Abstract
Despite advances in antimicrobials, vaccination and public health measures, infectious diseases remain a leading cause of morbidity and mortality worldwide. With the increase in antimicrobial resistance and the emergence of new pathogens, there remains a need for new and more accurate diagnostics, the ability to monitor adequate treatment response as well as the ability to predict prognosis for an individual. Transcriptional approaches using blood signatures have enabled a better understanding of the host response to diseases, leading not only to new avenues of basic research, but also to the identification of potential biomarkers for use in diagnosis, prognosis and treatment monitoring.
Keywords: immune response, infection, tuberculosis
1. Introduction
Since the 1990s, whole genome expression mRNA microarray technology has been available as a tool to researchers. It involves profiling the expression levels of thousands of genes simultaneously and is increasingly being used to advance our understanding of the complex transcriptional response that occurs as a consequence of a disease process [1–3]. Modern microarray platforms are capable of reliably and reproducibly measuring the expression of over 40 000 mRNA transcripts, which can encompass all of the known functional human genome. By comparing between cohorts of individuals, or by sequentially sampling over a time course, it is possible to delineate a comparative transcriptional response to a given perturbation. Depending on the scale of the response, many hundreds or thousands of significantly differentially regulated genes may be identified. To comprehend the biological relevance of these complex data, bioinformatics tools have been developed that use known relationships between genes and their biological functions. With these tools, it is possible to determine the most biologically significant groups of differentially regulated genes as well as key transcriptional regulatory pathways/networks perturbed upon a given stimulus/disease [4–7].
2. Gene expression profiling using peripheral blood
The early gene expression studies involving human peripheral blood focused on cancer and autoimmune conditions [2,8]. The use of peripheral blood for whole genome expression studies in disease has a number of advantages. Peripheral blood is easily accessible, whereas the site of primary infection in certain diseases may not be easy to access, and it is possible to obtain reproducible blood transcriptional profiles from volumes as small as that obtained from a finger prick [9–11]. Blood represents a reservoir where there is a dynamic exchange of chemokines, cytokines and cells trafficking to and from sites of active disease and the lymphatic system. These cells include neutrophils, basophils, eosinophils, T cells and B cells; however, the cellular composition of the blood can vary depending on the ethnic background [12] of the patient as well as the scale and specificity of the host response to a disease. Different immune cells may have different baseline levels of gene expression as well as different transcriptional amplification programmes [13,14]. The interpretation of blood-derived transcriptional signatures therefore has to be made in the context of the cellular composition of the blood being sampled. Analysis of a subset of cells such as peripheral blood mononuclear cells (PBMCs) has been used as an attempt to control for this variable, although this excludes from analysis granulocytes such as neutrophils, which make up the greatest proportion of the immune cells found within blood and are important innate immune system effector cells. Furthermore, neutrophils have been seen to be important drivers of the blood transcriptional signature in certain diseases [15,16]. The superiority or suitability of PBMCs or whole blood leucocytes (globin RNA reduced) for transcriptional signatures in disease has yet to be rigorously compared. In addition, expression analysis of the subsets of blood leucocytes has also been an informative approach [14–17]. In some cases, this approach may be necessary to reveal signatures expressed in a discrete cell population that is present only in low numbers in the peripheral blood [18], or where the sensitivity of specific gene expression profiles may be masked if a unique profile is present in the cells found in low frequency in human blood [19]. An alternative approach is to apply computational deconvolution methods; this generates data for both the heterogeneous sample (whole blood, PBMCs) and its specific cell subsets [20].
During inflammatory, autoimmune diseases, cancers or infectious diseases the changes observed in a transcriptional blood signature can represent changes in cell numbers, which may be reduced as a consequence of apoptosis or migration of cells from the blood to other tissues. Conversely, increased cell numbers may result from proliferation of cells or migration of cells from the tissues or bone marrow into the blood. Alternatively, the transcriptional changes may be due to discrete changes in transcription within a specific cellular population as a consequence of perturbations resulting from the disease state. As an example, early in the host response to a pathogen, pattern recognition receptors, which are expressed on a wide variety of cell types including cells of the innate immune system, recognize the highly conserved microbial products of pathogens [21]. Binding of these receptors results in activation of signalling pathways and via transcriptional regulators the induction of transcriptional programmes [22]. Different pattern recognition receptors can recognize different microbial products and activate specific transcriptional programmes, enabling a transcriptional response appropriate to the pathogen. These directly activated immune cells can traffic from the site of infection and enter the blood stream either directly or via the lymphatic system. Overall, therefore, the transcriptional signatures observed in the blood in response to a disease may be as a consequence of changes in absolute cell numbers, changes in the proportions of cell types as well as changes in transcription within cell populations.
The specificity of a transcriptional signature for a particular disease may lie in the combination of activated pathways and transcription programmes rather than in uniquely activated disease-specific genes. Tools have been designed to characterize this transcriptional response, such as gene set enrichment analysis, transcriptional networks and modular approaches that can identify combinations of transcriptional programmes associated with diseases [23]. Algorithmic approaches such as the molecular distance to health or variations on this principle have used gene expression data to assess severity of disease or quantify disease response to treatment [15,24,25].
3. Infectious diseases
Gene expression studies in infectious diseases have been used to identify transcriptional signatures that differentiate between bacterial and viral infections [14,26,27], bacterial meningitis [28], acute febrile [29] and viral illnesses [30,31], as well as specific disease pathogens such as Burkholderia pseudomallei [24,32], dengue virus [33–35], human immunodeficiency virus [36,37], Mycobacterium leprae [38], Staphylococcus aureus [39], Streptococcus pneumoniae [40], Salmonella enterica [41] and Mycobacterium tuberculosis [15–17,25,42–49].
One of the challenges in infectious diseases is to identify the causative agent early in the disease, so that appropriate therapy can be initiated early or inappropriate therapy avoided. The traditional approach involves identification of the pathogen itself either by direct visualization, culture, nucleic acid amplification of pathogen-specific DNA or by measuring a specific antibody-mediated response. These approaches are not feasible for all infectious diseases, owing to either the inaccessibility of the tissue or difficulty in culturing the pathogen, or because the generation of an antibody-specific response is too slow to guide initial therapy. An ability to identify a transcriptional signature specific for an infectious disease based on the early host response would, therefore, be advantageous for use in diagnosis as well as revealing information about the early host response. Studies have shown that host blood transcriptional signatures can differentiate between different infectious diseases [14,27,29]. These signatures reveal that the broad response to viral infections includes well recognized, upregulated viral response elements including interferon-inducible genes [14,27], and can discriminate not only between bacterial and viral infection [27] but also between different viral infections [14,30].
Gene transcription profiling during early infectious disease has been used to identify biomarkers that can potentially be used to predict outcome as well as give a better understanding of the potential deficiencies or exaggerated transcriptional responses that correlate with poor outcome. In HIV, pathways involved in apoptosis, development, cell cycle and DNA damage were found to be differentially regulated in patients with rapid disease progression compared with those with slow disease progression [36]. In dengue infection, transcriptional signatures have enabled identification of differences in children with dengue virus fever from those who developed the more serious dengue haemorrhagic fever (DHF) [33,34]. Transcriptional differences could be seen in patients who subsequently developed DHF, prior to the development of the clinical features, including reduced expression of interferon response genes and differences in complement expression [33,34]. These studies have enabled the generation of hypotheses for further experiments to investigate the previously poorly understood aetiology of DHF, and in addition have identified potential early prognostic biomarkers [34].
Blood-derived gene expression signatures have also been seen to correlate with effective vaccination as measured by the quantity of neutralizing antibodies in yellow fever, influenza and pneumococcal vaccines [9,19,50]. These studies have not only identified putative biomarkers for predicting successful vaccination, but also enabled the description of the temporal kinetics of the transcriptional response to vaccination. Furthermore, they have identified the difference in transcriptional response between the influenza and pneumococcal vaccines. The influenza vaccine induced a greater anti-viral response as seen by the induction of interferon genes, compared with the pneumococcal vaccine, which induced more of an inflammatory response [9].
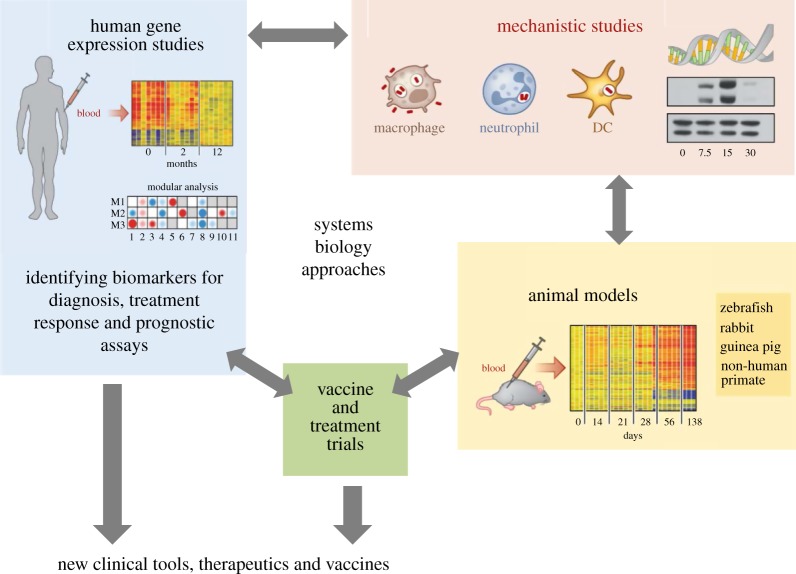
Transcriptional signatures and systems biology approaches in infectious diseases can therefore feed into many different aspects of research, including translational clinical studies as well as basic immunological work (figure 1).
Figure 1.
Using a systems biology approach in infectious disease research. Adapted from O‘Garra et al. [51].
4. Tuberculosis: background of some of the current difficulties
Mycobacterium tuberculosis, the causative organism of tuberculosis, is estimated to have infected one-third of the world's population. In 2011, it was responsible for approximately 8.6 million cases of active tuberculosis and 1.4 million deaths [52]. M. tuberculosis is mainly spread via the aerosol route, and the outcome from inhalation of M. tuberculosis depends on a variety of factors: environmental and sociological factors, virulence of the M. tuberculosis strain and the capability of the host immune response [51]. The World Health Organization STOP TB strategy to halve the prevalence of tuberculosis by 2015 recognizes the need for new diagnostics, drugs and vaccines [53]. Development of these, however, is hampered by our incomplete understanding of the immune response to M. tuberculosis [51]. CD4+ T cells and the cytokines tumour necrosis factor, interleukin-12 and interferon (IFN)-γ have been shown to be critical in the control of M. tuberculosis, perturbations in these factors in animal models and humans being detrimental to the host [51]. These factors alone are not sufficient for an adequate host response however, and while numerous other factors have been identified, it remains unclear which combination of factors constitute a protective host immune response and what factors determine whether an individual goes on to develop tuberculosis [51].
Approximately 5–10% of those infected will develop active tuberculosis disease within the first year. The remaining 90% have clinically asymptomatic latent tuberculosis that carries an approximate 10% lifetime risk of progressing to active tuberculosis. Latent tuberculosis is defined as evidence of immunological exposure to M. tuberculosis with no symptoms of active clinical disease [54]. The host immune response is critical in maintaining this clinically latent state; perturbations in the immune system, either iatrogenic by immune modulating drugs or through diseases that compromise the immune system such as HIV infection, can lead to progression from latent to active tuberculosis [55,56].
In the non-HIV-infected individual, it is not possible currently to predict the outcome from M. tuberculosis infection. If it were, treatment of latent tuberculosis infection could be better targeted to those at the highest risk. Additionally, improved diagnostic tests for active tuberculosis would assist greatly in clinical management, enabling earlier diagnosis and hence prompt treatment initiation (which would also reduce the risk of onward transmission to others), as well as avoiding misdiagnosis of active tuberculosis and thus minimizing adverse events owing to unnecessary anti-tuberculosis treatment, which are common with current regimens [57].
Currently, there is only one licensed preventive vaccine against M. tuberculosis, the bacillus Calmette–Guérin (BCG) vaccine [58]. Its efficacy is not optimal and it has been shown to offer variable protection against the most common form of the disease—adult pulmonary tuberculosis [59–61]. A new, more effective vaccine is therefore required. Twelve vaccines are currently in phase 1 or phase 2 trials, although vaccine development is hampered by our poor understanding of what constitutes a protective immune response. The immunological readouts in current use do not provide correlates of a protective host response [62,63]. This may in part explain the inability of one of the most promising vaccine candidates to provide any addition protection above BCG vaccination in a phase 2 trial, despite demonstrating improved protection in animal models and induction of antigen-specific TH1 and TH17 cells in infants [64–66].
5. Tuberculosis peripheral blood gene expression studies
There has been a number of transcriptomic studies investigating the host response to M. tuberculosis infection; predominantly these have focused on the most common form of the disease—adult pulmonary tuberculosis (table 1).
Table 1.
Summary of blood transcriptional profiling studies in tuberculosis. HC, healthy controls; LTB, latent tuberculosis; OD, other diseases; PBMC, peripheral blood mononuclear cells; TB, tuberculosis; TLR, Toll-like receptors. Modified from Berry et al. [67].
| geographical region | year | sample | study design | key pathways | reference |
|---|---|---|---|---|---|
| South Africa, Malawi | 2013 | whole blood | TB versus OD TB versus LTB (HIV positive and negative) |
— | [49] |
| UK | 2013 | whole blood | TB versus OD TB treatment |
interferon signalling, role of pattern recognition receptors, antigen presentation | [16] |
| South Africa | 2013 | whole blood | TB treatment | complement; B-cell markers; CD64 | [48] |
| Germany | 2012 | whole blood | TB versus OD | interferon signalling; complement; TLR signalling; Fcγ-receptor-mediated phagocytosis | [47] |
| South Africa | 2012 | whole blood | TB treatment | — | [25] |
| Indonesia | 2012 | PBMC | TB versus HC TB treatment |
interferon signalling | [17] |
| The Gambia | 2011 | whole blood | TB versus LTB | JAK–STAT pathway; interferon signalling; TLR | [46] |
| USA and Brazil | 2011 | whole blood | TB versus LTB versus HC | interferon signalling | [45] |
| South Africa | 2011 | whole blood | TB versus LTB versus HC | — | [44] |
| UK and South Africa | 2010 | whole blood | TB versus LTB versus HC TB versus OD TB treatment |
interferon signalling | [15] |
| South Africa | 2007 | whole blood | TB versus LTB | — | [43] |
| Germany | 2007 | PBMC | TB versus LTB | — | [42] |
Several studies have been designed to attempt to identify a transcriptional signature that can differentiate active pulmonary disease from latent infection and/or healthy controls. Differentiating active tuberculosis disease from latently infected and healthy controls has enabled the identification of many immunological pathways that may be relevant to the pathogenesis of active tuberculosis such as type 1 and type 2 interferon signalling, Toll-like receptor signalling and also T- and B-cell function gene expression [15,17,25,42,43,45–48].
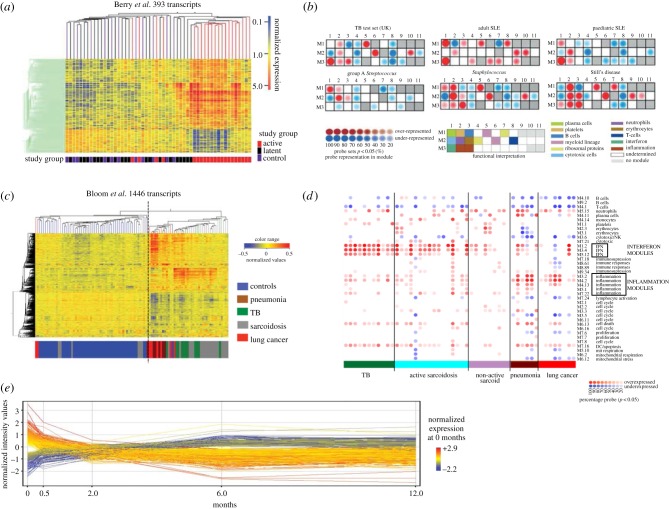
Early studies have identified interferon-inducible, inflammatory and chemokine genes as being differentially expressed between active pulmonary tuberculosis patients and controls. However, these studies involved small numbers of patients and had no independent test or validation sets [42,43]. In 2010, work involving patients from both the UK and South Africa identified a neutrophil-driven interferon signature present in active disease that was absent in the majority of healthy and latently infected individuals, and which correlated with the extent of radiographic lung disease and was diminished with anti-tuberculosis therapy (figure 2a). This transcriptional signature was validated with independent test and validation sets [15] and has subsequently been confirmed in studies involving patients from different countries (South Africa, USA, China, The Gambia, Germany and Indonesia) by other groups, using different microarray platforms and different microarray analytical approaches [17,45–47,68]. Type I interferon signalling had been previously largely underappreciated [69,70] and is now the focus of further work to determine the importance of this type I interferon signalling and how it influences the outcome following M. tuberculosis infection [51]. Approximately 10% of the latently infected individuals in the Berry et al. study [15] had a signature of active tuberculosis. It is unclear, at the present time, whether these individuals represent subclinical disease, incipient conversion from latent to active disease or another process. Further work is required to identify transcriptional signatures of latent M. tuberculosis infection that can predict those at risk of progression, and this will require longitudinal follow-up.
Figure 2.
Transcriptional signatures in tuberculosis. (a) A 393 transcript signature was able to broadly distinguish active TB from latently infected and healthy controls (from Berry et al. [15]). (b) A modular approach is able to identify the key transcriptional differences between TB and other inflammatory diseases (modified from Berry et al. [15]). (c) A 1446 transcript signature reveals that pulmonary granulomatous diseases display similar transcriptional signatures that are distinct from pneumonia and lung cancer (modified from Bloom et al. [16]). (d) Modular approach showing the differences between active sarcoidosis, TB compared with pneumonias and lung cancer (modified from Bloom et al. [16]). (e) A 664 transcript signature is seen to change on treatment by as early as two weeks (modified from Bloom et al. [25]). SLE, systemic lupus erythematosus; TB, tuberculosis.
A meta-analysis of eight publicly available tuberculosis microarray datasets undertaken by Joosten et al. [71] found that after integration of the data from these separate studies interferon signalling was no longer a dominant pathway as had been described in many of the studies when the data had been analysed in isolation. Instead, there was enrichment for genes associated with myeloid cell inflammation, and TREM1 signalling was now found to be the most significant pathway [71].
To develop a diagnostic test based on gene expression levels, a transcriptional signature that can distinguish between active tuberculosis and other diseases is needed. An 86 transcript signature was able to differentiate between pulmonary tuberculosis and selected other diseases [15]. Using a modular approach, it was possible to identify distinct modular patterns for the different inflammatory diseases (group A Streptococcus, Staphylococcus, adult and paediatric systemic lupus erythematosus and Still's disease) and that tuberculosis had over-representation of interferon, inflammatory and myeloid lineage modules (figure 2b) [15]. However, further studies comparing tuberculosis with sarcoidosis as well as melioidosis revealed an overlap with the interferon-dominated pulmonary tuberculosis transcriptional signature [32,47,72]. These signatures have shown that diseases with similar pathophysiology may share similar transcriptional profiles—sarcoidosis, melioidosis and tuberculosis are all granulomatous diseases that can affect the lungs. However, when studies were designed that directly compared sarcoidosis and tuberculosis, blood transcriptional signatures could differentiate sarcoidosis from tuberculosis, and revealed an increased interferon-inducible response in tuberculosis in terms of the number of genes upregulated as well as in the magnitude of the response. Conversely, increased eukaryotic initiation factor 2 signalling was detected in patients with sarcoidosis [16]. One study revealed increased metabolic activity and antimicrobial defence response in tuberculosis when compared with sarcoidosis [47]. These studies demonstrate not only that diseases with similar pathological findings affecting the same organ can be distinguished using blood transcriptional signatures, but also that we can gain novel immunological insights from these blood transcriptional studies.
The specificity of any future diagnostic test based on transcriptional signatures will depend on its ability to accurately discriminate tuberculosis from a large number of different diseases. Importantly, it will be necessary to distinguish patients who are co-infected with HIV or not, as well as with comorbidities such as diabetes. Bloom et al. [16] set out to derive transcriptional signatures that could differentiate between active tuberculosis and other diseases (figure 2c). They included diseases that could clinically mimic tuberculosis, and by use of a modular approach to characterize the different diseases they again revealed the strong interferon response found in patients with active sarcoidosis and tuberculosis, but, in contrast, revealed an inflammatory nature of the blood transcriptome of patients with lung cancer or pneumonia (figure 2d) [16]. Previous studies had excluded HIV-infected participants in order to first define tuberculosis in the absence of any co-infection or co-morbidities. Additionally, there was concern that the reduced CD4+ T cells, as well an HIV transcriptional signature itself, would confound any potential analysis. More recently, Kaforou et al. [49] have published a large case–control study involving both HIV-infected and HIV-uninfected individuals. This study, by prospectively recruiting patients presenting with symptoms of TB who on further investigation were diagnosed with another disease, generated a large clinically relevant and diverse ‘other diseases’ cohort to act as a comparison group. In both their own datasets and an independent validation dataset, the derived transcriptional signatures were able to differentiate with a high degree of sensitivity and specificity between tuberculosis and healthy controls as well as tuberculosis and other diseases in both HIV-infected and HIV-uninfected individuals [49]. A transcriptional signature for use as a diagnostic will need to be able to diagnose TB from these real-world alternative diagnoses, and in addition the derivation of a signature that can distinguish patients in the context of HIV co-infection is of great importance as in several parts of the world a large number of tuberculosis cases are co-infected with HIV. This study is the first one to identify a transcriptional signature that is able to differentiate those that are M. tuberculosis infected from those non-infected despite HIV co-infection.
A transcriptional signature that reflects response to effective treatment has implications for the development of a clinical test that can determine a successful treatment response. Currently, sputum smear and culture testing after two months of therapy is the best predictor of relapse following treatment completion, but this is limited as sputum is not always accessible. A test that could identify treatment failure or success earlier would have implications for trials of novel drugs and regimens as well as aiding clinical practice by assisting in the identification of drug resistance or non-compliance earlier than is currently possible. Berry et al. [15] first demonstrated that significant transcriptional changes could be detected after two months following initiation of successful therapy. Two recently published studies following patients recruited from South Africa longitudinally showed that a transcriptional signature of active pulmonary tuberculosis quickly diminished with successful treatment [25,48]. Transcripts (1261) were seen to significantly fall in expression by one week. Among them were complement genes (C1q, C2 and derpin G1), which the authors hypothesized was related to the rapid reduction in mycobacterial load that was driving complement production [48]. A separate longitudinal study derived a 664 transcript signature that significantly changed over the course of treatment as well as by two weeks—this 664 transcript list was enriched for genes involved in interferon signalling [25].
6. Future
Gene expression microarray studies have advanced the immunological knowledge of the host response to infectious diseases, opening up avenues for further research as well as identification of potential biomarkers for use in diagnostic and prognostic tests as well as evaluations of response to treatment or vaccination.
RNA-Seq (also referred to as whole transcriptome shotgun sequencing) is an exciting developing new technology [1,73,74] that is starting to become available as a diagnostic approach to infectious and other diseases. RNA-Seq is a tag-based, high-throughput approach in which sequences are mapped against a reference genome thereby eliminating background signals and the need for statistical normalization. The technology has an advantage over microarray in that it additionally allows the detection of novel transcripts rather than reliance on known probe-based sequences. To date, the use of RNA-Seq for diagnosis in tuberculosis has been mostly restricted to looking at small sample numbers (in bovine tuberculosis [75]), and to the best of our knowledge, no reports are available for human tuberculosis. Currently, the cost, challenges of statistical data analysis and the logistics of large data storage currently make microarray a more practical option for analysis approaches that involve large samples/patient numbers. However, ongoing developments in next-generation sequencing, such as the recent United States Food and Drug Administration approval of the Illumina MiSeqD system for clinical use [76], make this a very exciting area for the future diagnosis of human disease.
Integration of the data from the complex host transcriptional signatures, host clinical phenotypes as well as pathogen genotype/gene expression using systems biology tools that continue to be developed and refined will enable researchers to better synthesize and understand the large volume of complex data being generated. This integrated analysis together with work in experimental models will provide a better understanding of the immunological response to infectious disease (figure 1). Gene expression data benefit from being widely available as an open resource for researchers. Many studies have already taken advantage of publicly available datasets to reanalyse data and integrate it to advance immunological knowledge [71,77] as well as to test or derive transcriptional signatures [16,30,32,47].
The identification of specific transcriptional responses for a disease will further our understanding of the host immune response and also aid in the development of specific tests that can be used in clinical practice. Further studies need to be undertaken to broaden the number of infectious diseases profiled, including co-infection studies, so that the transcriptional signatures may be better defined and be more applicable to ‘real-world’ situations. Furthermore, these transcriptional signatures will then need to be tested in large-scale prospective clinical trials to assess their ability to perform in large diverse populations. The ultimate use of transcriptionally derived signatures in the clinical management of infectious disease may not be as a standalone diagnostic, but rather used to support and aid in clinical diagnosis/management alongside other tools. For example, in active tuberculosis, a blood signature could support the current smear test, M. tuberculosis culture, clinical symptoms and imaging such as chest radiographs or cross-sectional imaging. For treatment response, it could be used alongside monitoring of clearance of M. tuberculosis from the sputum.
References
- 1.Pascual V, Chaussabel D, Banchereau J. 2010. A genomic approach to human autoimmune diseases. Annu. Rev. Immunol. 28, 535–571. ( 10.1146/annurev-immunol-030409-101221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaussabel D, Pascual V, Banchereau J. 2010. Assessing the human immune system through blood transcriptomics. BMC Biol. 8, 84 ( 10.1186/1741-7007-8-84) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mejias A, Ramilo O. 2013. Transcriptional profiling in infectious diseases: ready for prime time? J. Infect. 68, S94–S99. ( 10.1016/j.jinf.2013.09.018) [DOI] [PubMed] [Google Scholar]
- 4.Khatri P, Sirota M, Butte AJ. 2012. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput. Biol. 8, e1002375 ( 10.1371/journal.pcbi.1002375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Germain RN, Meier-Schellersheim M, Nita-Lazar A, Fraser ID. 2011. Systems biology in immunology: a computational modeling perspective. Annu. Rev. Immunol. 29, 527–585. ( 10.1146/annurev-immunol-030409-101317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zak DE, Aderem A. 2009. Systems biology of innate immunity. Immunol. Rev. 227, 264–282. ( 10.1111/j.1600-065X.2008.00721.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slonim DK. 2002. From patterns to pathways: gene expression data analysis comes of age. Nat. Genet. 32, 502–508. ( 10.1038/ng1033) [DOI] [PubMed] [Google Scholar]
- 8.Golub TR, et al. 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286, 531–537. ( 10.1126/science.286.5439.531) [DOI] [PubMed] [Google Scholar]
- 9.Obermoser G, et al. 2013. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity 38, 831–844. ( 10.1016/j.immuni.2012.12.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. 2003. Individuality and variation in gene expression patterns in human blood. Proc. Natl Acad. Sci. USA 100, 1896–1901. ( 10.1073/pnas.252784499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohr S, Liew CC. 2007. The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol. Med. 13, 422–432. ( 10.1016/j.molmed.2007.08.003) [DOI] [PubMed] [Google Scholar]
- 12.Rougemont A, Boisson ME. 1975. Letter: racial differences in leucoyte count. Br. Med. J. 2, 684–685. ( 10.1136/bmj.2.5972.684-c) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovén J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. 2012. Revisiting global gene expression analysis. Cell 151, 476–482. ( 10.1016/j.cell.2012.10.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu X, Yu J, Crosby SD, Storch GA. 2013. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc. Natl Acad. Sci. USA 110, 12 792–12 797. ( 10.1073/pnas.1302968110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berry MP, et al. 2010. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977. ( 10.1038/nature09247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloom CI, et al. 2013. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS ONE 8, e70630 ( 10.1371/journal.pone.0070630) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ottenhoff TH, et al. 2012. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS ONE 7, e45839 ( 10.1371/journal.pone.0045839) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JC, et al. 2011. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J. Clin. Invest. 121, 4170–4179. ( 10.1172/JCI59255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakaya HI, et al. 2011. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 12, 786–795. ( 10.1038/ni.2067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen-Orr SS, Gaujoux R. 2013. Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr. Opin. Immunol. 25, 571–578. ( 10.1016/j.coi.2013.09.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medzhitov R, Horng T. 2009. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 9, 692–703. ( 10.1038/nri2634) [DOI] [PubMed] [Google Scholar]
- 22.Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA. 2002. Human macrophage activation programs induced by bacterial pathogens. Proc. Natl Acad. Sci. USA 99, 1503–1508. ( 10.1073/pnas.022649799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaussabel D, et al. 2008. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity 29, 150–164. ( 10.1016/j.immuni.2008.05.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pankla R, Buddhisa S, Berry M, Blankenship DM, Bancroft GJ, Banchereau J, Lertmemongkolchai G, Chaussabel D. 2009. Genomic transcriptional profiling identifies a candidate blood biomarker signature for the diagnosis of septicemic melioidosis. Genome Biol. 10, R127 ( 10.1186/gb-2009-10-11-r127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloom CI, et al. 2012. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS ONE 7, e46191 ( 10.1371/journal.pone.0046191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaussabel D, Allman W, Mejias A, Chung W, Bennett L, Ramilo O, Pascual V, Palucka AK, Banchereau J. 2005. Analysis of significance patterns identifies ubiquitous and disease-specific gene-expression signatures in patient peripheral blood leukocytes. Ann. NY Acad. Sci. 1062, 146–154. ( 10.1196/annals.1358.017) [DOI] [PubMed] [Google Scholar]
- 27.Ramilo O, et al. 2007. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 109, 2066–2077. ( 10.1182/blood-2006-02-002477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lill M, Koks S, Soomets U, Schalkwyk LC, Fernandes C, Lutsar I, Taba P. 2013. Peripheral blood RNA gene expression profiling in patients with bacterial meningitis. Front. Neurosci. 7, 33 ( 10.3389/fnins.2013.00033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popper SJ, Watson VE, Shimizu C, Kanegaye JT, Burns JC, Relman DA. 2009. Gene transcript abundance profiles distinguish Kawasaki disease from adenovirus infection. J. Infect. Dis. 200, 657–666. ( 10.1086/603538) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaas AK, et al. 2009. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 6, 207–217. ( 10.1016/j.chom.2009.07.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mejias A, et al. 2013. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med. 10, e1001549 ( 10.1371/journal.pmed.1001549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koh GC, Schreiber MF, Bautista R, Maude RR, Dunachie S, Limmathurotsakul D, Day NP, Dougan G, Peacock SJ. 2013. Host responses to melioidosis and tuberculosis are both dominated by interferon-mediated signaling. PLoS ONE 8, e54961 ( 10.1371/journal.pone.0054961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ubol S, Masrinoul P, Chaijaruwanich J, Kalayanarooj S, Charoensirisuthikul T, Kasisith J. 2008. Differences in global gene expression in peripheral blood mononuclear cells indicate a significant role of the innate responses in progression of dengue fever but not dengue hemorrhagic fever. J. Infect. Dis. 197, 1459–1467. ( 10.1086/587699) [DOI] [PubMed] [Google Scholar]
- 34.Nascimento EJ, et al. 2009. Gene expression profiling during early acute febrile stage of dengue infection can predict the disease outcome. PLoS ONE 4, e7892 ( 10.1371/journal.pone.0007892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loke P, Hammond SN, Leung JM, Kim CC, Batra S, Rocha C, Balmaseda A, Harris E. 2010. Gene expression patterns of dengue virus-infected children from Nicaragua reveal a distinct signature of increased metabolism. PLoS Neglected Trop. Dis. 4, e710 ( 10.1371/journal.pntd.0000710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang ZN, et al. 2013. Transcriptomic analysis of peripheral blood mononuclear cells in rapid progressors in early HIV infection identifies a signature closely correlated with disease progression. Clin. Chem. 59, 1175–1186. ( 10.1373/clinchem.2012.197335) [DOI] [PubMed] [Google Scholar]
- 37.Gupta A, Nagilla P, Le HS, Bunney C, Zych C, Thalamuthu A, Bar-Joseph Z, Mathavan S, Ayyavoo V. 2011. Comparative expression profile of miRNA and mRNA in primary peripheral blood mononuclear cells infected with human immunodeficiency virus (HIV-1). PLoS ONE 6, e22730 ( 10.1371/journal.pone.0022730) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Teles RM, et al. 2013. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science 339, 1448–1453. ( 10.1126/science.1233665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ardura MI, et al. 2009. Enhanced monocyte response and decreased central memory T cells in children with invasive Staphylococcus aureus infections. PLoS ONE 4, e5446 ( 10.1371/journal.pone.0005446) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu K, Chen L, Kaur R, Pichichero M. 2012. Transcriptome signature in young children with acute otitis media due to Streptococcus pneumoniae. Microbes Infect. Inst. Pasteur 14, 600–609. ( 10.1016/j.micinf.2012.01.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson LJ, et al. 2009. Transcriptional response in the peripheral blood of patients infected with Salmonella enterica serovar Typhi. Proc. Natl Acad. Sci. USA 106, 22 433–22 438. ( 10.1073/pnas.0912386106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobsen M, Repsilber D, Gutschmidt A, Neher A, Feldmann K, Mollenkopf HJ, Ziegler A, Kaufmann SH. 2007. Candidate biomarkers for discrimination between infection and disease caused by Mycobacterium tuberculosis. J. Mol. Med. 85, 613–621. ( 10.1007/s00109-007-0157-6) [DOI] [PubMed] [Google Scholar]
- 43.Mistry R, et al. 2007. Gene-expression patterns in whole blood identify subjects at risk for recurrent tuberculosis. J. Infect. Dis. 195, 357–365. ( 10.1086/510397) [DOI] [PubMed] [Google Scholar]
- 44.Maertzdorf J, Repsilber D, Parida SK, Stanley K, Roberts T, Black G, Walzl G, Kaufmann SH. 2011. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 12, 15–22. ( 10.1038/gene.2010.51) [DOI] [PubMed] [Google Scholar]
- 45.Lesho E, et al. 2011. Transcriptional responses of host peripheral blood cells to tuberculosis infection. Tuberculosis 91, 390–399. ( 10.1016/j.tube.2011.07.002) [DOI] [PubMed] [Google Scholar]
- 46.Maertzdorf J, Ota M, Repsilber D, Mollenkopf HJ, Weiner J, Hill PC, Kaufmann SH. 2011. Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PLoS ONE 6, e26938 ( 10.1371/journal.pone.0026938) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maertzdorf J, Weiner J, 3rd, Mollenkopf HJ, Network TB, Bauer T, Prasse A, Muller-Quernheim J, Kaufmann SH. 2012. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc. Natl Acad. Sci. USA 109, 7853–7858. ( 10.1073/pnas.1121072109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cliff JM, et al. 2013. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J. Infect. Dis. 207, 18–29. ( 10.1093/infdis/jis499) [DOI] [PubMed] [Google Scholar]
- 49.Kaforou M, et al. 2013. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case–control study. PLoS Med. 10, e1001538 ( 10.1371/journal.pmed.1001538) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Querec TD, et al. 2009. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 10, 116–125. ( 10.1038/ni.1688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MPR. 2013. The immune response in tuberculosis. Annu. Rev. Immunol. 31, 475–527. ( 10.1146/annurev-immunol-032712-095939) [DOI] [PubMed] [Google Scholar]
- 52.Annonymous. 2012. Global tuberculosis report 2012. Geneva, Switzerland: World Health Organisation.
- 53.Annonymous. 2010 The global plan to Stop TB 2011–2015. Geneva, Switzerland: World Health Organisation.
- 54.Vynnycky E, Fine PE. 2000. Lifetime risks, incubation period, and serial interval of tuberculosis. Am. J. Epidemiol. 152, 247–263. ( 10.1093/aje/152.3.247) [DOI] [PubMed] [Google Scholar]
- 55.Keane J, Gershon S, Wise RP, Mirabile-Levens E, Kasznica J, Schwieterman WD, Siegel JN, Braun MM. 2001. Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N. Engl. J. Med. 345, 1098–1104. ( 10.1056/NEJMoa011110) [DOI] [PubMed] [Google Scholar]
- 56.Havlir DV, Barnes PF. 1999. Tuberculosis in patients with human immunodeficiency virus infection. N. Engl. J. Med. 340, 367–373. ( 10.1056/NEJM199902043400507) [DOI] [PubMed] [Google Scholar]
- 57.Breen RA, et al. 2006. Adverse events and treatment interruption in tuberculosis patients with and without HIV co-infection. Thorax 61, 791–794. ( 10.1136/thx.2006.058867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pitt JM, Blankley S, McShane H, O'Garra A. 2013. Vaccination against tuberculosis: how can we better BCG? Microb. Pathog. 58, 2–16. ( 10.1016/j.micpath.2012.12.002) [DOI] [PubMed] [Google Scholar]
- 59.Colditz GA, Brewer TF, Berkey CS, Wilson ME, Burdick E, Fineberg HV, Mosteller F. 1994. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. J. Am. Med. Assoc. 271, 698–702. ( 10.1001/jama.1994.03510330076038) [DOI] [PubMed] [Google Scholar]
- 60.Fine PE. 1995. Variation in protection by BCG: implications of and for heterologous immunity. Lancet 346, 1339–1345. ( 10.1016/S0140-6736(95)92348-9) [DOI] [PubMed] [Google Scholar]
- 61.Mangtani P, et al. 2013. Protection by BCG against tuberculosis: a systematic review of randomised controlled trials. Clin. Infect. Dis. 58, 470–480. ( 10.1093/cid/cit790) [DOI] [PubMed] [Google Scholar]
- 62.Kagina BM, et al. 2010. Specific T cell frequency and cytokine expression profile do not correlate with protection against tuberculosis after bacillus Calmette–Guerin vaccination of newborns. Am. J. Respir. Crit. Care Med. 182, 1073–1079. ( 10.1164/rccm.201003-0334OC) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaufmann SH. 2012. Tuberculosis vaccine development: strength lies in tenacity. Trends Immunol. 33, 373–379. ( 10.1016/j.it.2012.03.004) [DOI] [PubMed] [Google Scholar]
- 64.Tameris MD, et al. 2013. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet 381, 1021–1028. ( 10.1016/S0140-6736(13)60177-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scriba TJ, et al. 2011. Dose-finding study of the novel tuberculosis vaccine, MVA85A, in healthy BCG-vaccinated infants. J. Infect. Dis. 203, 1832–1843. ( 10.1093/infdis/jir195) [DOI] [PubMed] [Google Scholar]
- 66.Verreck FA, et al. 2009. MVA.85A boosting of BCG and an attenuated, phoP deficient M. tuberculosis vaccine both show protective efficacy against tuberculosis in rhesus macaques. PLoS ONE 4, e5264 ( 10.1371/journal.pone.0005264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berry MP, Blankley S, Graham CM, Bloom CI, O'Garra A. 2013. Systems approaches to studying the immune response in tuberculosis. Curr. Opin. Immunol. 25, 579–587. ( 10.1016/j.coi.2013.08.003) [DOI] [PubMed] [Google Scholar]
- 68.Lu C, et al. 2011. Novel biomarkers distinguishing active tuberculosis from latent infection identified by gene expression profile of peripheral blood mononuclear cells. PLoS ONE 6, e24290 ( 10.1371/journal.pone.0024290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, Musser JM, Barry CE, 3rd, Freedman VH, Kaplan G. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc. Natl Acad. Sci. USA 98, 5752–5757. ( 10.1073/pnas.091096998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manca C, Tsenova L, Freeman S, Barczak AK, Tovey M, Murray PJ, Barry C, Kaplan G. 2005. Hypervirulent M. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak-Stat pathway. J. Interferon Cytokine Res. 25, 694–701. ( 10.1089/jir.2005.25.694) [DOI] [PubMed] [Google Scholar]
- 71.Joosten SA, Fletcher HA, Ottenhoff TH. 2013. A helicopter perspective on TB biomarkers: pathway and process based analysis of gene expression data provides new insight into TB pathogenesis. PLoS ONE 8, e73230 ( 10.1371/journal.pone.0073230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koth LL, Solberg OD, Peng JC, Bhakta NR, Nguyen CP, Woodruff PG. 2011. Sarcoidosis blood transcriptome reflects lung inflammation and overlaps with tuberculosis. Am. J. Respir. Crit. Care Med. 184, 1153–1163. ( 10.1164/rccm.201106-1143OC) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. 2014. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS ONE 9, e78644 ( 10.1371/journal.pone.0078644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. ( 10.1038/nrg2484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Churbanov A, Milligan B. 2012. Accurate diagnostics for bovine tuberculosis based on high-throughput sequencing. PLoS ONE 7, e50147 ( 10.1371/journal.pone.0050147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sheridan C. 2014. Milestone approval lifts Illumina's NGS from research into clinic. Nat. Biotechnol. 32, 111–112. ( 10.1038/nbt0214-111) [DOI] [PubMed] [Google Scholar]
- 77.Sambarey A, Prashanthi K, Chandra N. 2013. Mining large-scale response networks reveals ‘topmost activities’ in Mycobacterium tuberculosis infection. Sci. Rep. 3, 2302 ( 10.1038/srep02302) [DOI] [PMC free article] [PubMed] [Google Scholar]