Abstract
Once referred to as “short-axon” neurons by Cajal, GABA (gamma-amino butyric acid)-ergic interneurons are essential components of the neocortex. They are distributed throughout the cortical laminae and are responsible for shaping circuit output through a rich array of inhibitory mechanisms. Numerous fate-mapping and transplantation studies have examined the embryonic origins of the diversity of interneurons that are defined along various parameters such as morphology, neurochemical marker expression and physiological properties, and have been extensively reviewed elsewhere. Here, we focus on discussing two recent studies that have, for the first time, examined the production and organization of neocortical interneurons originated from individual progenitors, i.e. with clonal resolution, and provided important new insights into the cellular processes underlying the development of inhibitory interneurons in the neocortex.
Introduction
GABAergic interneurons comprise ~20% of the neuronal population in the neocortex. In addition to providing local inhibition and shaping circuit output, they are crucial in generating network oscillations, thereby further influencing the response of the circuit to incoming signals. Born in the ventral regions of the developing telencephalon, neocortical interneurons are a diverse cell population and have been extensively characterized based on their morphological, neurochemical, and physiological properties [1–7]. Previous fate-mapping and transplantation studies have suggested that, similar to the dorsally-derived excitatory neurons, the developmental history (i.e. place and time of birth) of inhibitory interneurons has a strong influence on their subtype specification and distribution in the mature neocortex [8–16]. In the case of excitatory neurons, lineage history not only contributes to the spatial/structural organization, but also influences the functional development of the neocortex as excitatory cells derived from the same progenitor cell exhibit preferential (both electrical and chemical synapse-based) connectivity and similar physiological properties amongst each other in comparison to nearby non-lineage related cells [17–19].
However, unlike neocortical excitatory neurons, virtually nothing is known about the lineage development of interneurons at the single progenitor cell level. Thus, fundamental questions about the production, specification, and organization of interneurons that underlie the construction of functional neocortical circuits remain largely open. For instance, do individual progenitors produce the same subtype or different subtypes of interneurons? How heterogeneous are the progenitors spatially and temporally regarding their proliferative behavior and neuronal output? Does the lineage relationship of interneurons influence their spatial and functional organization? Considering the incredible diversity of interneuron subtypes that carry out distinct essential functions in the neocortex, lineage analysis of interneuron progenitors can certainly help unravel how diverse components of this fundamental cell population are produced and assembled structurally and functionally, by following the behavior of one progenitor cell at a time.
Answering these questions requires clonal analysis of interneuron production and organization. By exploiting retrovirus-mediated gene transfer in conjunction with mouse genetics, Brown et al. (2011) [20] and Ciceri et al. (2013) [21] specifically labeled individual progenitor cells in the embryonic medial ganglionic eminence (MGE) and preoptic area (PoA), which are responsible for producing more than 70% of neocortical interneurons, and analyzed the behavior of individual progenitors and their progeny from birth to maturation.
In the first study, the specificity is achieved by taking advantage of the exquisite fidelity of the subgroup A avian sarcoma leukosis virus (ASLV)-receptor interaction [22]. TVA, the cognate receptor for ASLV, was selectively expressed in the progenitor cells in the MGE and PoA by crossing R26LSL-TVAiLacZ, a knock-in mouse line that expresses TVA in a Cre recombinase-dependent manner [23], with an Nkx2.1-Cre transgenic mouse line [15]. Nkx2.1 encodes a homeobox transcription factor specifically expressed in the progenitor cells in the MGE and the PoA [10, 15, 24]. Dividing progenitors expressing TVA at the ventricular zone (VZ) surface were then labeled by performing in utero intraventricular injection of low-titer RCAS (replication-competent ASLV long terminal repeat with splice acceptor) retrovirus expressing fluorescent proteins at embryonic day (E) 11 to 12 [20].
In the second study, the specificity is achieved by using retroviruses carrying a reversed and double-floxed cDNA sequence encoding a fluorescent protein. Thus, while the retroviruses infect progenitors indiscriminately, only those expressing Cre recombinase are capable of inverting the cDNA sequence for expression by recombination and are subsequently labeled [21]. It is worth noting, however, that the Cre-mediated inversion is reversible so that in the continued presence of Cre, the cDNA sequence can become reversed again, and therefore its expression is not stable until additional recombination to permanently delete a set of flox sequences [21].
Clonal production of neocortical interneurons
While progenitors in the VZ of the dorsal telencephalon responsible for producing neocortical excitatory neurons have been extensively characterized, progenitors in the ventral telencephalon including the MGE are less understood. By labeling and characterizing individual progenitors, both studies demonstrated that progenitors in the VZ of the MGE and PoA are radial glial cells in nature. They exhibited the defining morphological characteristics including a cell body in the VZ, a short process reaching the ventricular surface with a large endfoot and a long fine radial process directed toward the pial surface [20, 21]. They expressed astrocyte-specific glutamate transporter (GLAST) and brain-lipid-binding protein (BLBP), two proteins known to be specifically expressed in radial glial cells, as well as the neural progenitor marker, Nestin [20].
Clonal clusters labeled in the MGE and PoA at the embryonic stages mostly contained a single mitotic radial glial progenitor (RGP) and a few short-process cells that closely associated with the long radial glial process (Figure 1, left). The progressive increase in the number of short-process cells suggests that the RGP undergoes asymmetric divisions to self-renew, and simultaneously produce short-process cells. Indeed, using time-lapse imaging in organotypic slice cultures, Brown et al. directly observed asymmetric divisions of RGPs. RGPs displayed interkinetic nuclear migration and divided at the VZ surface. After mitosis, while one of the two daughter cells remained as a bipolar RGP, the other daughter cell initially moved along the renewed RGP away from the VZ and then detached and migrated tangentially towards the neocortex. In some asymmetric divisions, the resulting non-radial glial daughter cell reentered the cell cycle and divided again in the subventricular zone (SVZ), likely representing intermediate progenitors (IPs) in the region.
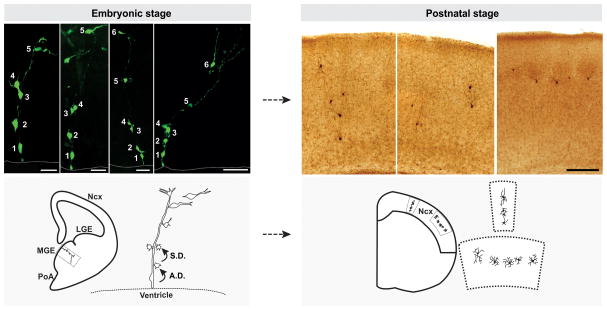
Figure 1. Clonal production and organization of neocortical interneurons.
(Left) In the embryonic MGE, RGPs divide asymmetrically at the VZ surface to self-renew and to simultaneously produce differentiating interneurons or IPs, which then divide symmetrically in the SVZ to produce differentiating interneurons. The progeny (cell 2–6) of the same RGP (cell 1) are initially closely associated with the mother RGP and organized into radially aligned clonal clusters. As development proceeds, the early-born cells progressively move away from the VZ, acquire the characteristic morphological and biophysical features of differentiating interneurons, and migrate tangentially towards the neocortex. Broken lines represent the VZ surface. (Right) After arriving at their destination in the neocortex, inhibitory interneuron clones do not randomly disperse, but form spatially organized vertical or horizontal clusters. Images of raw data are shown at the top and schematics are shown at the bottom. Scale bars (from left to right): 25 μm, 25 μm, 50 μm, 50 μm and 250 μm. Ncx, neocortex; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; PoA, preoptic area; A.D., asymmetric division; S.D., symmetric division. Adapted from Brown et al. [20].
These divisions in the SVZ appeared to be symmetric, producing two daughter cells with similar morphology and cellular behavior. Interestingly, simultaneous or synchronous symmetric divisions in the SVZ were frequently observed within a clone, suggesting that IPs in the MGE undergo multiple rounds of symmetric division. In comparison, IPs in the dorsal telencephalon mostly undergo one round of symmetric division [25]. The relative size of the SVZ compared to the VZ in the ventral telencephalon is substantially larger than that in the dorsal telencephalon [26]. Related to this, while the divisions in the dorsal telencephalon occur mostly at the VZ surface, substantial divisions in the ventral telencephalon take place in the SVZ. Together, these observations suggest that synchronous symmetric IP divisions may be a crucial feature of interneuron neurogenesis in the MGE.
MGE-originated interneurons exhibit a birth date-dependent inside-out distribution in the neocortex [27]. Within a clone, asymmetric divisions of the RGP generate interneurons at different times, whereas synchronous symmetric divisions of the IP generate interneurons around the same time. Therefore, progenitor division patterns may fundamentally influence the distribution and differentiation of clonally related neocortical interneurons. A direct comparison between RGP lineages and IP lineages will provide crucial insights into this; however, this requires the identification and characterization of IP-specific markers in the MGE. It has been postulated that cyclins D1 and D2 are expressed in distinct progenitor niches, with cyclin D1 predominantly found in the VZ (likely in RGPs), and cyclin D2 in the SVZ (likely in IPs) of the cerebral cortex and the ganglionic eminence (GE) [28]. Other potential candidates for IP lineages in the MGE include Mash1/Asc1 and Dlx1/5 [29–32].
In embryonic stages, the daughter cells including IPs and differentiating interneurons appeared closely associated with the radial glial process of the RGP initially. Brown et al noticed that the cells located farthest away from the VZ often acquired the typical bipolar morphology of tangentially migrating interneurons [20]. Moreover, they were positive for TuJ1, a differentiated neuronal marker, and possessed Na+ conductance. These observations raised the possibility that associating with the mother radial glial fiber is critical for proper interneuron differentiation and that acquiring differentiating interneuron properties is a prerequisite for commencing tangential migration. Should this be the case, the morphology and property of radial glial fibers may also play crucial roles in regulating neocortical interneuron differentiation and distribution.
Clustering of clonally related neocortical interneurons
Ventrally derived interneurons embark on a long, tangential journey to reach their destination in the neocortex. Previous studies suggest that this migration process appears to be random [33, 34]. Unexpected from what would be predicted from a random migration, both studies showed that interneurons labeled at the clonal density do not randomly disperse, but form spatially isolated clusters in the neocortex (Figure 1, right) [20, 21].
In Brown et al. study, the authors relied on the known fact of the spatial clustering of excitatory neuron clones in the neocortex, and systematically compared and contrasted the distribution of clonally related neocortical excitatory neurons and interneurons labeled using the same strategy. After serial sectioning and three-dimensional reconstruction to recover all labeled neurons in the neocortex, the nearest neighbor distance (NND) analysis was applied, which reports the spacing pattern of the data points within the datasets. The authors found that clonally related interneurons exhibited shorter NNDs than randomly simulated datasets, suggesting a clustering feature in their distribution. Using the same spatial criteria that reliably identify the vertical (within 300 μm2 spanning all the layers from the pia to the white matter) and horizontal clusters of sparsely labeled clonally related excitatory neurons, the authors showed that clonally related interneurons also form vertical or horizontal clusters consisting of three or more interneurons. While horizontal clusters are typically restricted to one or two adjacent layers, vertical clusters often span two or more layers. Inferring from the birth date-dependent distribution, prominent horizontal clusters most probably arise from clones with predominant IP symmetric divisions that produce interneurons at similar time, whereas prominent vertical clusters likely arise from clones with robust consecutive RGP asymmetric divisions that produce interneurons at different times. Notably, the size and distribution of clusters were clearly variable, suggesting a significant degree of heterogeneity in lineage progression, migration, differentiation and maturation of individual clones. In addition, extensive neonatal death may also further sculpt clonally related interneuron clusters [35].
In Ciceri et al. study, the same NND analysis was used to demonstrate the clustering of clonally related neocortical interneurons. The authors further characterized the clusters identified based on agglomerative hierarchical clustering methods, where interneurons were grouped according to proximity relationships. The number of clusters in the experimental datasets was then calculated using a threshold distance value that maximized the difference between the numbers of clusters observed in the experimental datasets and the mean number of clusters of randomly simulated data points. While the biological significance of this analytic process was not immediately clear, the threshold distance obtained with this method was about 300 μm, comparable to that in Brown et al. study. However, this threshold distance was applied in all dimensions, including the radial/vertical direction (i.e. not including all the layers). The average cluster size was significantly smaller than that of Brown et al. and there were many 2-cell clusters (13 out of 20). While some clusters spanned two non-adjacent or more than three layers, most clusters were located within the same or two adjacent layers. Furthermore, the authors showed that all intralaminar clusters were isochronic, whereas all clusters that spanned several layers were heterochronic. This birth date profile of the clusters is consistent with the possibility that intralaminar clusters probably represent clonally related interneurons originating from symmetric IP division(s) and interlaminar clusters likely represent those arising from asymmetric RGP divisions. Moreover, the smaller intralaminar clusters may represent a fraction of the clones produced by RGP asymmetric divisions as well as IP symmetric divisions, as the cluster detection method in this study does not include any cells more than 300 μm apart even if they are distinctly located within a limited distance, e.g. radially in non-adjacent layers.
Notably, Ciceri et al. implied that the lineage relationship might not be the only factor contributing to interneuron clustering in the neocortex, as they suggested some mixing of clones. While it is possible that two nearby clones labeled at the embryonic stages remain closely distributed in the mature neocortex, it appeared rare that two adjacent clones were labeled simultaneously by low-titer retrovirus [20]. The nature of any non-lineage factors that may affect interneuron clustering in the neocortex is currently unclear. A previous study suggested that excitatory neurons can influence the laminar distribution of interneuron populations in the neocortex [36].
Importantly, by using different Cre lines, Ciceri et al. demonstrated that lineage-dependent clustering appears to be a general feature of neocortical interneurons, including those derived from the caudal ganglionic eminence (CGE). While the precise processes that lead to cluster formation among clonally related interneurons remain to be determined, it is tempting to think that the migration of clonally related interneurons is coordinated to maintain their spatial organization from embryonic origins to final destination in the neocortex.
Subtype composition of neocortical interneuron clonal clusters
Clustering of clonally related neocortical interneurons allows for the first time to examine the progeny fate potential of individual progenitors. MGE-derived neocortical interneurons predominantly express neurochemical marker parvalbumin (PV) or somatostatin (SOM) [5, 10, 14, 37]. In Brown et al. study, about 25% (19 out of 75) of isolated vertical clusters contained interneurons expressing either only PV (~17%) or only SOM (~8%). The remaining 75% contained both PV+ and SOM+ interneurons [20]. In comparison, in Ciceri et al. study, about 53% (75 out of 143) of clusters contained interneurons expressing either only PV (~48%) or only SOM (~5%). The remaining 47% of clusters contained both PV+ and SOM+ interneurons [21].
Both studies observed a significant number of interneuron clusters containing both PV+ and SOM+ interneurons, suggesting that individual RGPs in the MGE and PoA are capable of generating more than one interneuron subtype. Previous transplantation studies showed both spatial and temporal bias of interneuron subtypes derived from the MGE [8, 13, 16], indicating the heterogeneity of MGE progenitors at different time points and along the dorsal-ventral axis. It is worth noting, however, that the transplantation procedure might have pushed progenitors unintentionally out of the cell cycle towards differentiation, thereby potentially reducing the diversity arising from consecutive asymmetric divisions of RGPs. Interestingly, Ciceri et al. observed more than twice as many clusters containing PV+ only or SOM+ only interneurons compared to Brown et al. This difference may reflect that the generally smaller clonal clusters in Ciceri et al. are potential partial clones originating from symmetric IP division(s), which may occupy similar layer and develop into similar subtypes.
Notably, Ciceri et al. reported that MGE progenitors labeled at E11.5 gave rise to PV+ and SOM+ interneurons in a 3:1 proportion. This is not very consistent with a previous fate-mapping study where in both E10.5 and E12.5 labeling, the number of PV+ interneurons is no more than twice that of SOM+ interneurons [11]. Although the 3:1 ratio matches the final proportion of these two interneuron subtypes reported in the rat prefrontal cortex in one study [38], more recent experiments have shown that PV+ and SOM+ comprise ~40% and ~30% of the total neocortical GABAergic interneurons, respectively [39]. In addition, it has been shown that whereas PV+ interneurons are continuously produced throughout the neurogenic period, SOM+ interneurons are generated relatively earlier [11, 16], which would predict that E11.5 progenitors should produce a proportion of PV:SOM-expressing cells lower than 3:1. Related to this, Ciceri et al. showed that the fraction of clonal clusters labeled in PV-Cre (66%) or SOM-Cre (54%) mice with three or more interneurons was substantially larger than that in Nkx2.1-Cre (35%) mice. As a result, the average size of clonal clusters labeled in PV-Cre (2.9±0.1 cells) mice was significantly larger than that labeled in Nkx2.1-Cre (2.5±0.2 cells) mice. Considering that the Nkx2.1 lineage produces both PV+ and SOM+ interneurons, it is intriguing that the clonal clusters labeled by Nkx2.1-Cre are on average smaller than those labeled by PV-Cre.
Future directions
The first two clonal studies have substantially expanded our knowledge of neocortical interneuron production and organization. It is evident from these studies that interneuron progenitors are heterogeneous in their proliferative behavior, which may fundamentally contribute to the diversity in progeny cell type (or subtype) and their localization. In the case of dorsally-derived excitatory neurons, it has long been thought that the fate potential of a largely common progenitor pool changes over time to generate different layers of excitatory neurons in a temporally defined manner. Interestingly, this prevailing model of a progressive restriction of the neurogenic potential of RGPs was recently challenged; Franco et al. showed that a fate-restricted RGP population expressing transcription factor CUX2 gives rise to only superficial-layer excitatory neurons [40]. This observation has led the way to a model, as reflected in Ciceri et al., where multiple fate-restricted progenitor pools exist within the germinal zones of the developing ventral telencephalon, and generate interneurons destined for different layers of the neocortex [41, 42]. Notably, contrary to Franco et al., a recent study showed that the CUX2-expressing RGPs do produce both deep- and superficial-layer excitatory neurons in the neocortex [43].
How about neocortical interneuron genesis? Based on the analyses of RGP behavior and lineage progression, we suggest the following model. A majority of RGPs divide actively from the beginning of neurogenesis (Figure 2A, red colored); they might keep proliferating through the entire period of neurogenesis, generating interneurons for all cortical laminae, or they might be progressively depleted after several rounds of division generating neocortical interneurons located mostly in deep layers. In contrast, some RGPs may be less mitotically active in the beginning and start dividing later on (Figure 2A, blue colored); they would preferentially generate neocortical interneurons localized in superficial or any selective layers. The relative proportion and/or fate potential of different populations of RGPs could change during neurogenesis. Within individual clones, there are interneurons generated directly by the RGP through asymmetric divisions and those generated by the IP through one or more rounds of symmetric division [20]. Those generated at different times likely occupy different cortical layers [20], whereas those generated concurrently probably occupy one or two adjacent cortical layers and might also develop into similar subtypes [21].
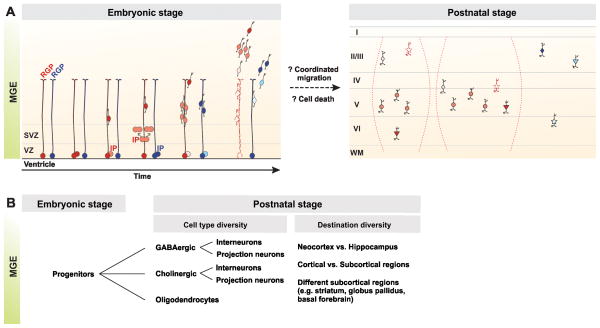
Figure 2. A model of clonal origins of neocortical interneuron diversity and distribution.
(A) RGPs in the VZ of the MGE are likely heterogeneous in proliferative behavior and neurogenesis potential. A majority of RGPs (red colored) actively divide starting at the beginning of neurogenesis. They undergo asymmetric divisions to give rise to an interneuron or an IP, which divides symmetrically one or more rounds to produce a number of interneurons concurrently. As time proceeds, these RGPs may progressively be depleted or be maintained to generate more interneurons (broken lines and symbols), resulting in an interneuron lineage that form a vertical or horizontal cluster (in broken parentheses) with distinct subtypes (represented by shapes) in the mature neocortex, likely via coordinated migration. In comparison, some RGPs (blue colored) only become mitotically active at the late stage of neurogenesis. They may generate a lineage of a few interneurons that are restricted in defined layer(s). Cell death may further sculpt individual lineages in composition and distribution. (B) MGE progenitors produce neurons (including GABAergic interneurons and projection neurons, as well as cholinergic interneurons and projection neurons) and oligodendrocytes that are distributed in the neocortex, as well as the hippocampus and subcortical regions, e.g. the striatum, globus pallidus, and basal forebrain. It remains unclear whether individual progenitors produce the same or distinct cell types that are distributed in one or different brain regions.
It is important to point out that progenitors in the VZ of the ventral telencephalon including the MGE produce neurons and oligodendrocytes that are not only distributed in the neocortex, but also in the hippocampus and many subcortical regions, e.g. the striatum, the globus pallidus and the basal forebrain (Figure 2B) [15, 24, 44–46]. At the clonal level, however, whether neocortical, hippocampal and subcortical interneurons are generated by distinct RGPs is unclear. Similarly, whether individual RGPs can give rise to both neurons and oligodendrocytes remains unknown. MGE progenitors also produce cholinergic neurons [15]. Considering the progeny heterogeneity, both in terms of cell type/subtype specification and the place they ultimately inhabit, it is likely that the progenitor dynamics in the MGE are much more complex than our current understanding.
To experimentally address these fundamental questions, it is necessary to perform systematic clonal analysis of the ventral telencephalon progenitors with exquisite spatial and temporal control as well as a better resolution of division patterns. For instance, recently developed mosaic analysis with double markers (MADM) is a powerful genetic method which, when combined with CreER transgenic mouse lines, allows for simultaneous labeling of two daughter cells of a dividing progenitor with two distinct colors for lineage tracing in a spatially and temporally defined manner [47, 48].
Furthermore, the molecular underpinning of progenitor heterogeneity and lineage progression remains to be determined. In the Drosophila nervous system, it has been elegantly demonstrated that the rich diversity of cell types can be achieved as neuronal progenitors change over time by producing successive waves of regulatory proteins [49, 50]. The mouse subpallial progenitor zone is heterogeneous and can be divided into at least 18 domains based on the combinatorial expression of several transcription factors [9]. Rapid progress in single cell transcriptome analysis enables systematic investigation of the gene expression profile of individual progenitors or clones to reveal molecular signatures that define distinct lineages originated from the ventral telencephalon [51, 52].
Last but not the least, the physiological implications of the clustering of clonally related interneurons in the neocortex has not been explored. For instance, do electrical and/or chemical synapses preferentially form between clonally related interneurons within individual clusters? Do sister interneurons share common presynaptic input and/or have common postsynaptic targets, which would indicate that clonally related inhibitory interneurons act together in a network? These studies will likely provide fundamental insights into the assembly and operation of functional circuits in the neocortex.
Highlights.
Neocortical interneurons are produced as spatially organized clonal units in MGE.
Clonally related interneurons form spatially isolated clusters in mature neocortex.
Individual clonal clusters can contain more than one interneuron subtype.
Progenitor division modes may influence interneuron diversity and distribution.
Acknowledgments
We apologize to the authors whose work we could not cite owing to space limitations. We thank Dr. Z. Josh Huang, Dr. Stewart A. Anderson, Dr. Xinjun Zhang and Ryan Insolera for critical reading of the manuscript. Our research was supported by grants from the National Institutes of Health (R01DA024681 and P01NS048120), the McKnight Foundation and the SimonsFoundation (to S. -H.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Ascoli GA, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 2008;9(7):557–68. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeFelipe J, et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat Rev Neurosci. 2013;14(3):202–16. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fishell G, Rudy B. Mechanisms of inhibition within the telencephalon: "where the wild things are". Annu Rev Neurosci. 2011;34:535–67. doi: 10.1146/annurev-neuro-061010-113717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maass W, Natschlager T, Markram H. Fading memory and kernel properties of generic cortical microcircuit models. J Physiol Paris. 2004;98(4–6):315–30. doi: 10.1016/j.jphysparis.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 5.Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7(9):687–96. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- 6.Freund TF. Interneuron Diversity series: Rhythm and mood in perisomatic inhibition. Trends Neurosci. 2003;26(9):489–95. doi: 10.1016/S0166-2236(03)00227-3. [DOI] [PubMed] [Google Scholar]
- 7.McBain CJ, Fisahn A. Interneurons unbound. Nat Rev Neurosci. 2001;2(1):11–23. doi: 10.1038/35049047. [DOI] [PubMed] [Google Scholar]
- 8.Butt SJ, et al. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48(4):591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 9.Flames N, et al. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27(36):9682–95. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogarty M, et al. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci. 2007;27(41):10935–46. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyoshi G, et al. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27(29):7786–98. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taniguchi H, Lu J, Huang ZJ. The spatial and temporal origin of chandelier cells in mouse neocortex. Science. 2013;339(6115):70–4. doi: 10.1126/science.1227622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wonders CP, et al. A spatial bias for the origins of interneuron subgroups within the medial ganglionic eminence. Dev Biol. 2008;314(1):127–36. doi: 10.1016/j.ydbio.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Q, et al. Origins of cortical interneuron subtypes. J Neurosci. 2004;24(11):2612–22. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506(1):16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- 16.Inan M, Welagen J, Anderson SA. Spatial and temporal bias in the mitotic origins of somatostatin- and parvalbumin-expressing interneuron subgroups and the chandelier subtype in the medial ganglionic eminence. Cereb Cortex. 2012;22(4):820–7. doi: 10.1093/cercor/bhr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu YC, et al. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458(7237):501–4. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu YC, et al. Preferential electrical coupling regulates neocortical lineage-dependent microcircuit assembly. Nature. 2012;486(7401):113–7. doi: 10.1038/nature10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, et al. Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature. 2012;486(7401):118–21. doi: 10.1038/nature11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Brown KN, et al. Clonal production and organization of inhibitory interneurons in the neocortex. Science. 2011;334(6055):480–6. doi: 10.1126/science.1208884. This study is the first specific clonal analysis of GABAergic neocortical interneurons. The authors found that neocortical interneurons are produced as spatially organized clonal units during neurogenesis and clonally related interneurons form discrete clusters in the mature neocortex, with vertical or horizontal organization. Moreover, clonally related interneurons express the same or distinct neurochemical markers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Ciceri G, et al. Lineage-specific laminar organization of cortical GABAergic interneurons. Nat Neurosci. 2013;16(9):1199–210. doi: 10.1038/nn.3485. This study confirms the clustering property of clonally related neocortical interneurons. In addition, the authors demonstrate that this property is common to different classes of interneurons, independent of their origin. Moreover, they found that individual interneuron clonal clusters are typically distributed in one or two adjacent layers and consisted of iscronically generated neurons. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert JM, et al. The receptor for the subgroup A avian leukosis-sarcoma viruses binds to subgroup A but not to subgroup C envelope glycoprotein. J Virol. 1994;68(9):5623–8. doi: 10.1128/jvi.68.9.5623-5628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seidler B, et al. A Cre-loxP-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors. Proc Natl Acad Sci U S A. 2008;105(29):10137–42. doi: 10.1073/pnas.0800487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20(16):6063–76. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noctor SC, et al. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7(2):136–44. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 26.Hansen DV, et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013 doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valcanis H, Tan SS. Layer specification of transplanted interneurons in developing mouse neocortex. J Neurosci. 2003;23(12):5113–22. doi: 10.1523/JNEUROSCI.23-12-05113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glickstein SB, Alexander S, Ross ME. Differences in cyclin D2 and D1 protein expression distinguish forebrain progenitor subsets. Cereb Cortex. 2007;17(3):632–42. doi: 10.1093/cercor/bhk008. [DOI] [PubMed] [Google Scholar]
- 29.Bulfone A, et al. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci. 1993;13(7):3155–72. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126(3):525–34. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- 31.Petryniak MA, et al. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55(3):417–33. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long JE, et al. Dlx1&2 and Mash1 transcription factors control MGE and CGE patterning and differentiation through parallel and overlapping pathways. Cereb Cortex. 2009;19(Suppl 1):i96–106. doi: 10.1093/cercor/bhp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka DH, et al. Multidirectional and multizonal tangential migration of GABAergic interneurons in the developing cerebral cortex. Development. 2006;133(11):2167–76. doi: 10.1242/dev.02382. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka DH, et al. Random walk behavior of migrating cortical interneurons in the marginal zone: time-lapse analysis in flat-mount cortex. J Neurosci. 2009;29(5):1300–11. doi: 10.1523/JNEUROSCI.5446-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Southwell DG, et al. Intrinsically determined cell death of developing cortical interneurons. Nature. 2012;491(7422):109–13. doi: 10.1038/nature11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lodato S, et al. Excitatory projection neuron subtypes control the distribution of local inhibitory interneurons in the cerebral cortex. Neuron. 2011;69(4):763–79. doi: 10.1016/j.neuron.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Q, et al. Sonic hedgehog signaling confers ventral telencephalic progenitors with distinct cortical interneuron fates. Neuron. 2010;65(3):328–40. doi: 10.1016/j.neuron.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawaguchi Y, Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex. 1997;7(6):476–86. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- 39.Rudy B, et al. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71(1):45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franco SJ, et al. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337(6095):746–9. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartolini G, Ciceri G, Marin O. Integration of GABAergic interneurons into cortical cell assemblies: lessons from embryos and adults. Neuron. 2013;79(5):849–64. doi: 10.1016/j.neuron.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 42.Marin O. Brain development: The neuron family tree remodelled. Nature. 2012;490(7419):185–6. doi: 10.1038/490185a. [DOI] [PubMed] [Google Scholar]
- 43.Guo C, et al. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron. 2013;80(5):1167–74. doi: 10.1016/j.neuron.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butt SJ, et al. The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron. 2008;59(5):722–32. doi: 10.1016/j.neuron.2008.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kessaris N, et al. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9(2):173–9. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKinsey GL, et al. Dlx1&2-dependent expression of Zfhx1b (Sip1, Zeb2) regulates the fate switch between cortical and striatal interneurons. Neuron. 2013;77(1):83–98. doi: 10.1016/j.neuron.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tasic B, et al. Extensions of MADM (mosaic analysis with double markers) in mice. PLoS One. 2012;7(3):e33332. doi: 10.1371/journal.pone.0033332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zong H, et al. Mosaic analysis with double markers in mice. Cell. 2005;121(3):479–92. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 49•.Bayraktar OA, Doe CQ. Combinatorial temporal patterning in progenitors expands neural diversity. Nature. 2013;498(7455):449–55. doi: 10.1038/nature12266. This study reports that five transcription factors are sequentially expressed in individual Drosophila medulla neuroblasts as they age. This precise temporal patterning together with Notch-dependent fate choice control the neuronal fate of the progeny. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Li X, et al. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 2013;498(7455):456–62. doi: 10.1038/nature12319. This paper demonstrates that Drosophila intermediate progenitors (INPs) sequentially express three transcription factors which are required for generating distinct types of neurons over time. Similarly, Drosophila type II neuroblasts also sequentially express a different set of transcription factors to produce distinct progeny along development. Those two temporal axes act combinatorially to achieve the neural diversity in the Drosophila central compex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–82. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 52.Tang F, Lao K, Surani MA. Development and applications of single-cell transcriptome analysis. Nat Methods. 2011;8(4 Suppl):S6–11. doi: 10.1038/nmeth.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]