance to models aimed at predict gene expression form sequence. Here too, a systematic study that will place the site of all human TFs in each orientation and within the same sequence background can provide much insight.
Combinatorial co-regulation by different TFs
The architecture of promoters is often composed of binding-sites for many TFs working together to orchestrate a transcriptional output. TFs can cooperate with each other by direct physical interactions, forming homodimers, heterodimers, or larger transcriptional complexes41. Many TFs belongs to families that bind their target genes as dimers, such as bZIP, bHLH, or nuclear hormone receptor families42. Interaction among TFs can be specific to context and condition, resulting in a unique transcriptional program. Since many of these interactions are synergistic, we cannot simply sum up the effects that were measured for individual TFs. Dissecting the effect on expression of TF combinations is therefore a great challenge.
Studies focused on a small number of TFs had shown that for the same TF, different partners have different effects on transcription, resulting in distinct functional pathways43,44. By systematically mapping all combinatorial protein-protein interactions for 1222 TFs in human, 762 interactions were found45. However, out of these interactions, we cannot tell which affect transcription and by what mechanism. Analysis of Chip-seq data of 119 factors36 found a statistical enrichment for specific TF combinations, which may suggest that these TFs work together. Interestingly, the same TF was found to co-associate with different partners in different contexts such as gene-proximal and distal regions.
Even with the recent progress in mapping TFs interactions at the protein level and studying their co-association when binding the genome, we do not have a systematic view of how different combinations of TFs affect gene expression. This requires direct expression measurements of different constructs containing various combinations of TF binding-sites. Such measurements, in comparison to measurements of each TF separately, could answer which of the TF-pairs work synergistically, whether they act to increase or decrease expression, and to what extent. However, the space of TF combinations is vast, and systematically assaying it is challenging even with the recent advances in throughput.
The effect of binding site number
Multiple binding sites for the same TF, also known as homotypic clusters of TFBSs, are statistically enriched in proximal promoters and distal enhancers. Conservation of such site clusters between vertebrate and invertebrates suggests that homotypic clustering is a general organization principle of cis-regulatory regions46. Using multiple binding sites for the same TF as a mean to regulate expression may have several mechanistic advantages. These include lateral diffusion of a TF binding along a regulatory region47–49, high-affinity cooperative binding50, and functional redundancy51,52. In order to integrate information about the number of sites when predicting gene expression, we need to know the function that relates TFBS number to expression. Does expression increase linearly with the number of binding sites? What is the range in which adding a binding-site still has a substantial effect? Are these behaviors general or TF-specific?
Few studies set out to investigate the effect of binding-site number for one specific TF12,39. Increasing number of sites for Bicoid led to greater-than-additive increase in expression11. The same study showed that Bicoid bound to a strong site promotes occupancy of an adjacent weak site by cooperative DNA binding12. By parallel measurements of thousands of synthesized promoters, a recent study in yeast designed synthetic promoters to systematically test the dependence of expression on the number of sites29. The consensus site for Gcn4 was planted in all possible combinations of 1–7 sites at 7 predefined locations within two different promoter sequences (128 sequences for each context). Examining the average expression of all possible locations for each number of sites resulted in a clear relationship between the number of sites and the average expression, which accurately fits a logistic function, and in which expression increases with addition of each of the first four sites but then mostly saturates. Such saturation in expression was also observed in small-scale studies12,39. However, it is unclear whether this saturation is at the level of binding or activation of transcription. Intriguingly, comparing the expression of individual promoters with specific combinations of site locations suggests that the relative distance between two sites significantly affects expression. As one example, when located in close vicinity, the TF molecules may sterically occlude each other.
Future studies are needed to study the effect of binding site number on other TFs, to quantitatively characterize the effect of the distance between multiple binding-sites, and to understand which biological mechanism underlie the sigmoidal behavior.
The effect of TFBS distance from the TSS and DNA helical repeats
When addressing the question of how the relative location of a TF binding-site affects transcription, one should remember that the DNA is a three-dimensional molecule and that changing the distance has a geometric aspect. For example, two regulatory elements that are five nucleotides apart from each other are also located on opposite sides of the DNA double helix. Therefore, we should decipher the effects on expression of both the absolute distance of the TFBS from the TSS, and the relative angle between the TFBS and the TSS on the DNA double helix.
A computational genome-wide analysis of TF motifs and gene expression data addressed the first parameter and computed the effect of absolute distance on gene expression2. Depending on the TF identity, expression can reach its maximal values when the motif is either within 150 bp from the start codon, at intermediate distance of 150–300 bp, or at long-range distances of 300–450bp. The second parameter of relative angle on the DNA double helix was addressed by changing the distance between regulatory elements in promoters with small increments. Insertion of a spacer sequence between TFBS and the TSS yielded an interesting expression pattern10,39, with spacers with 5bp multiplicity (5, 15, 25) having lower expression than spacers with 10bp multiplicity (10, 20, 30), resulting in a periodic function composed of consecutive peaks whose period is ~10bp. It was suggested that efficient initiation of transcription requires a stereospecific alignment between some promoter elements such as the TFBS and the TATA-box. It appears that the assembly of proteins bound to various promoter elements can interact with each other when located on the same side of the DNA helix. However, while some studies support the helical spacing idea, others did not reproduce the periodic phenotype. In those cases, increasing the distance between a TFBS and the TSS resulted in a gradual decrease in expression with no periodic behavior53. Utilizing the high throughput technology to systematically investigate the effect of binding site location on expression significantly increased the number of the tested TFBSs, the number of different promoter contexts and the resolution of the distance increments29. This higher resolution revealed that even small 1–7 bp changes in site location could have major effects on expression. Periodicity in expression was obtained for only one of the designed promoter backgrounds, in which sliding the tested TF site resulted in expression being a ~10bp periodic function of site location and the function persisted for 6 consecutive peaks. Notably, this periodicity was significant in only one of the two tested promoter contexts for the same TF, suggesting that some contexts have more flexibility in the way in which they can generate stereospecific alignments if these are indeed required.
These findings leave many open questions regarding the effect of TFBS location on expression. In which cases is stereo-alignment required for transcriptional activation? How does it depend on the nature of the TF and the context? Are there cases in which interacting with the DNA helix from opposite sides is advantageous (e.g., in the case of independent regulators that should not interact with each other but may sterically occlude each other)?
The effect of epigenetic context and nucleosomes
In addition to TF binding sites, the transcriptional activity of a gene depends on the local composition and organization of its chromatin environment. Silencing mechanisms such as DNA methylation, nucleosome positioning and histone modifications add epigenetic regulation onto DNA without changing the genetic information. Thus, knowing the composition of TF sites, their geometry and combinatorial interactions is not enough in order to predict gene expression, and the chromatin structure should also be incorporated. For example, a weak binding-site may drive higher expression levels than a strong site if the latter is embedded within a silenced region. The importance of epigenetic context was recently demonstrated in genome-wide enhancer measurements, which revealed that the same DNA sequence may or may not act as an enhancer depending on its genomic locus and epigenetic environment14. Polycomb group (PcG) proteins, DNA-methyltransferases and histone modifiers are recruited to specific target genes to ensure well-timed and spatially restricted gene expression patterns54,55. The identification of DNA elements required for the targeting of these silencing complexes is key to our understanding of epigenetic dynamic processes. In addition to complexes-mediated silencing, DNA sequences have intrinsic preferences for nucleosome formation. The DNA segments that are wrapped by nucleosomes are less accessible to most trans-acting factors such as RNA-polymerase and transcription factors. In order to decipher the effect of nucleosomes on gene expression, we should know the DNA sequence preferences for nucleosome binding, how it affects the accessibility of neighboring cis-regulatory elements such as TF binding-site and TSS, and how this effect depends on distance.
In the past several decades, it was shown that the affinity of nucleosomes for different DNA sequences varies greatly, spanning a range of 5,000-fold between the weakest and strongest binding56,57. This range is thought to reflect differences in the ability of DNA sequence to bend around the histone octamer into nucleosome structure58. Two sequence features were shown to play an opposite role in determining nucleosome affinity. The first, which has a positive effect on affinity, consist of ~10 bp periodicities of specific dinucleotides59,60. The second feature, which has a negative effect on nucleosome formation, consists of homopolymeric stretches of deoxyadenosine nucleotides, referred to as poly(dA:dT) tracts. These disfavoring nucleosome sequences were found to be strongly associated with nucleosome depletion both in vivo and in vitro (Reviewed in Segal & Widom61), and enrichment of poly(dA:dT) tracts in promoters suggests that they play a regulatory role in gene transcription. Indeed, studies of individual genes in yeast showed that poly(dA:dT) tracts stimulate transcription by increasing the accessibility of a nearby TF binding site62,63. A recent study in yeast tested the effect of poly(dA:dT) on expression by measuring 70 designed promotes, in which the length, composition and distance from TF binding site of poly(dA:dT) tracts were systematically varied64. Notably, manipulating only poly(dA:dT) tracts led to significant effects on gene expression that resemble TF binding site alteration.
Advances in technology enabled the design and measurements of 777 synthetic promoters dedicated to deciphering the effect of nucleosomes on expression29. Adding strong TF binding-sites instead of poly(dA:dT) tracts had a similar effect on expression, suggesting an alternative mechanism for nucleosome eviction. In this mechanism, nucleosomes are competed out by trans-acting factor rather than intrinsic physical properties of the DNA sequence. Investigation of nucleosome positions in three primary human cell types found that the positioning signal for nucleosomes is different from the 10-bp dinucleotide periodicity observed in other eukaryotes65. It is composed of strong G/C cores in the center of the nucleosome (dyad) that are flanked by A/T repelling sequences. Another difference is the GC content in promoter regions. In contrast to fly and yeast, where AT-rich promoters have intrinsic sequence signals for nucleosome eviction, human promoters are enriched for CpG islands, intrinsically favorable for nucleosome formation. However, as in other organisms, promoters of active genes have a nucleosome-free region (NFR) of about 150bp. These observations suggest that CpG-rich segments in mammalian promoters override intrinsic signals of high nucleosome affinity to become active. High-resolution maps of nucleosome footprints reveal that both CpG and non-CpG promoters are subjected to similar epigenetic regulation. These observations support the idea that in addition to intrinsic nucleotides composition other mechanisms are employed to determine nucleosome positioning in mammalians 66.
The mechanism by which CpG-rich sequences evict nucleosome is still poorly understood. In addition, not much is known about the sequence features that determine higher order chromatin structure and the recruitment of silencing complexes such as polycomb. Since higher order structure has a predominant effect of gene expression in higher eukaryotes, deriving such an understanding should improve our ability to predict expression form sequence.
Do different organisms follow the same grammatical rules?
The letters composing the language of the DNA (A,C,G,T) are conserved among all organisms. So is the basic principle of transcription regulation representing a specific interaction between a transcription factor and the DNA molecule. However, different organisms differ in the set of grammatical rules they “choose” to employ for gene expression regulation. For example: homotypic clusters of TFBSs are enriched in enhancers and promoters of human and fly but less so in yeast67, suggesting that while mammalian genomes have evolved to tune expression by multiplying binding-sites for the same TF, the evolution of yeast may had taken another strategy to regulate expression. Another example is the signal for nucleosome eviction. While yeast and fly utilize intrinsic physical properties of AT-rich tracts, mammalian genomes likely rely on a different mechanism to achieve the same eviction of nucleosomes from their CpG-rich promoters. In addition, different organisms employ various epigenetic strategies to shut down the expression of specific target genes. Some epigenetic silencing complexes such as the Sir protein complex in yeast and Polycomb in fly and mammalian share many features55,68. However, DNA methylation at CpG dinucleotides, which is commonly associated with gene silencing in mammalian, is not a predominant mechanism in fly and yeast55,69. Although genomes differ in the extent to which they utilize different features of the transcriptional grammar, an intriguing question is whether the behavior of each feature is universal across organisms. For example, will AT-rich tracts inhibit nucleosome formation even in the CpG-rich promoters found in mammalian genomes? Since the physical and biochemical characteristic of TFs and DNA molecules do not change between organisms, it is tempting to hypothesize that the principle rules governing transcription should be maintained, but this awaits further experimentation. As one example, although multiplicity of TFBSs is not a prevalent feature of yeast promoters, increasing binding site number led to an increase in expression29 in a similar fashion as observed in mammalian39.
Can we solve the grammar of transcription solely by increasing the throughput of the number of sequences designed and measured?
Recent advances in technology provide en efficient mean to design, construct and accurately measure the effect of thousands regulatory sequences on expression. Further reductions in DNA synthesis and sequencing costs in combination with increased quality and length of the synthesized oligos can lead to an era in which experiments will no longer be the major limiting factor. Researchers should soon be able to systematically test the effect of various elements on gene expression and by that increase our understanding of the relative contribution of each regulatory element to expression. However, it is unclear whether overcoming this experimental barrier will be sufficient for deciphering the grammar of transcriptional regulation. The main challenge of this approach is its inherent limitation to study regulatory elements that we already know. Many novel insights into transcriptional regulation were achieved by the discovery of new regulatory mechanisms such as DNA methylation, ncRNA, and nucleosomes. For example, the incorporation of nucleosomes and, specifically, poly(dA:dT) tracts into the computational models improved their performance in predicting expression from sequence64. This discovery of “hidden factors” in the DNA sequence is essential to our progress in understanding the grammar of transcription, and such an understanding cannot be achieved solely by increasing the throughput of sequence measurements. We thus believe that successful approaches for deciphering regulatory grammar should combine powerful synthetic-based approaches for dissecting the effect of known regulatory elements as well as exploratory unbiased approaches that have the potential of uncovering previously unknown regulatory mechanisms.
References
- 1.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–51. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen DH, D’Haeseleer P. Deciphering principles of transcription regulation in eukaryotic genomes. Mol Syst Biol. 2006;2:0012. doi: 10.1038/msb4100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–98. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 4.Segal EYB, Simon I, Friedman N, Koller D. From Promoter Sequence to Expression: A Probabilistic Framework. Proc. 6th Inter. Conf. on Research in Computational Molecular Biology (RECOMB); Washington, DC. 2002. [Google Scholar]
- 5.Morin R, et al. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 6.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 7.Crawford GE, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–31. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaulton KJ, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–9. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dostie J, Dekker J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, et al. Requirement of stereospecific alignments for initiation from the simian virus 40 early promoter. Nature. 1986;319:121–6. doi: 10.1038/319121a0. [DOI] [PubMed] [Google Scholar]
- 11.Giniger E, Ptashne M. Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc Natl Acad Sci U S A. 1988;85:382–6. doi: 10.1073/pnas.85.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burz DS, Rivera-Pomar R, Jackle H, Hanes SD. Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 1998;17:5998–6009. doi: 10.1093/emboj/17.20.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold CD, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–7. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 15.Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat Methods. 2006;3:917–22. doi: 10.1038/nmeth937. [DOI] [PubMed] [Google Scholar]
- 16.Blanco J, Girard F, Kamachi Y, Kondoh H, Gehring WJ. Functional analysis of the chicken delta1-crystallin enhancer activity in Drosophila reveals remarkable evolutionary conservation between chicken and fly. Development. 2005;132:1895–905. doi: 10.1242/dev.01738. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz YB, et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012;22:2188–98. doi: 10.1101/gr.138156.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herold M, Bartkuhn M, Renkawitz R. CTCF: insights into insulator function during development. Development. 2012;139:1045–57. doi: 10.1242/dev.065268. [DOI] [PubMed] [Google Scholar]
- 19.Chiang DY, Nix DA, Shultzaberger RK, Gasch AP, Eisen MB. Flexible promoter architecture requirements for coactivator recruitment. BMC Mol Biol. 2006;7:16. doi: 10.1186/1471-2199-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ligr M, Siddharthan R, Cross FR, Siggia ED. Gene expression from random libraries of yeast promoters. Genetics. 2006;172:2113–22. doi: 10.1534/genetics.105.052688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinkhabwala A, Guet CC. Uncovering cis regulatory codes using synthetic promoter shuffling. PLoS One. 2008;3:e2030. doi: 10.1371/journal.pone.0002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gertz J, Siggia ED, Cohen BA. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 2009;457:215–8. doi: 10.1038/nature07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox RS, 3rd, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinney JB, Murugan A, Callan CG, Jr, Cox EC. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc Natl Acad Sci U S A. 2010;107:9158–63. doi: 10.1073/pnas.1004290107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlabach MR, Hu JK, Li M, Elledge SJ. Synthetic design of strong promoters. Proc Natl Acad Sci U S A. 2010;107:2538–43. doi: 10.1073/pnas.0914803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patwardhan RP, et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol. 2009;27:1173–5. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–70. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melnikov A, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–7. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharon E, et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol. 2012;30:521–30. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc Natl Acad Sci U S A. 2012;109:19498–503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger MF, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–76. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–3. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jolma A, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–39. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Kheradpour P, et al. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–11. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lidor Nili E, et al. p53 binds preferentially to genomic regions with high DNA-encoded nucleosome occupancy. Genome Res. 2010;20:1361–8. doi: 10.1101/gr.103945.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitfield TW, et al. Functional analysis of transcription factor binding sites in human promoters. Genome Biol. 2012;13:R50. doi: 10.1186/gb-2012-13-9-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.FitzGerald PC, Shlyakhtenko A, Mir AA, Vinson C. Clustering of DNA sequences in human promoters. Genome Res. 2004;14:1562–74. doi: 10.1101/gr.1953904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu M, et al. GA-binding protein-dependent transcription initiator elements. Effect of helical spacing between polyomavirus enhancer a factor 3(PEA3)/Ets-binding sites on initiator activity. J Biol Chem. 1997;272:29060–7. doi: 10.1074/jbc.272.46.29060. [DOI] [PubMed] [Google Scholar]
- 40.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walhout AJ. Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome Res. 2006;16:1445–54. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- 42.Reece-Hoyes JS, et al. A compendium of Caenorhabditis elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 2005;6:R110. doi: 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakiri L, Matsuo K, Wisniewska M, Wagner EF, Yaniv M. Promoter specificity and biological activity of tethered AP-1 dimers. Mol Cell Biol. 2002;22:4952–64. doi: 10.1128/MCB.22.13.4952-4964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravasi T, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–52. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gotea V, et al. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010;20:565–77. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JG, Takeda Y, Matthews BW, Anderson WF. Kinetic studies on Cro repressor-operator DNA interaction. J Mol Biol. 1987;196:149–58. doi: 10.1016/0022-2836(87)90517-1. [DOI] [PubMed] [Google Scholar]
- 48.Khoury AM, Lee HJ, Lillis M, Lu P. Lac repressor-operator interaction: DNA length dependence. Biochim Biophys Acta. 1990;1087:55–60. doi: 10.1016/0167-4781(90)90120-q. [DOI] [PubMed] [Google Scholar]
- 49.Coleman RA, Pugh BF. Evidence for functional binding and stable sliding of the TATA binding protein on nonspecific DNA. J Biol Chem. 1995;270:13850–9. doi: 10.1074/jbc.270.23.13850. [DOI] [PubMed] [Google Scholar]
- 50.Hertel KJ, Lynch KW, Maniatis T. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol. 1997;9:350–7. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- 51.Somma MP, Pisano C, Lavia P. The housekeeping promoter from the mouse CpG island HTF9 contains multiple protein-binding elements that are functionally redundant. Nucleic Acids Res. 1991;19:2817–24. doi: 10.1093/nar/19.11.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papatsenko DA, et al. Extraction of functional binding sites from unique regulatory regions: the Drosophila early developmental enhancers. Genome Res. 2002;12:470–81. doi: 10.1101/gr.212502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He X, Hohn T, Futterer J. Transcriptional activation of the rice tungro bacilliform virus gene is critically dependent on an activator element located immediately upstream of the TATA box. J Biol Chem. 2000;275:11799–808. doi: 10.1074/jbc.275.16.11799. [DOI] [PubMed] [Google Scholar]
- 54.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 55.Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011;12:123–35. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- 56.Gencheva M, et al. In Vitro and in Vivo nucleosome positioning on the ovine beta-lactoglobulin gene are related. J Mol Biol. 2006;361:216–30. doi: 10.1016/j.jmb.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 57.Thastrom A, Bingham LM, Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J Mol Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 58.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–50. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 59.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191:659–75. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 60.Field Y, et al. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985;82:8419–23. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–9. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raveh-Sadka T, et al. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat Genet. 2012;44:743–50. doi: 10.1038/ng.2305. [DOI] [PubMed] [Google Scholar]
- 65.Valouev A, et al. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–20. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kelly TK, et al. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012;22:2497–506. doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wunderlich Z, Mirny LA. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 2009;25:434–40. doi: 10.1016/j.tig.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pirrotta V, Gross DS. Epigenetic silencing mechanisms in budding yeast and fruit fly: different paths, same destinations. Mol Cell. 2005;18:395–8. doi: 10.1016/j.molcel.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 69.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]