Abstract
The alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors are important glutamatergic receptors mediating fast excitatory synaptic transmission in the brain. The regulation of the four subunits of AMPA receptors, GluA1-4, is poorly understood. Excitatory synaptic transmission is highly energy-demanding, and this energy is derived mainly from the oxidative pathway. Recently, we found that specificity factor regulates all subunits of cytochrome c oxidase (COX), a critical energy-generating enzyme. COX is also regulated by nuclear respiratory factor 1 (NRF-1), which transcriptionally controls the Gria2 (GluA2) gene of AMPA receptors. The goal of the present study was to test our hypothesis that Sp-factors (Sp1, Sp3, and/or Sp4) also regulate AMPA subunit genes. If so, we wish to determine if Sp-factors and NRF-1 function via a complementary, concurrent and parallel, or a combination of complementary and concurrent/parallel mechanism. By means of multiple approaches, including electrophoretic mobility shift and supershift assays, chromatin immunoprecipitation, promoter mutations, real-time quantitative PCR, and western blot analysis, we found that Sp4, but not Sp1 or Sp3, regulates the Gria2, but not Gria 1, 3, or 4, subunit gene of the AMPA receptor in a concurrent and parallel manner with NRF-1. Thus, Sp4 and NRF-1 both mediate the tight coupling between neuronal activity and energy metabolism at the transcriptional level.
Keywords: AMPA receptor, gene regulation, GluA2, Sp4, specificity protein 4, transcription factor
1. Introduction
Glutamate is the major excitatory neurotransmitter in the brain, and the alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors are a major class of glutamatergic receptors (for review see [1]). AMPA receptors are heterotetramers composed of various combinations of the GluA1, GluA2, GluA3, and GluA4 subunits [2]. They are widely expressed in the brain, where they mediate the majority of ionotropic, ligand-gated, fast excitatory synaptic transmission [1]. As such, AMPA receptors play crucial roles in normal neuronal activity, including synaptic plasticity, synaptic scaling, homeostatic synaptic plasticity, learning and memory, as well as in synaptogenesis and the formation of neuronal circuitry [3-6]. Most AMPA receptors in the cerebral neocortex and hippocampus contain the GluA2 subunit, usually in combination with the GluA1 subunit [7, 8]. The GluA2 subunit is unique, as its inclusion in the heterotetramer renders AMPA receptors significantly less permeable to Ca2+ [9]. In the adult, GluA3 expression is much lower than those of GluA1 or GluA2 [7, 10]. The GluA4 subunit is present mainly during development [11].
We have shown previously that the expression of GluA2 is governed by neuronal activity [12, 13]. Excitatory glutamatergic neuronal activity is highly energy-demanding, with most of this energy utilized to extrude excess cations that enter through glutamatergic receptors (for reviews see [14]). Thus, at the cellular level, an intimate link exists between excitatory activity and the energy requirements of neurons. Recently, we found that this link extends to the molecular level, in that the same transcription factor, nuclear respiratory factor 1 (NRF-1), co-regulates genes that mediate energy metabolism and neuronal activity. Specifically, NRF-1 regulates the expression of all 13 subunits of the energy-generating enzyme, cytochrome c oxidase (COX), as well as the expression of the GluA2 subunit of the AMPA receptor [15, 16].
Specificity protein (Sp) is a family of zinc-finger transcription factors that binds to GC-rich DNA motifs (for review see [17]). Sp1, Sp3, and Sp4 are three members of the Sp family that compete for the same cis- motifs: the GC box ‘GGGCGG’ with high affinity, or the GT (‘GGGTGG’) and CT (‘CCCTCC’) boxes with significantly lower affinities [17, 18]. While the expressions of Sp1 and Sp3 are ubiquitous, that of Sp4 is restricted mainly to neurons and testicular cells [19]. Recently, we found that, like NRF-1, Sp factors also regulate all 13 subunits of COX [20]. The question arises as to whether Sp factors also couple energy metabolism to neuronal activity by regulating specific subunits of the AMPA receptor. If so, do they operate with NRF-1 via a complementary (regulating different subunits), concurrent and parallel (regulating the same subunit in the same direction), or a combination of complementary and concurrent/parallel mechanism? The goal of the present study was to test our hypothesis that Sp1, Sp3, and Sp4 also mediate the transcriptional coupling of synaptic transmission and energy metabolism.
2. Material and Methods
All experiments and animal procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health Publications No. 80-23, revised 1996), and all protocols were approved by the Medical College of Wisconsin Animal Care and Use Committee (approval can be provided upon request). All efforts were made to minimize the number of animals used and their suffering.
2.1. Cell Culture
The mouse neuroblastoma (N2a) cell line was acquired from ATCC (Manassas, VA, USA) and grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (50 units/mL), and streptomycin (100 μg/mL) at 37°C with 5% CO2.
Cultures of murine or rat primary visual cortical neurons were carried out as described previously [21]. Briefly, neonatal mouse or rat pups (1-day-old) were killed by decapitation. Visual cortical tissue was removed, trypsinized, and triturated. Individual neurons were then plated in poly-L-lysine-coated, 35 mm dishes at a density of 200,000 cells/dish and maintained in Neurobasal-A media supplemented with B27. To suppress the proliferation of glial cells, Ara-C (Sigma, St Louis, MO, USA) was added.
2.2. In silico analysis of promoters of murine AMPA receptor subunit genes
DNA sequences surrounding the transcription start points (TSPs) of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunit genes (Gria1-4) were derived from the NCBI mouse genome database (Gria1 GenBank ID: NC_000077.6, Gria2 GenBank ID: NC_000069.6, Gria3 GenBank ID: NC_000086.7, and Gria4 GenBank ID: NC_000075.6). Computer-assisted search for the typical Sp1 binding motif (‘GGGCGG’), the atypical Sp1 binding motifs (‘GGGTGG’ and ‘CCCTCC’), or their complements, was conducted on sequences encompassing 1 kb upstream and 1 kb downstream of the TSP of each gene. These motifs were applicable to Sp1, Sp3, or Sp4.
To determine the degree of conservation of the Sp binding motif among species, NCBI's Ensembl interface was used to align promoters of AMPA receptor subunits from mice, rats, and humans.
2.3. Electrophoretic mobility shift and supershift assays
To determine if Sp1, Sp3, or Sp4 bound in vitro to putative Sp binding elements in the promoter regions of AMPA receptor subunit genes, electrophoretic mobility shift assays (EMSA) were carried out as described previously with a few modifications [22]. Briefly, oligonucleotide probes containing putative Sp binding sequences on each AMPA receptor subunit promoter (identified from in silico analysis), were synthesized (Table 1), annealed, and labeled with [α-32P] dATP (50 μCi/200 ng; Perkin-Elmer, Shelton, CT, USA) using Klenow fragment (Invitrogen). Nuclear extract from mouse primary visual cortical tissue and HeLa cells were isolated as described previously [23, 24]. Ten μg of either mouse cortical and/or HeLa nuclear extract were incubated with 2 μg of calf thymus DNA and with each labeled EMSA probe. Supershift assays were performed with 1 μg of Sp4, 3, or 1 specific antibody (Sp4, V-20, SC645; Sp3, H-225, SC13018; Sp1, H-225, SC14027; all from Santa Cruz Biotechnology (SCBT), Santa Cruz, CA, USA) and incubated with the probe/nuclear extract mixture for 20 min. The Sp4, Sp3, and Sp1 antibodies were tested for specificity using western blot analysis and showed two adjacent bands at the appropriate molecular weights. These bands corresponded to the phosphorylated and non-phosphorylated forms of Sp factors. For the cold competition experiment, nuclear extracts were incubated with 100-fold excess of unlabeled oligonucleotides. All reactions were loaded onto 4.5% polyacrylamide gel (58:1, Acrylamide:Bisacrylamide) and run for 3 h at 200 V in 0.25× Tris-borate-EDTA buffer. Results were visualized on a phosphoimager and exposed on film. The positive control probe, murine GM3 Synthase gene, is known to bind Sp1 and contains two Sp1 binding sites in a tandem repeat [25]. The negative controls were AMPA receptor subunit probes with mutated Sp binding sequences (Table 1).
Table 1.
EMSA Probes. Positions of probes are given relative to TSP. Putative Sp binding sites are underlined.
| Gene Promoter | Position | EMSA Sequence |
|---|---|---|
| Gria1 | -189/-172 | F: 5′ TTTTTGGAGAGGCAGGGGTGAT 3′ |
| R: 5′ TTTTATCACCCCTGCCTCTCCA 3′ | ||
| Gria2 | -89/-64 | F: 5′ TTTTGCGCTGTGCGGGGGAGGGGTGGGTGC 3′ |
| R: 5′ TTTTGCACCCACCCCTCCCCCGCACAGCGC 3′ | ||
| Gria3 | -87/-69 | F: 5′ TTTTCAATGAACCCGCCTTCCAG 3′ |
| R: 5′ TTTTCTGGAAGGCGGGTTCATTG 3′ | ||
| Gria4 | +507/+527 | F: 5′ TTTTGTAGCCCGGGCGGGGGTCGGC 3′ |
| R: 5′ TTTTGCCGACCCCCGCCCGGGCTAC 3′ | ||
| GM3 Synthase | -58/-38 | F: 5′ TTTTGCGCGACCCCGCCCCCGCCTA 3′ |
| R: 5′ TTTTTAGGCGGGGGCGGGGTCGCGC 3′ | ||
| Mutant Sp Gria2 | -89/-64 | F: 5′ TTTTGCGCTGTGTTTTTTTTTTTTGTGTGC 3′ |
| R: 5′ TTTTGCACACAAAAAAAAAAAACACAGCGC 3′ |
2.4. Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed on murine visual cortical tissue as described previously [24]. Briefly, murine visual cortical tissue was quickly removed, finely chopped, fixed in 2% formaldehyde, and resuspended in swelling buffer (85 mM KCl, 5 mM PIPES, pH 8.0, 1% Nonidet P-40, and protease inhibitors). The tissue was homogenized in a Dounce tissue homogenizer and centrifuged to isolate the nuclei. The nuclei were resuspended and sonicated in SDS lysis buffer (1% SDS, 50 mM Tris-HCl, pH 8.1, 10 mM EDTA). Immunoprecipitation was performed with 2 μg of Sp1, Sp3, or Sp4 antibodies. Two μg of anti-nerve growth factor receptor (NGFR) antibodies (sc-6188, SCBT) or ‘no antibody’ blanks were used as negative controls. Semi-quantitative PCR, utilizing primers (Table 2) surrounding the putative Sp binding sites identified through in silico analysis, was performed. Positive controls for Sp factor binding were GM3 Synthase (known to bind Sp1) and neurotrophin 3 (known to bind Sp4) [25, 26]. β-actin promoter (Actb) was the negative control (Table 2). PCR additives and cycling parameters used to improve the reproducibility and quality of ChIP are listed in Table 2. 2% agarose gels stained with ethidium bromide were used to visualize PCR products.
Table 2.
Primers and conditions used for ChIP analysis.
| Gene Promoter | Sequence | Amplicon Length | Cycling Conditions |
|---|---|---|---|
| Gria1 | F: 5′ GGGAGGTGAAGGGGACAGTGGG 3′ | 170 bp | 94°: 30s, 57°: 30s, 72°: 45s; 2.5% DMSO |
| R: 5′ CCCAGGAGCCAGCTCGGAGT 3′ | |||
| Gria2 | F: 5′ ATCGAATGCGCAAGACTGGA 3′ | 201 bp | 94°: 30s, 60°: 30s, 72°: 45s; 2.5% DMSO |
| R: 5′ GAAAGTCACAGAGAGGGGCA 3′ | |||
| Gria3 | F: 5′ CAGAAATCGCTTTGGGGAACC 3′ | 71 bp | 94°: 30s, 60°: 30s, 72°: 45s; 2.5% DMSO |
| R: 5′ ACTTCTGTCTTCACACCAATCTG 3′ | |||
| Gria4 | F: 5′ AGCCTCCCTGAAGACCTGGGC 3′ | 231 bp | 94°: 30s, 60°: 30s, 72°: 45s; 2.5% DMSO |
| R: 5′ GGCTCCCTTGCGCTCCCTTG 3′ | |||
| GM3 Synthase | F: 5′ CACCTACTTCTCGGCTGGAG 3′ | 198 bp | 94°: 30s, 57°: 30s, 72°: 45s. |
| R: 5′ AATTCAGCCCCGGACAGT 3′ | |||
| NT3 | F: 5′ GAGCAAACTCCAAAATGCCAGG 3′ | 219 bp | 94°: 30s, 57°: 30s, 72°: 45s |
| R: 5′ AAAGTTGCGCCGGGCTATCTC 3′ | |||
| β-Actin | F: 5′ GCTCTTTTCCAGCCTTCCTT 3′ | 187 bp | 94°: 30s, 57°: 30s, 72°: 45s. |
| R: 5′ CGGATGTCAACGTCACACTT 3′ |
2.5. Construction of luciferase reporter vectors and transfection in N2a cells
The Gria2 luciferase reporter construct was created by PCR amplification of the mouse Gria2 proximal promoter (primers listed in Table 3). The PCR product was digested with restriction enzymes (MluI and BgIII), and ligated into Promega's pGL3 luciferase vectors (E1751, Promega, Madison, WI, USA). The QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA, USA) was used to mutate putative Sp binding sites. Primers used to create Sp mutations are listed in Table 3. Sequencing was done to verify all constructs.
Table 3.
Primers used for promoter cloning and mutagenesis analysis. Mutated Sp binding sites are underlined.
| Gene Promoter | Position | Primer |
|---|---|---|
| Gria2 | -947/+152 | F: 5′ CAGACGCGTCCCAAGCAGGCTCGGTGTAATGA 3′ |
| R: 5′ CAGAGATCTGCTGTGGTCCCGGTGTCTGG 3′ | ||
| Mutant Sp Gria2 | -90/-52 | F: 5′ GGCGCTGTGCGTGTGAGTGTTGTGTGCGCGAGCTCGCCG 3′ |
| R: 5′ CGGCGAGCTCGCGCACACAACACTCACACGCACAGCGCC 3′ |
N2a cells, plated at 60% confluency in 24-well plates, were transfected with either 0.6 μg of the Gria2 reporter construct or 0.6 μg of the Gria2 reporter construct with mutated Sp binding site. pRL-TK (0.06 μg), a vector with a thymidine kinase promoter that constitutively expresses renilla luciferase, was added to each well and served as an internal control for the assay. KCl stimulation (at a final concentration of 20 mM) was performed for 5 h as described previously [27]. Cells were harvested 48 h after transfection and the lysates were measured for luciferase activity. The average from six independent transfections for each promoter construct is reported.
2.6. Plasmid construction of Sp1, Sp3, and Sp4 shRNA, transfection, and KCl treatment
Sp1 silencing was carried out using a combination of three to five Sp1-specific hairpin RNA (shRNA) (sc-29488, SCBT). Specific shRNA sequences for silencing murine Sp3 or Sp4 were chosen from the RNAi Consortium's Public TRC Cloning Database at the Broad Institute (Table 4). The sequences were cloned into Addgene's pLKO.1 TRC cloning vector (Plasmid 10878, Addgene, Cambridge, MA, USA). The pLKO.1 non-mammalian shRNA control vector (SHC002, Sigma), which contains a scrambled shRNA sequence that targets no known mammalian genes, was used as the control.
Table 4.
Sp3 and Sp4 shRNA sequences. shRNA sequences cloned into the pLKO.1 vector for silencing of Sp3 and Sp4
| Gene | Hairpin Sequence | |
|---|---|---|
| Sp3 | 1 | 5′ CCGG—ATGAGAAACTGTTGGTATTTA—CTCGAG—TAAATACCAACAGTTTCTCAT—TTTTTG 3′ |
| 5′ AATTCAAAAA—ATGAGAAACTGTTGGTATTTA—CTCGAG—TAAATACCAACAGTTTCTCAT 3′ | ||
| 2 | 5′ CCGG—TTACCTTTGTACCAATCAATA—CTCGAG—TATTGATTGGTACAAAGGTAA—TTTTTG 3′ | |
| 5′ AATTCAAAAA—TTACCTTTGTACCAATCAATA—CTCGAG—TATTGATTGGTACAAAGGTAA 3′ | ||
| Sp4 | 1 | 5′ CCGG—CCAGTAACAATCACTAGTGTT—CTCGAG—AACACTAGTGATTGTTACTGG—TTTTTG 3′ |
| 5′ AATTCAAAAA—CCAGTAACAATCACTAGTGTT—CTCGAG—AACACTAGTGATTGTTACTGG 3′ | ||
| 2 | 5′ CCGG—CTGGACAACAGCAGATTATTA—CTCGAG—TAATAATCTGCTGTTGTCCAG—TTTTTG 3′ | |
| 5′ AATTCAAAAA—CTGGACAACAGCAGATTATTA—CTCGAG—TAATAATCTGCTGTTGTCCAG 3′ |
N2a cells at 60% confluence in 6-well dishes were transfected the day after plating with either the pLKO.1 non-mammalian control vector (3 μg), or the Sp1, Sp3, or Sp4 shRNA vector constructs (3 μg). TurboGFP (1 μg) vector was added to all wells to monitor transfection and selection efficiency. Transfection was performed using 5 μl of JetPrime transfection reagent (PolyPlus Transfection, Illkirch, France) per well. To select for purely transfected cells, 5 μg/mL of puromycin was added 1.5 days after transfection. Transfection efficiency was around 75% but the addition of puromycin effectively yielded 100% transfected cells. KCl stimulation was performed at a final concentration of 20 mM for 5 h as previously described [27].
Transfection of murine or rat primary neuronal cultures was carried out 5 days post-plating with Sp1, Sp3, or Sp4 shRNA constructs (2 μg) or the pLKO.1 non-mammalian control (2 μg) using Neurofect transfection reagent per 6-well plate according to the manufacturer's instructions (Genlantis, San Diego, CA). TurboGFP (0.5 μg) vector was added to each well for transfection visualization and selection efficiency. Transfection efficiency was around 40 – 50% before selection. Puromycin selection, however, effectively yielded 100% transfected cells. KCl stimulation was performed on rat visual cortical neurons as described for N2a cells above.
2.7. Sp1, Sp3, and Sp4 over-expression and TTX treatment
cDNA clones for human Sp1, Sp3, and Sp4 (Open Biosystems, Lafayette, CO, USA) were cloned into pcDNA Dest40 vectors (Gateway Multisite Cloning kit, Invitrogen) according to the manufacturer's instructions and as described previously [20].
Transfection procedure for N2a cells and primary neuronal culture was similar to that described above. Either 1.5 μg of Sp1, Sp3, or Sp4 over-expression vector, or 1 μg of the pcDNA3.1 empty vector and 0.5 μg of turboGFP vector were used for both N2a cells and primary neuronal cultures. Three days of TTX treatment (0.4 μM) commenced the day after plating as previously described [27]. Primary neuronal cultures and N2a cells with TTX were harvested 4 days after transfection, whereas those without TTX were harvested 2 days after transfection.
2.8. RNA isolation and cDNA synthesis
TRIZOL (Invitrogen) was used to isolate total RNA according to the manufacturer's instructions. DNase I treatment was done on 1 μg of total RNA, and cDNA was synthesized with iScript cDNA synthesis kit (170-8891, BioRad, Hercules, CA, USA).
2.9. Real-time quantitative PCR
The Cepheid Smart Cycler Detection system (Cepheid, Sunnyvale, CA, USA) and/or the iCycler System (BioRad) were used with the IQ SYBR Green SuperMix (170-8880, BioRad) to perform real-time quantitative PCR (RTqPCR). The sequences of primers used for analysis are listed in Table 5. Primer sequences were the same for mouse and rat. RTqPCR conditions were optimized to yield an amplification efficiency of 95% -105%. Products were run on agarose gel to verify the correct amplification length. Melt curve analyses of each PCR reaction was performed to verify the formation of a single desired PCR product. The 2-ΔΔCT method was used to quantify the relative amount of transcripts, with β-actin and Gapdh acting as the internal controls for N2a cells and primary neurons, respectively.
Table 5. Real-time qPCR Primers.
| Gene | Primer |
|---|---|
| Gria1 | F: 5′ GAGCAACGAAAGCCCTGTGA 3′ |
| R: 5′ CCCTTGGGTGTCGCAATG 3′ | |
| Gria2 | F: 5′ AAAGAATACCCTGGAGCACAC 3′ |
| R: 5′ CCAAACAATCTCCTGCATTTCC 3′ | |
| Gria3 | F: 5′ TTCGGAAGTCCAAGGGAAAGT 3′ |
| R: 5′ CACGGCTTTCTCTGCTCAATG 3′ | |
| Gria4 | F: 5′ GGCTCGTGTCCGCAAGTC 3′ |
| R: 5′ TTCGCTGCTCAATGTATTCATTC 3′ | |
| Sp1 | F: 5′ CTCCAGACCATTAACCTCAGTG 3′ |
| R: 5′ ATCATGTATTCCATCACCACCAG 3′ | |
| Sp3 | F: 5′ TCAGGCACAGACAGTGACCCCT 3′ |
| R: 5′ AGCGTGAGTGTCTGAACAGGCG 3′ | |
| Sp4 | F: 5′ TTGCAGCAAGGCCAGCAGACC 3′ |
| R: 5′ GCTTCTTCTTTCCTGGTTCACTGCT 3′ | |
| Actb | F: 5′ GTGACGTTGACATCCGTAAAGA 3′ |
| R: 5′ GCCGGACTCATCGTACTCC 3′ | |
| Gapdh | F: 5′ AGGTCGGTGTGAACGGATTTG 3′ |
| R: 5′ GGGGTCGTTGATGGCAACA 3′ |
2.10. Western blot analysis
Sample buffer (12.5 mM EDTA, 50 mM Tris-HCl pH 6.8, 1% SDS, 10% glycerol) was used to harvest control, shRNA, and over-expression protein samples. Equal protein amounts of each sample was loaded onto 10% SDS-PAGE gel and electrophoretically transferred onto polyvinylidene difluoride membranes. Membranes were blocked with 5% non-fat dry milk and incubated in primary antibodies against Sp1 (1:1000; Santa Cruz), Sp3 (1:1000; Santa Cruz), Sp4 (1:1000; Santa Cruz), GluA1 (1:500; Ab1504, Millipore Chemicon, Billerica, MA, USA), GluA2 (1:200; 75-002 clone L21/32, UC Davis/NIH NeuroMab Facility, Davis, CA, USA), GluA3 (1:50; sc-7613, Santa Cruz) and GluA4 (1:50; sc-7614, Santa Cruz). The loading control was β-actin (1:5000; Sigma). Secondary antibodies were from Vector Laboratories (Burlingame, CA, USA) and ECL reagent was used to visualize protein position and intensity. Gel Doc from BioRad (Hercules, CA, USA) was used to perform quantitative analyses of relative changes.
2.11. Statistical analysis
Analysis of variance (ANOVA) was applied to determine significance among group means. The Student's t-test was then applied to determine significance between two groups. P-values of 0.05 or less were considered significant.
3. Results
3.1. In silico promoter analysis of AMPA receptor subunit genes
Computer assisted search for the typical Sp binding motif, the GC-box (GGGCGG), in the DNA sequence 1 kb upstream and 1 kb downstream of the transcription start site (TSP) of the AMPA receptor subunit genes revealed the motif on the Gria3 and Gria4, but not the Gria1 or Gria2 promoter region. The Gria2 promoter contained the atypical Sp binding motif, the GT-box (GGGTGG). The Gria1 promoter contained GC rich regions, in which sequences resembling the Sp binding motif were present (see Table 1 for binding motifs).
3.2. In vitro binding of Sp proteins to AMPA receptor subunit promoters
The ability of Sp1, Sp3, and Sp4 to bind candidate sites in vitro was determined by electrophoretic mobility shift (EMSA) and supershift assays. The positive control for Sp binding was the GM3 synthase promoter [25], which contained two Sp1 binding sites (2 GC-boxes) in tandem. When incubated with HeLa nuclear extract, GM3 synthase formed specific DNA/Sp1, DNA/Sp3, and DNA/Sp4 shift and super shift complexes (Figure 1, lane 6 for Sp1/Sp3/Sp4 shift, lanes 8, 9, and 10 for Sp1, Sp3, and Sp4 supershifts, respectively). As Sp1 is capable of homotypic interactions that lead to multimeric complexes [17], the Sp1 supershift reaction may be detecting two Sp1 proteins bound to the tandem GC-boxes (Figure 1, lane 8, Sp1 Supershift*). When incubated with mouse visual cortical tissue nuclear extract, GM3 synthase formed specific DNA/Sp3 and DNA/Sp4 shift and supershift complexes (Figure 1, lane 1 for Sp3/Sp4 shift, lanes 4 and 5 for Sp3 and Sp4 supershift, respectively) but did not form a DNA/Sp1 shift or supershift complex (Figure 1, lanes 1 and 3 respectively). The lack of a strong Sp4 supershift with HeLa nuclear extract as compared to mouse visual cortical nuclear extract is consistent with our previous observations of different levels of Sp4 abundance in the two nuclear extracts and with its reported neuronal distribution [19, 28]. Similarly, the lack of Sp1 shift or supershift with mouse cortical nuclear extract as compared to HeLa nuclear extract was due to the lower abundance of Sp1 in mouse cortical nuclear extract as compared to HeLa nuclear extract [28]. The shift bands for visual cortical and HeLa nuclear extract were competed out with the addition of cold competitors (Figure 1, lanes 2 and 7, respectively).
Figure 1.
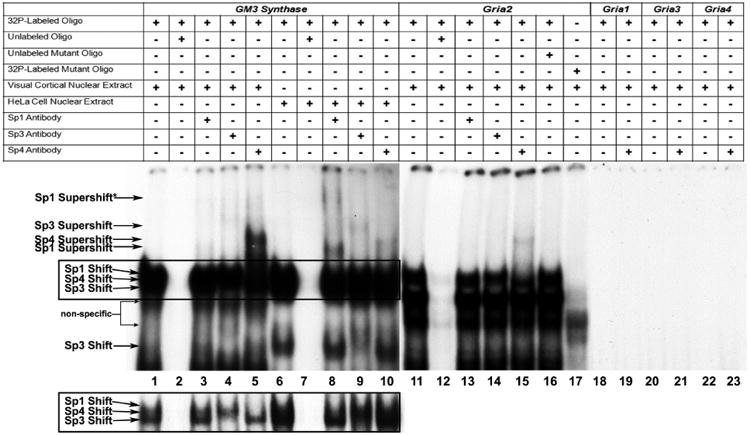
In vitro binding of Sp factors to the GM3 Synthase and Gria2 promoters using EMSA and supershift assays. All lanes contain 32P-labeled oligonucleotides and are labeled by a “+” or a “−“sign depending on whether they also contain mouse visual cortical or HeLa nuclear extract, excess oligos or mutant oligos (unlabeled), and Sp1, Sp3, or Sp4 antibodies. Sp1, Sp3, or Sp4 shift, supershift, and non-specific complexes are indicated by arrows. The positive control for Sp factor binding was GM3 Synthase. Specific Sp1, 3, or 4 shift bands were revealed upon incubation with cortical (lanes 1-5) or HeLa (lanes 5-10) nuclear extract (lanes 1 and 6, respectively). Excess unlabeled competitor competed the shift bands (lanes 2 and 7). The addition of Sp1 antibody did not yield a supershift band with cortical nuclear extract (lane 3) but gave two specific supershift bands with HeLa nuclear extract (lane 8), corresponding to the presence of tandem Sp binding. The addition of Sp3 antibody yielded a faint specific supershift band with cortical nuclear extract (lane 4) and a stronger supershift band with HeLa nuclear extract (lane 9). The addition of Sp4 antibody yielded a very strong supershift band with cortical nuclear extract (lane 5) and a much fainter supershift band with HeLa nuclear extract (lane 10). The relative levels of the Sp1, Sp3, and Sp4 supershift bands in mouse cortical and HeLa nuclear extract corresponds to the relative levels of these transcription factors in these tissue and cell types.
As Sp1, 3, and 4 all recognize the same cis motif, their combined binding yielded a rather thick shift band. However, with shorter gel exposure (boxed lanes 1 to 10 highlighted in the lower left), the individual shift bands can sometimes be dissociated. As shown in lane 1 in the inset, cortical nuclear extract did not reveal any detectable Sp1 shift band, but did show distinct Sp3 and Sp4 bands. The Sp3 shift band disappeared when it was supershifted (lane 4). Likewise, the Sp4 shift band disappeared when it was supershifted (lane 5). With HeLa nuclear extract, the rather abundant presence of both Sp1 and Sp3 caused the bands to merge even with lighter exposure (lane 6). However, the upper portion (Sp1) disappeared when supershifted with Sp1 antibody (lane 8), and the lower portion (Sp3) became lighter when supershifted with Sp3 antibody (lane 9). No change was detectable with anti-Sp4 antibody (lane 10), indicating that HeLa nuclear extract did not contain detectable Sp4.
Incubation of cortical nuclear extract with the Gria2 probe yielded a specific Sp4 shift band (lane 11) which was competed out with the addition of excess cold probes (lane 12). The addition of Sp4 anitbody yielded a specific supershift band (lane 15), but the addition of Sp1 and Sp3 antibodies did not (lanes 13 and 14). The addition of excess unlabeled mutant Gria2 probes did not compete out the shift reaction (lane 16). Labeled Gria2 probes with mutant Sp sites did not yield specific Sp shift bands (lane 17). Labeled Gria1, Gria3, and Gria4 probes did not yield specific Sp shift (lanes 18, 20, and 22, respectively) or Sp4 supershift bands (lanes 19, 21, and 23, respectively).
When mouse visual cortical nuclear extract was incubated with AMPA receptor subunit probes containing putative Sp sites, Gria2 gave positive shift and supershift bands against Sp factors. Specifically, a shift band was observed (Figure 1, lane 11) that was competed out with the addition of unlabeled competitors (Figure 1, lane 12). A supershift band against Sp4 was present (Figure 1, lane 15) but there were no supershift bands against Sp1 or Sp3 (Figure 1, lanes 13 and 14, respectively). The addition of unlabeled probe with mutated Sp binding site did not compete out the Sp4 shift band (Figure 1, lane 16) and shift reactions with mutated Sp site did not reveal Sp binding (Figure 1, lane 17). Gria1, Gria3, and Gria4 did not give positive shift (Figure 1, lanes 18, 20, and 22, respectively) nor Sp4 supershift bands (Figure 1, lanes 19, 21, and 23, respectively).
3.3. In vivo interaction of Sp factors with AMPA receptor subunit genes in murine visual cortex
Interactions of Sp factors with AMPA receptor subunit gene promoters were verified in vivo by chromatin immunoprecipitation (ChIP) assays of mouse visual cortical nuclear extract. As seen in Figure 2, PCR products from all 0.5% and 0.1% input DNA reactions revealed specific bands. There were enriched bands for GM3 Synthase and Neurotrophin 3 positive controls in the Sp4 immunoprecipitated samples. An enriched band was present for Gria2, but not for the β-actin negative control nor for the Gria1, Gria3, and Gria4 promoters. No enrichment was found for samples immunoprecipitated with NGFR nor in the “no antibody” negative controls. There was also no enrichment of DNA in the Sp1 or Sp3 immunoprecipitated samples for any of the tested regions (data not shown).
Figure 2.
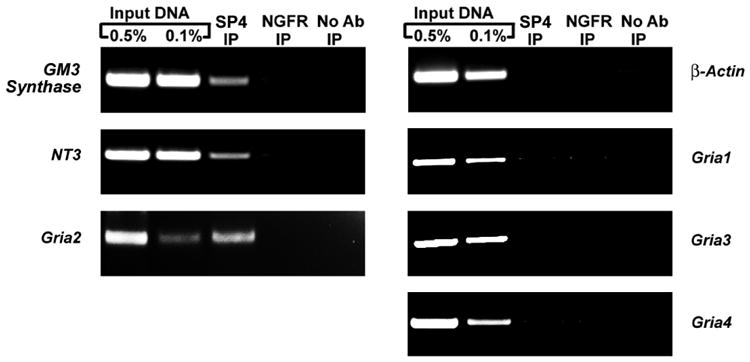
Sp4 interactions with AMPA receptor subunit promoters in mouse visual cortical tissue. Precipitation of chromatin was carried out from mouse visual cortical tissue nuclear extract with anti Sp4 antibodies (Sp4 IP lane), anti-nerve growth factor receptor p75 antibody (negative control, NGFR IP lane), or no antibody (negative control, No Ab lane). 0.5% (input 0.5% IP lane) and 0.1% (input 0.1% IP lane) of input chromatin served as control reactions for PCR. Positive controls for Sp4 binding were GM3 synthase and Neurotrophin 3, whereas p-Actin was the negative control. Sp4 interacted with Gria2, but not with Gria1, Gria3, or Gria4.
3.4. Effect of mutated Sp binding site on Gria2 promoter
The wild type Gria2 promoter as well as the Gria2 promoter with mutated Sp binding site were transfected separately into N2a cells. Analysis revealed a significant 56% decrease in the activity of Gria2 promoter containing the mutated Sp motif as compared to the wild type control (P < 0.001, Figure 3).
Figure 3.

Relative luciferase activity of the wild type Gria2 promoter (wt) and the Gria2 promoter with mutated Sp binding site (mut). Mutating the Sp binding site on Gria2 resulted in a significant decrease in luciferase activity as compared to the wild type promoter. KCl depolarization significantly increased Gria2 wild type promoter activity, but could not increase activity in the Gria2 promoter with mutated Sp site. N = 6 for each construct. ***= P < 0.001; X = NS. Both mutant and wild type + KCl were compared to the wild type. The mutant + KCl was compared to the mutant.
3.5. Effect of mutated Sp binding sites on the response of the Gria2 promoter to KCl stimulation
We have shown previously that the KCl-mediated increase in neuronal activity up-regulates the Gria2 promoter. To determine if Sp factor binding is necessary for this activity-mediated up-regulation, Gria2 control promoters or Gria2 promoters with mutated Sp site were transfected into N2a cells and subjected to depolarizing KCl (20 mM) treatment. As shown in figure 3, N2a cells transfected with the control Gria2 promoter and subjected to KCl stimulation showed a 47% increase in promoter activity (P < 0.001). This increase was abolished by mutating the Sp binding site (Figure 3), confirming a requirement for Sp binding in the activity-induced up-regulation of the Gria2 promoter.
3.6. Effect of silencing Sp1, Sp3, and Sp4 by RNA interference on AMPA receptor subunits in N2a cells
To determine whether a knockdown of Sp1, Sp3, or Sp4 had an effect on the expression of AMPA receptor subunits, respective Sp proteins were silenced in N2a cells by means of 2 - 5 specific plasmid shRNA vectors. Quantitative real-time PCR indicated that silencing Sp4 decreased Sp4 mRNA levels by 68% (P < 0.001, Figure 4A) and Sp4 protein levels by 56% (P < 0.01, Figure 4C-D). Sp4 silencing also decreased Gria2 mRNA levels by 62% (P < 0.001; Figure 4B) and protein levels by 41% (P < 0.05, Figure 4E-F). mRNA levels of Gria1, Gria3, and Gria4, however, did not change significantly with Sp4 silencing (Figure 4B), nor did the protein level of GluA1 (Figure 4E-F), GluA3 or GluA4 (Supplementary Figure 1A-B).
Figure 4.
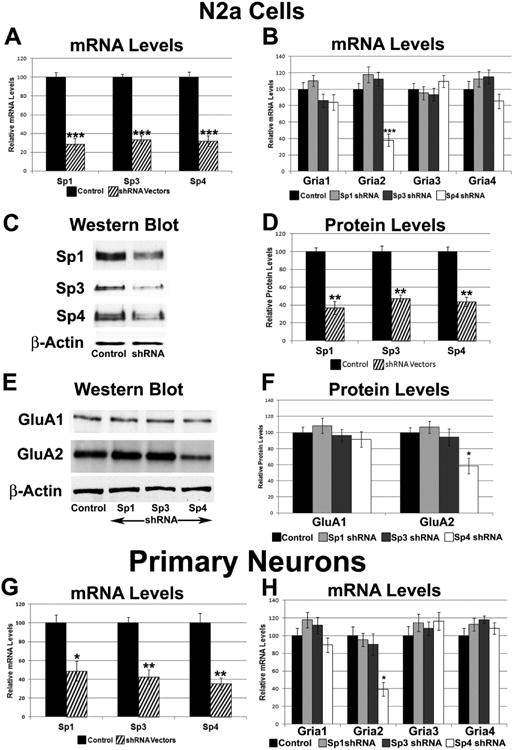
Effect of silencing of Sp1, Sp3, or Sp4 on the expression of the AMPA receptor subunit genes using RNA interference. (A) N2a cells transfected with Sp1, Sp3, or Sp4 shRNA showed significant down-regulation of Sp1, Sp3, and Sp4 transcripts, respectively. N = 6. (B) In N2a cells, Gria2 transcript levels were decreased with silencing of Sp4, but not with silencing of either Sp3 or Sp1. N = 6. (C-D) In N2a cells, protein levels of Sp1, Sp3, and Sp4 decreased significantly with shRNA against Sp1, Sp3, and Sp4, respectively. A representative western blot for β-actin, the loading control, is shown. N = 3. (E-F) Sp4 silencing reduced the protein levels of GluA2 but not that of GluA1. Protein levels of GluA1 and GluA2 did not change significantly with Sp1 or Sp3 silencing. The loading control was β-actin. N = 3. (G) shRNA against Sp1, Sp3, or Sp4 showed a significant down-regulation in the transcript levels of Sp1, Sp3, and Sp4, respectively. N = 3. (H) Transcript levels of Gria2, but not Gria1, Gria3, and Gria4, decreased with Sp4 shRNA in primary neurons. Transcript levels of Gria1-4 did not change significantly with Sp1 or Sp3 shRNA in primary neurons. N = 3. *= P < 0.05, **= P < 0.01, and ***= P < 0.001 when compared to controls.
Silencing of Sp1 resulted in a 71% decrease in its mRNA levels (P < 0.001, Figure 4A) and a 63% decrease in its protein levels (P < 0.01, Figure 4C-D). Silencing of Sp3 resulted in a 66% decrease in its mRNA levels (P < 0.001, Figure 4A) and a 52% decrease in its protein levels (P < 0.01, Figure 4C-D). However, mRNA levels of Gria1, Gria3, and Gria4 (Figure 4B), or protein levels of GluA1 and GluA2 (Figure 4E-F), GluA3 and GluA4 (Supplementary Figure 1A-B) were not changed significantly with Sp1 or Sp3 silencing.
3.7. Effect of silencing Sp1, Sp3, and Sp4 by RNA interference on AMPA receptor subunits in primary neurons
To determine whether the lack of an effect seen with silencing Sp1 and Sp3 was specific to N2a cells, Sp1, Sp3, and Sp4 were each silenced in cultured primary mouse cortical neurons. Gapdh served as the internal control. Sp4 silencing resulted in a 64% decrease in its mRNA levels (P < 0.01, Figure 4G). Gria2's mRNA levels were also decreased significantly by 61% (P < 0.05; Figure 4H). However, Gria1, Gria3, and Gria4 transcripts were not changed significantly with Sp4 silencing (Figure 4H).
Silencing of Sp1 decreased its mRNA levels by 51% (P < 0.01, Figure 4G) and silencing of Sp3 decreased its mRNA levels by 57% (P < 0.01, Figure 4G). However, mRNA levels of Gria1-4 were not changed significantly by the silencing of either Sp1 or Sp3 (Figure 4H).
3.8. Effect of over-expressing Sp1, Sp3, and Sp4 on AMPA receptor subunits in N2a cells
To determine whether over-expressing Sp1, Sp3, or Sp4 had an effect on the expression of the AMPA receptor subunits, plasmids expressing Sp1, Sp3, or Sp4 were transfected into N2a cells. β-actin served as the internal control. Sp4 over-expression increased its mRNA levels approximately 30-fold (P < 0.001, Figure 5A) and its protein levels by 132% (P < 0.01, Figure 5C-D). The mRNA and protein levels of GluA2 were also increased by 135% and 47%, respectively (P < 0.001, Figure 5B and P < 0.05, Figure 5E-F, respectively). On the other hand, mRNA levels of Gria1, Gria3, and Gria4 did not change significantly with Sp4 over-expression (Figure 5B), and neither did the protein level of GluA1 (Figure 5E-F), nor of GluA3 and GluA4 (Supplementary Figure 1C-D).
Figure 5.
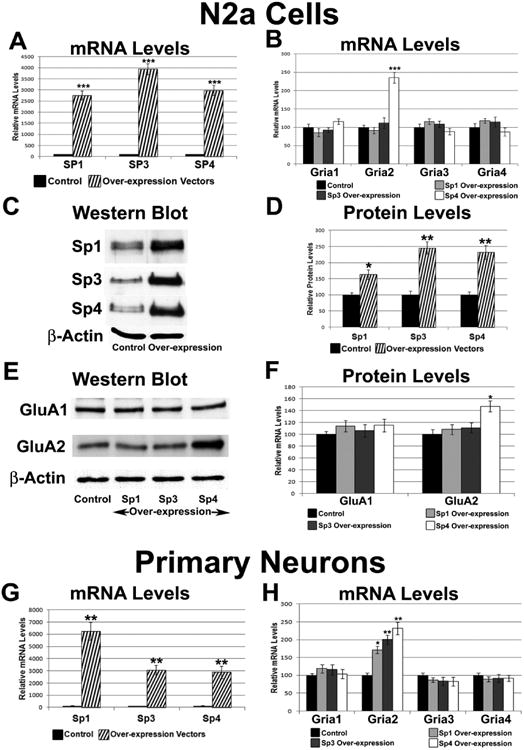
Effect of over-expressing Sp1, Sp3, or Sp4 on the transcript levels of AMPA receptor subunit genes. (A) N2a cells transfected with Sp1, Sp3, and Sp4 expression vectors revealed an up-regulation of Sp1, Sp3, and Sp4 transcripts, respectively. N = 6. (B) Sp4 over-expression, but not Sp1 or Sp3 over-expression, increased Gria2 transcript levels. N = 6. (C-D) In N2a cells, protein levels of Sp1, Sp3, and Sp4 were increased significantly with over-expression of Sp1, Sp3, and Sp4, respectively. A representative blot of the loading control, β-actin, is shown. N = 3. (E-F) In N2a cells, the protein level of GluA2, but not that of GluA1, was increased significantly with over-expression of Sp4. Protein levels of GluA1 and GluA2 did not change significantly with over-expressing Sp1 or Sp3. The loading control was β-actin. N = 3. (G) Over-expression of Sp1, Sp3, or Sp4 in primary neurons increased transcript levels of Sp1, Sp3, and Sp4, respectively. N = 3. (H) Sp4, Sp3, and Sp1 over-expression increased Gria2 transcript levels, but not Gria1, Gria3 and Gria4 transcript levels, in primary neurons. N = 3. *= P < 0.05, **= P < 0.01, and ***= P < 0.001 when compared to controls.
Sp1 over-expression led to a significant 27-fold increase in its mRNA levels (P < 0.001, Figure 5A) and a significant 63% increase in its protein levels (P < 0.01, Figure 5C-D). Sp3 over-expression led to a significant 39-fold increase in its mRNA levels (P < 0.001, Figure 5A) and a significant 146% increase in its protein levels (P < 0.05, Figure 5C-D). However, mRNA levels of Gria1-4 (Figure 5B) or protein levels of GluA1 and GluA2 (Figure 5E-F), or of GluA3 and GluA4 (Supplementary Figure 1C-D) did not change significantly with over-expression of Sp1 or Sp3.
3.9. Effect of over-expressing Sp1, Sp3, and Sp4 on AMPA receptor subunits in primary neurons
Cultured mouse cortical neurons were transfected with Sp1, Sp3, or Sp4 expression vectors to determine whether the lack of an effect seen with over-expression of Sp1 and Sp3 was restricted to N2a cells. Gapdh served as the internal control. Sp4 over-expression resulted in a 29-fold increase in its mRNA levels (P < 0.01, Figure 5G). Gria2 mRNA levels also increased significantly by 131% with Sp4 over-expression (P < 0.01; Figure 5H). However, Gria1, Gria3, and Gria4 mRNA levels were not changed significantly (Figure 5H).
Over-expression of Sp1 resulted in a significant 63-fold increase in its mRNA levels (P < 0.01, Figure 5G) and over-expression of Sp3 resulted in significant 31-fold increase in its mRNA levels (P < 0.01, Figure 5G). Gria2 mRNA levels increased significantly with Sp1 and Sp3 over-expression (P < 0.05 and P < 0.01, respectively; Figure 5H), but those of Gria1, Gria3, or Gria4 did not (Figure 5H).
3.10. Silencing Sp4 abolished KCl-induced transcript up-regulation of Gria2 in N2a cells
Gria2 mRNA levels are up-regulated in response to neuronal activity induced by physiological concentrations of KCl [15]. To determine whether Sp1, Sp3, or Sp4 was necessary for this up-regulation, control N2a cells and cells transfected with the respective shRNAs against these proteins were subjected to 20 mM KCl for 5 h. While Gria2 transcript levels were significantly increased by 110% (P < 0.001) with KCl-induced depolarization, silencing of only Sp4 abolished this increase (Figure 6A). Silencing of Sp1 or Sp3 did not prevent the KCl-induced increase of Gria2 transcript (Figure 6A). KCl treatment increased transcript levels of Gria1, Gria3, and Gria4 significantly (P < 0.001; Figure 6A), but levels of these subunits were not significantly changed with silencing Sp1, Sp3, or Sp4 (Figure 6A).
Figure 6.

Effect of changes in neuronal activity on Gria1, 2, 3, and 4 transcript levels in the absence or presence of Sp1, Sp3, or Sp4 silencing or over-expression in N2a cells. (A) Transcript levels of all AMPA receptor subunit genes were increased by 5 h of 20 mM KCl treatment. KCl is a depolarizing agent that increases neuronal activity. Silencing Sp4 did not allow for transcript levels of Gria2 to increase with KCl, but had no effect on the KCl-induced increase in the transcript levels of Gria1, Gria3, and Gria4. Silencing Sp1 and Sp3 did not prevent the KCl-induced up-regulation of Gria1-4 transcript levels. N = 6. (B) Transcript levels of all AMPA receptor subunit genes were decreased with 3 days of 0.4 μM TTX treatment. TTX decreases neuronal activity by blocking action potentials. Sp4 over-expression rescued the TTX-mediated down-regulation of Gria2 transcript levels, but did not rescue the TTX-mediated down-regulation of the Gria1, Gria3, and Gria4 transcripts. Sp1 or Sp3 over-expression could not rescue the TTX-mediated down-regulation of Gria1-4 transcript levels. N = 6. *= P < 0.05, **= P < 0.01, and ***= P < 0.001 when compared to controls. ### = P < 0.001 and X = non-significant when compared to KCl or TTX treatment alone.
3.11. Over-expression of Sp4 rescued tetrodotoxin-induced transcript reduction of Gria2 in N2a cells
Gria2 mRNA levels are known to be down-regulated in response to a decrease in neuronal activity induced by the action potential blocker TTX [15, 29]. To determine if Sp factors could rescue the decrease in AMPA transcript levels induced by the TTX-mediated decrease in neuronal activity, control N2a cells and cells transfected with Sp1, Sp3, or Sp4 over-expression vectors were subjected to 0.4 μM TTX-treatment for 3 days. TTX significantly decreased Gria2 transcript levels by 44% (P < 0.01; Figure 6B) but the decrease was abolished by Sp4 over-expression (P < 0.001 as compared to TTX alone; Figure 6B). Gria1, Gria3, and Gria4 transcript levels decreased significantly with TTX treatment (P < 0.01, P < 0.001, and P < 0.001, respectively; Figure 6B), but they were not rescued by Sp4 over-expression (Figure 6B). Over-expressing Sp1 or Sp3 could not rescue the TTX-mediated decrease in Gria1-4 transcript levels (Figure 6B).
3.12. Silencing Sp4 abolished KCl-induced transcript up-regulation of Gria2 in primary neurons
Transcript levels of Sp4, Gria1, Gria2, Gria3, and Gria4 in rat visual cortical neurons were all significantly up-regulated by 20 mM KCl for 5 h (P < 0.05 - 0.01; Fig. 7A). When neurons were transfected with Sp4 shRNA, transcript levels of Sp4 and Gria2 were significantly down-regulated to 44% and 45%, respectively (P < 0.01 and P < 0.05, respectively), whereas those of Gria1, Gria3, and Gria4 were not affected (Fig. 7A). Likewise, silencing Sp4 abolished the increase (Figure 7A) in transcript levels of Sp4 and Gria2 induced by KCl depolarization (P < 0.01), but did not affect those of Gria1, Gria3, and Gria4 (Fig. 7A).
Figure 7.

Effect of changes in neuronal activity on Gria1, 2, 3, and 4 transcript levels in the absence or presence of Sp4 silencing or over-expression in rat visual cortical neurons. (A) Transcript levels of Sp4 and all AMPA receptor subunit genes were increased by 5 h of 20 mM KCl treatment. Silencing Sp4 reduced its own transcript levels as well as that of Gria2, but not those of Gria1, 3, or 4. Sp4 silencing also did not allow for transcript levels of Sp4 itself and Gria2 to increase with KCl, but it had no effect on KCl-induced up-regulation of Gria1, Gria3, and Gria4 transcripts. N = 3. (B) Transcript levels of Sp4 and all AMPA receptor subunit genes were decreased with 3 days of 0.4 μM TTX treatment. Sp4 over-expression significantly increased its own transcript levels as well as that of Gria2, but not those of Gria1, 3, or 4. Sp4 over-expression also rescued the TTX-mediated down-regulation of Gria2 transcript levels, but not those of Gria1, Gria3, and Gria4 transcripts. N = 3. *= P < 0.05, **= P < 0.01, and ***= P < 0.001 when compared to controls. # = P < 0.05, ## = P < 0.01, ### = P < 0.001 and X = non-significant when compared to KCl or TTX treatment alone.
3.13. Over-expression of Sp4 rescued tetrodotoxin-induced transcript reduction of Gria2 in primary neurons
Transcript levels of Sp4, Gria1, Gria2, Gria3, and Gria4 in rat visual cortical neurons were all significantly down-regulated by 0.4 μM TTX-treatment for 3 days (P < 0.05; Fig. 7B). Over-expressing Sp4 significantly up-regulated Sp4 and Gria2 transcripts by 163% and 237%, respectively (P < 0.01 and P < 0.001, respectively), but did not affect those of Gria1, Gria3, and Gria4 (Fig. 7B). Sp4 over-expression also rescued Sp4 and Gria2 transcripts from being down-regulated by TTX (P < 0.05 and P < 0.001, respectively, as compared to TTX alone), but had no effect on those of Gria1, Gria3, and Gria4 (Fig. 7B).
3.14. Homology of the Sp Binding Site
The functional Gria2 Sp binding site is conserved among mice, rats, and humans (Figure 8).
Figure 8.
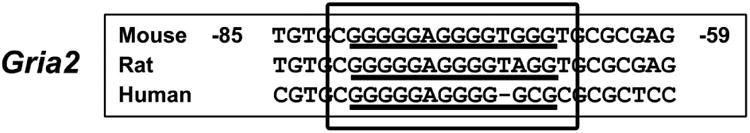
Aligned partial sequences of the Gria2 promoters from mice, rats, and humans showed conserved Sp binding sites.
4. Discussion
Using multiple molecular and biochemical approaches, including in silico analysis, electrophoretic mobility shift and supershift assays, ChIP, RNA interference and over-expression assays in both N2a cells and rodent primary cortical neurons, we document for the first time that it is Sp4, and not Sp1 or Sp3, transcription factor that regulates the GluA2 (Gria2), and not GluA1 (Gria1), GluA3 (Gria3), or GluA4 (Gria4), subunit of the AMPA receptor in neurons. Furthermore, a knockdown of Sp4 did not allow for the activity-mediated increase of Gria2, whereas over-expression of Sp4 rescued the TTX-mediated down-regulation of Gria2. The Sp binding motif on the Gria2 promoter is conserved among mice, rats, and humans.
AMPA receptors are excitatory glutamatergic neurotransmitter receptors that mediate most of the excitatory neurotransmission in the central nervous system (for review see [1]). These receptors are heterotetrameric proteins composed of GluA1-4 subunits. The majority of AMPA receptors in the adult cortex and hippocampus contain the GluA2 subunit in GluA1/GluA2 heterotetramers [2, 7, 8]. The expression of GluA3 is lower than that of GluA1, and the GluA4 subunit is developmentally expressed [7, 8, 11]. Our current study indicates that Sp4 regulates only the GluA2 subunit of AMPA receptors.
Sp4 is in the specificity protein/Krüppel-like (SP/KLF) family of zinc finger transcription factors that bind to GC-rich sequences of DNA (for review see [17]). Of the 25 members in the SP/KLF family, Sp4 shares critical amino acid homology in its DNA binding domain with specificity proteins 1 (Sp1) and 3 (Sp3), with all three factors competing for the same cis-elements [17, 18]. However, unlike Sp1 and Sp3, whose expression is ubiquitous, Sp4 is found primarily in the brain and testes [19, 30].
The murine brain has a greater abundance of Sp4 than either HeLa or N2a cells, whereas the opposite is true for Sp1 and Sp3 [28]. Furthermore, in primary neurons, mRNA and protein levels of Sp4 are greater than those of Sp1 [31]. In glial cells, however, Sp1 and Sp3 are the major Sp factors present, with negligible expression of Sp4 [31]. Neuronal expression of Sp4 increases with development and Sp4-null male mice do not breed despite having a normal reproductive system, suggesting that the deficit lies in the nervous system [19]. Hypomorphic Sp4 mice have vacuolization in the hippocampal gray matter and deficits in both sensorimotor gating and contextual memory. Restoration of Sp4 rescues all observable molecular, histological, and behavioral abnormalities in these mice [32].
The Sp cis-motif was previously identified on the Gria2 promoter [33], but consistent with the importance of Sp4 in neurons is our finding that it is Sp4, not Sp1 or Sp3, that regulates the expression of Gria2 in neurons through this motif. Our results of a positive regulation of Gria2 after over-expression of Sp1 and Sp3 in primary neurons may be due to the absence of smaller isoforms of Sp1 and Sp3 in primary neurons [28]. The smaller Sp3 isoforms are known to act as transcriptional repressors and the same may be true for Sp1 [31]. Their absence in primary neurons may allow over-expression of the exogenous full length Sp3 or Sp1 to activate expression of target genes. N2a cells, on the other hand, possess the smaller isoforms of Sp3 and Sp1 [28], and so they did not exhibit positive regulation of Gria2 with an over-expression of exogenous full length Sp3 or Sp1.
Of the four AMPA subunits, GluA1 to 4, it is remarkable that Sp4 regulates only GluA2. This subunit is quite unique, in that it undergoes hydrolytic editing of its pre-mRNA in its transmembrane segment 2 from glutamine to arginine [9], thereby rendering GluA2-containing AMPA receptors less permeable to Ca2+; and GluA2 homotetrameric AMPA receptors are completely impermeable to Ca2+ [9, 34, 35]. Thus, GluA2 subunit determines the functional properties of AMPA receptors, including the I-V behavior of the channel and permeability to Ca2+ [36, 37]. GluA2-containing AMPA receptors also exhibit low single channel conductance as compared to AMPA receptors that do not contain GluA2 [37-40].
The special properties of the GluA2 subunit render it important for normal neuronal activities, including synaptic plasticity, learning, and memory (for reviews see [3, 5]). GluA2 is involved in bidirectional plasticity in the synapse as well as in AMPA receptor maintenance, internalization, and distance-dependent scaling [41-44]. Knocking down GluA2 blocks synaptic scaling [45]. GluA2 subunits are able to stimulate synaptic function and promote the growth and formation of dendritic spines in cultured cortical neurons [43]. This is thought to be through a direct interaction of GluA2's N-terminal domain with the cell-adhesion molecule, N-cadherin [43]. A mechanism for the maintenance of long-term potentiation (LTP), thought to underlie experience-dependent plasticity, learning, and memory, can be through an increase in GluA2-containing receptors in hippocampal synapses [46-48]. Knocking out Gria2 results in multiple behavioral abnormalities, including deficits in object exploration, grooming, eye-closure reflex, and spatial and non-spatial learning [49]. Thus, by regulating GluA2, it is likely that Sp4 plays an important role in mediating many of these neuronal and synaptic processes.
The GluA2 subunit responds to changes in neuronal activity. Specifically, physiological concentrations of KCl, a depolarizing agent that activates neuronal activity, increases both transcript and protein expression of GluA2 in cultured visual cortical neurons [13]. On the other hand, decreasing neuronal activity through TTX-induced impulse blockade down-regulates GluA2 mRNA and protein levels in these neurons [13]. Parallel to these neuronal activity-mediated changes is a change in transcript and protein levels of cytochrome c oxidase (COX) [13], a critical enzyme for energy generation in neurons [50]. In the supragranular and infragranular layers of the macaque visual cortex, GluA2 expression and COX activity levels are governed by visual input and neuronal activity [12]. Specifically, monocular impulse blockade induces a down-regulation of both GluA2 expression and COX activity in deprived ocular dominance columns [12]. Thus, both GluA2 and COX are tightly regulated at the cellular level by neuronal activity.
We have previously shown that Sp factors modulate COX mRNA and protein levels in neurons [20]. Sp4, by regulating GluA2 expression, couples neuronal activity and energy metabolism at the transcriptional level. GluA2 expression is also controlled by nuclear respiratory factor 1 (NRF-1) [15], a transcription factor that regulates all 13 COX subunits [16, 22, 29]. The present study indicates that Sp4 and NRF-1 employ the concurrent and parallel mechanism to regulate the GluA2, but not GluA1, GluA3, or GluA4 subunits.
A possible benefit of the concurrent and parallel mechanism is that the GluA2 subunit, important for AMPA receptor physiology and function, is regulated by multiple transcription factors. However, it is unlikely that these factors function redundantly. While both Sp4 and NRF-1 are positively regulated by neuronal activity [27, 28, 51], NRF-1 can interact directly with an upstream coactivator, peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α), to induce target gene expression [52, 53], whereas Sp factors are not known to interact with PGC-1 α directly. However, Sp factors can interact with host cell factor 1 (HCF-1) [54], a co-activator that reportedly interacts with PGC-1α [55], resulting in a possible indirect interaction between Sp factors and PGC-1α in the induction of target gene expression. Sp factors are also known to interact with a multitude of cellular proteins and undergo numerous post-translational modifications [30, 54]. Other contributing factors, such as the cellular environment, cell signaling, and varying amounts of neuronal activity may differentially recruit NRF-1 or Sp4 to regulate their common target genes, such as Gria2.
Supplementary Material
Highlights.
Sp4 regulates the expressions of AMPA receptor subunit GluA2, not GluA1, 3, or 4
Sp4 silencing prevented the KCl-induced up-regulation of GluA2 (Gria2) transcript
Sp4 over-expression rescued TTX-mediated down-regulation of Gria2 transcript
Sp4 regulates Gria2 in a concurrent and parallel manner with NRF-1 and NRF-2
Sp1 and Sp3 do not regulate the GluA1 -4 AMPA receptor subunits
Acknowledgments
This work is supported by NIH Grant R01 EY018441 and NIH/NEI Training Grant 1-T32-EY14537. Anusha Priya is a member of the MCW-MSTP, which is partially supported by a T32 grant from NIGMS, GM080202.
Abbreviations
- Sp
Specificity protein transcription factor
- Gria
Gene name for AMPA receptor subunit
Footnotes
All authors have no conflict of interest to declare.
Conflict of Interest: The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Isaac JT, Ashby MC, McBain CJ. The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron. 2007;54:859–871. doi: 10.1016/j.neuron.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Borges K, Dingledine R. AMPA receptors: molecular and functional diversity. Prog Brain Res. 1998;116:153–170. doi: 10.1016/s0079-6123(08)60436-7. [DOI] [PubMed] [Google Scholar]
- 3.Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 4.Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka H, Grooms SY, Bennett MV, Zukin RS. The AMPAR subunit GluR2: still front and center-stage. Brain Res. 2000;886:190–207. doi: 10.1016/s0006-8993(00)02951-6. [DOI] [PubMed] [Google Scholar]
- 7.Wenthold RJ, Petralia RS, Blahos J, II, Niedzielski AS. Evidence for multiple AMPA receptor complexes in hippocampal CA1/CA2 neurons. J Neurosci. 1996;16:1982–1989. doi: 10.1523/JNEUROSCI.16-06-01982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig AM, Blackstone CD, Huganir RL, Banker G. The distribution of glutamate receptors in cultured rat hippocampal neurons: postsynaptic clustering of AMPA-selective subunits. Neuron. 1993;10:1055–1068. doi: 10.1016/0896-6273(93)90054-u. [DOI] [PubMed] [Google Scholar]
- 9.Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 10.Petralia RS, Wenthold RJ. Light and electron immunocytochemical localization of AMPA-selective glutamate receptors in the rat brain. J Comp Neurol. 1992;318:329–354. doi: 10.1002/cne.903180309. [DOI] [PubMed] [Google Scholar]
- 11.Zhu JJ, Esteban JA, Hayashi Y, Malinow R. Postnatal synaptic potentiation: delivery of GluR4-containing AMPA receptors by spontaneous activity. Nat Neurosci. 2000;3:1098–1106. doi: 10.1038/80614. [DOI] [PubMed] [Google Scholar]
- 12.Wong-Riley MT, Jacobs P. AMPA glutamate receptor subunit 2 in normal and visually deprived macaque visual cortex. Vis Neurosci. 2002;19:563–573. doi: 10.1017/s0952523802195022. [DOI] [PubMed] [Google Scholar]
- 13.Bai X, Wong-Riley MT. Neuronal activity regulates protein and gene expressions of GluR2 in postnatal rat visual cortical neurons in culture. J Neurocytol. 2003;32:71–78. doi: 10.1023/a:1027380315902. [DOI] [PubMed] [Google Scholar]
- 14.Wong-Riley MT. Bigenomic regulation of cytochrome C oxidase in neurons and the tight coupling between neuronal activity and energy metabolism. Adv Exp Med Biol. 2012;748:283–304. doi: 10.1007/978-1-4614-3573-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhar SS, Liang HL, Wong-Riley MT. Nuclear respiratory factor 1 co-regulates AMPA glutamate receptor subunit 2 and cytochrome c oxidase: tight coupling of glutamatergic transmission and energy metabolism in neurons. J Neurochem. 2009;108:1595–1606. doi: 10.1111/j.1471-4159.2009.05929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhar SS, Ongwijitwat S, Wong-Riley MT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 18.Letovsky J, Dynan WS. Measurement of the binding of transcription factor Sp1 to a single GC box recognition sequence. Nucleic Acids Res. 1989;17:2639–2653. doi: 10.1093/nar/17.7.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Supp DM, Witte DP, Branford WW, Smith EP, Potter SS. Sp4, a member of the Sp1-family of zinc finger transcription factors, is required for normal murine growth, viability, and male fertility. Dev Biol. 1996;176:284–299. doi: 10.1006/dbio.1996.0134. [DOI] [PubMed] [Google Scholar]
- 20.Dhar SS, Johar K, Wong-Riley MT. Bigenomic transcriptional regulation of all thirteen cytochrome c oxidase subunit genes by specificity protein 1. Open Biol. 2013;3:120176. doi: 10.1098/rsob.120176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ongwijitwat S, Wong-Riley MT. Functional analysis of the rat cytochrome c oxidase subunit 6A1 promoter in primary neurons. Gene. 2004;337:163–171. doi: 10.1016/j.gene.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 22.Ongwijitwat S, Wong-Riley MT. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene. 2005;360:65–77. doi: 10.1016/j.gene.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 23.Abmayr SM, Yao T, Parmely T, Workman JL. Preparation of nuclear and cytoplasmic extracts from mammalian cells. Curr Protoc Mol Biol. 2006;Chapter 12 doi: 10.1002/0471142727.mb1201s75. Unit 12 11. [DOI] [PubMed] [Google Scholar]
- 24.Priya A, Johar K, Wong-Riley MT. Nuclear respiratory factor 2 regulates the expression of the same NMDA receptor subunit genes as NRF-1: both factors act by a concurrent and parallel mechanism to couple energy metabolism and synaptic transmission. Biochim Biophys Acta. 2013;1833:48–58. doi: 10.1016/j.bbamcr.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia T, Zeng G, Gao L, Yu RK. Sp1 and AP2 enhance promoter activity of the mouse GM3-synthase gene. Gene. 2005;351:109–118. doi: 10.1016/j.gene.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Ishimaru N, Tabuchi A, Hara D, Hayashi H, Sugimoto T, Yasuhara M, Shiota J, Tsuda M. Regulation of neurotrophin-3 gene transcription by Sp3 and Sp4 in neurons. J Neurochem. 2007;100:520–531. doi: 10.1111/j.1471-4159.2006.04216.x. [DOI] [PubMed] [Google Scholar]
- 27.Dhar SS, Wong-Riley MT. Coupling of energy metabolism and synaptic transmission at the transcriptional level: role of nuclear respiratory factor 1 in regulating both cytochrome c oxidase and NMDA glutamate receptor subunit genes. J Neurosci. 2009;29:483–492. doi: 10.1523/JNEUROSCI.3704-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Priya A, Johar K, Wong-Riley MT. Specificity Protein 4 functionally regulates the transcription of NMDA receptor subunits GluN1, GluN2A, and GluN2B. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbamcr.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ongwijitwat S, Liang HL, Graboyes EM, Wong-Riley MT. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Li L, He S, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochem Cell Biol. 2004;82:460–471. doi: 10.1139/o04-045. [DOI] [PubMed] [Google Scholar]
- 31.Mao X, Yang SH, Simpkins JW, Barger SW. Glutamate receptor activation evokes calpain-mediated degradation of Sp3 and Sp4, the prominent Sp-family transcription factors in neurons. J Neurochem. 2007;100:1300–1314. doi: 10.1111/j.1471-4159.2006.04297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Long JM, Geyer MA, Masliah E, Kelsoe JR, Wynshaw-Boris A, Chien KR. Reduced expression of the Sp4 gene in mice causes deficits in sensorimotor gating and memory associated with hippocampal vacuolization. Mol Psychiatry. 2005;10:393–406. doi: 10.1038/sj.mp.4001621. [DOI] [PubMed] [Google Scholar]
- 33.Myers SJ, Peters J, Huang Y, Comer MB, Barthel F, Dingledine R. Transcriptional regulation of the GluR2 gene: neural-specific expression, multiple promoters, and regulatory elements. J Neurosci. 1998;18:6723–6739. doi: 10.1523/JNEUROSCI.18-17-06723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 35.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 36.Jonas P, Racca C, Sakmann B, Seeburg PH, Monyer H. Differences in Ca2+ permeability of AMPA-type glutamate receptor channels in neocortical neurons caused by differential GluR-B subunit expression. Neuron. 1994;12:1281–1289. doi: 10.1016/0896-6273(94)90444-8. [DOI] [PubMed] [Google Scholar]
- 37.Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 38.Laezza F, Dingledine R. Voltage-controlled plasticity at GluR2-deficient synapses onto hippocampal interneurons. J Neurophysiol. 2004;92:3575–3581. doi: 10.1152/jn.00425.2004. [DOI] [PubMed] [Google Scholar]
- 39.Hestrin S. Different glutamate receptor channels mediate fast excitatory synaptic currents in inhibitory and excitatory cortical neurons. Neuron. 1993;11:1083–1091. doi: 10.1016/0896-6273(93)90221-c. [DOI] [PubMed] [Google Scholar]
- 40.Kamboj SK, Swanson GT, Cull-Candy SG. Intracellular spermine confers rectification on rat calcium-permeable AMPA and kainate receptors. J Physiol. 1995;486(Pt 2):297–303. doi: 10.1113/jphysiol.1995.sp020812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Passafaro M, Nakagawa T, Sala C, Sheng M. Induction of dendritic spines by an extracellular domain of AMPA receptor subunit GluR2. Nature. 2003;424:677–681. doi: 10.1038/nature01781. [DOI] [PubMed] [Google Scholar]
- 42.McCormack SG, Stornetta RL, Zhu JJ. Synaptic AMPA receptor exchange maintains bidirectional plasticity. Neuron. 2006;50:75–88. doi: 10.1016/j.neuron.2006.02.027. [DOI] [PubMed] [Google Scholar]
- 43.Saglietti L, Dequidt C, Kamieniarz K, Rousset MC, Valnegri P, Thoumine O, Beretta F, Fagni L, Choquet D, Sala C, Sheng M, Passafaro M. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron. 2007;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 44.Shipman SL, Herring BE, Suh YH, Roche KW, Nicoll RA. Distance-dependent scaling of AMPARs is cell-autonomous and GluA2 dependent. J Neurosci. 2013;33:13312–13319. doi: 10.1523/JNEUROSCI.0678-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gainey MA, Hurvitz-Wolff JR, Lambo ME, Turrigiano GG. Synaptic scaling requires the GluR2 subunit of the AMPA receptor. J Neurosci. 2009;29:6479–6489. doi: 10.1523/JNEUROSCI.3753-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu SQ, Cull-Candy SG. Synaptic activity at calcium-permeable AMPA receptors induces a switch in receptor subtype. Nature. 2000;405:454–458. doi: 10.1038/35013064. [DOI] [PubMed] [Google Scholar]
- 47.Liu SJ, Cull-Candy SG. Activity-dependent change in AMPA receptor properties in cerebellar stellate cells. J Neurosci. 2002;22:3881–3889. doi: 10.1523/JNEUROSCI.22-10-03881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plant K, Pelkey KA, Bortolotto ZA, Morita D, Terashima A, McBain CJ, Collingridge GL, Isaac JT. Transient incorporation of native GluR2-lacking AMPA receptors during hippocampal long-term potentiation. Nat Neurosci. 2006;9:602–604. doi: 10.1038/nn1678. [DOI] [PubMed] [Google Scholar]
- 49.Gerlai R, Henderson JT, Roder JC, Jia Z. Multiple behavioral anomalies in GluR2 mutant mice exhibiting enhanced LTP. Behav Brain Res. 1998;95:37–45. doi: 10.1016/s0166-4328(98)00002-3. [DOI] [PubMed] [Google Scholar]
- 50.Wong-Riley MT. Cytochrome oxidase: an endogenous metabolic marker for neuronal activity. Trends Neurosci. 1989;12:94–101. doi: 10.1016/0166-2236(89)90165-3. [DOI] [PubMed] [Google Scholar]
- 51.Zhang C, Wong-Riley MT. Depolarizing stimulation upregulates GA-binding protein in neurons: a transcription factor involved in the bigenomic expression of cytochrome oxidase subunits. Eur J Neurosci. 2000;12:1013–1023. doi: 10.1046/j.1460-9568.2000.00997.x. [DOI] [PubMed] [Google Scholar]
- 52.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 53.Vercauteren K, Gleyzer N, Scarpulla RC. PGC-1-related coactivator complexes with HCF-1 and NRF-2beta in mediating NRF-2(GABP)-dependent respiratory gene expression. J Biol Chem. 2008;283:12102–12111. doi: 10.1074/jbc.M710150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gunther M, Laithier M, Brison O. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol Cell Biochem. 2000;210:131–142. doi: 10.1023/a:1007177623283. [DOI] [PubMed] [Google Scholar]
- 55.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


