Abstract
Long noncoding RNAs (lncRNAs) are increasingly recognized to contribute to cellular development via diverse mechanisms during both health and disease. Here, we highlight recent progress on the study of lncRNAs that function in the development of blood cells. We emphasize lncRNAs that regulate blood cell fates through epigenetic control of gene expression, an emerging theme among functional lncRNAs. Many of these noncoding genes and their targets become dysregulated during malignant hematopoiesis, directly implicating lncRNAs in blood cancers such as leukemia. In a few cases, dysregulation of an lncRNA alone leads to malignant hematopoiesis in a mouse model. Thus, lncRNAs may be not only useful as markers for the diagnosis and prognosis of cancers of the blood, but also as potential targets for novel therapies.
Keywords: Hematopoiesis, Long noncoding RNAs (lncRNAs), Leukemia
Introduction
Mammalian genomes comprise thousands of protein-coding and noncoding genes. The latter can produce small structural and regulatory RNAs (including rRNAs, tRNAs, snoRNAs and microRNAs) as well as long noncoding RNAs (lncR-NAs), which are >200 nt yet do not function through an encoded protein product. lncRNAs mirror mRNAs in their biogenesis and regulation: most are Pol II transcripts that undergo splicing, capping and polyadenylation and are regulated by specific transcription factors (TFs) and chromatin modifiers [1, 2]. However, unlike mRNAs they are mainly found in the nucleus [2, 3] and rarely engage translating ribosomes in the cytoplasm [4, 5]. lncRNAs have been implicated in a variety of processes: they can recruit and regulate the activity of TFs and chromatin modifiers, organize chromosomal domains, and modulate mRNA splicing, translation, and degradation [6–8]. Such regulatory capacities are enabled by the versatility with which RNA molecules can interact with DNA, with other RNAs and with protein complexes [9, 10]. Crucial to this versatility in lncRNAs are specific features of primary sequence, secondary structure, and genomic positioning that experience different levels of evolutionary constraint [2, 11, 12].
Many lncRNAs are differentially expressed across tissues, developmental stages and physiological states [2, 13–15]. Some are required for life [16–20], and some are not but fulfill specialized functions in response to specific developmental or physiological cues [21–24]. Thus, lncR-NAs are increasingly recognized as important players during the development and functioning of mammalian cells and tissues. In this review, we summarize recent progress on the study of lncRNAs with roles during normal and malignant hematopoiesis. We highlight lncRNAs that regulate cell fate and differentiation decisions via epigenetic mechanisms, and discuss the clinical relevance of lncRNAs as diagnostic and prognostic biomarkers of hematopoietic malignancies and their potential as therapeutic targets.
lncRNAs during normal hematopoiesis
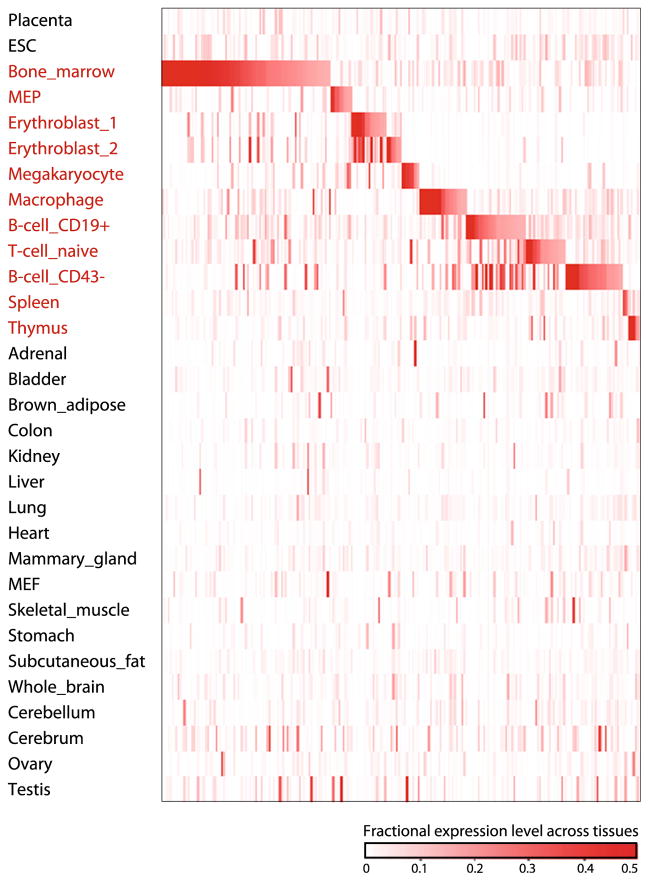
Hematopoiesis is a highly coordinated process comprising a hierarchy of well-defined cell differentiation states, making it an ideal system to identify lncRNAs that regulate cell fate. All hematopoietic effector cells—erythrocytes, myelocytes, and lymphocytes—derive from hematopoietic stem cells (HSCs) through a cascade of cell lineage specification, proliferation, and differentiation events. An analysis of the hematopoietic lineage expression of lncRNAs identified in the fetal liver of E14.5 mice (Fig. 1) reveals many with exquisite lineage specificity, suggesting that distinct collections of lncRNAs modulate the development or functioning of distinct blood cell types. Indeed, recent studies have described several lncRNAs associated with development or function of specific types of blood cells (Table 1; Fig. 2). In the next sections, we will discuss selected examples of functional lncRNAs that illustrate recent advances in the field.
Fig. 1.
Tissue specificity of mouse fetal liver lncRNAs. Relative abundance of lncRNAs (rows) expressed in the E14.5 mouse fetal liver across 30 primary cell and tissue types (columns) from the mouse ENCODE consortium. Color intensity represents the fractional gene-level expression across all tissues examined. Hematopoietic tissues are highlighted in red. Adapted from [44]
Table 1.
Examples of lncRNAs with roles in normal hematopoiesis
| Name | Cell type | Loss of function phenotype | Functional assays | Reference |
|---|---|---|---|---|
| H19 | HSC | Increased activation and proliferation; imparied repopulation ability | Mouse knockout models; transplantation assays | [29] |
| EGO | Eosinophils | Impaired expression of eosinophil differentiation genes | siRNA knockdown | [40] |
| HOTAIRM1 | Myeloid progenitors | Inhibition of the nearby HoxA1 and HoxA4 genes during granulocytic differentiation | siRNA and shRNA knockdown | [41] |
| LincRNA-EPS | Erythroblasts | Compromised differentiation; elevated apoptosis | shRNA knockdown; ectopic expression | [42] |
| DLEU2; elncRNA-EC1,3; lincRNA-EC2,4,5,8,9; alncRNA-EC1,2,3,7 | Erythroblasts | Impaired erythrocyte maturation | shRNA knockdown | [43] |
| NeST | T cells | Sensitivity to viral and bacterial pathogenesis in mice | Ectopic expression | [47, 48] |
| lincRNA-Cox2 | Dendritic cells; macrophages | Impaired gene regulation upon IFN stimulation | siRNA knockdown; overexpression | [52] |
| LincR-Ccr2-5′ AS | T cells | Downregulation of the nearby Ccr1, Ccr2, Ccr3, and Ccr5 genes | shRNA knockdown | [53] |
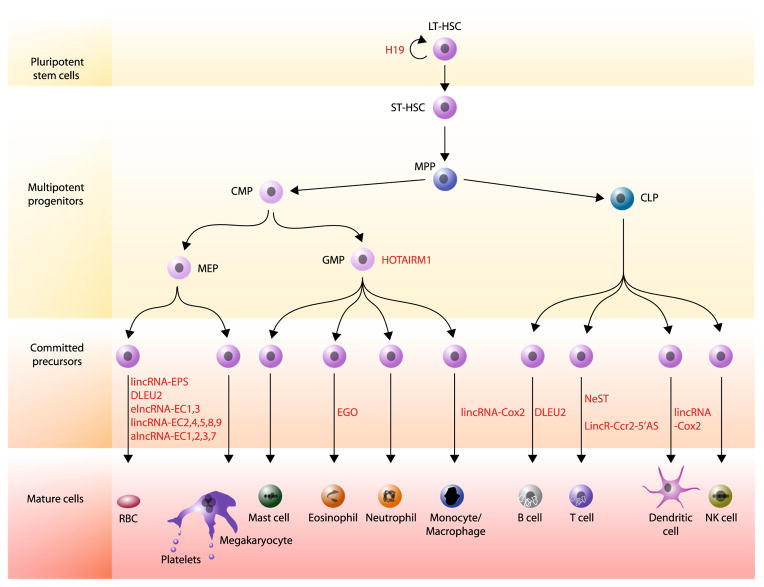
Fig. 2.
Involvement of lncRNAs in blood cell development. lncRNAs required for blood cell development, as determined through loss-of-function studies, are depicted next to the stage of hematopoietic development affected by their inhibition. LT-HSC, long-term hematopoietic stem cell; ST-HSC, short-term hematopoietic cell; MPP, multipotent progenitor; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; MEP, megakaryocyte/erythroid progenitor; GMP; granulocyte/monocyte progenitor; RBC, red blood cell; NK cell, natural killer cell
lncRNAs in hematopoietic stem cells
Self-renewal of hematopoietic stem cells (HSCs) is in part sustained by developmental and environmental signals from their niche, which act through developmentally conserved signaling pathways to promote proliferation while suppressing apoptosis and differentiation [25, 26]. During hematopoiesis, this self-renewing capability decreases as cells transition from multipotent stem cells to lineage-committed progenitors. The fact that lncRNAs have the capacity to modulate self-renewal of embryonic stem cells [27–29] suggests that they too may act in the circuitry controlling the HSC state.
Li and colleagues recently described the first example of lncRNA-mediated maintenance of adult HSC quiescence [30]. The H19 gene was in fact the first lncRNA to be identified, enriched in the embryonic fetal liver but downregulated after birth [31, 32]. The H19 transcript is a Pol II product that undergoes capping and polyadenylation, can be found in both nucleus and cytoplasm, and contains only small ORFs that do not template protein synthesis in vivo. During embryogenesis, H19 is transcribed from the maternal allele and regulates growth via imprinting in cis of Igf2, as well as by encoding miR-675, which down-regulates the receptor of the Igf2 ligand expressed from the paternal allele (Igf1r) [33–35]. H19 remains active in long-term HSCs and is gradually downregulated in short-term HSCs and multipotent progenitors. Deletion of H19 from the maternal allele results in increased HSC activation and proliferation and impairs repopulating ability. As expected, this effect is mediated by de-repression of maternal Igf2 expression and by increased Igf1r translation, resulting in increased signaling through the Igf1r. Overexpression of H19-derived mir-675 restores proper levels of Igf1r protein, and concomitant deletion of the Igf1r locus partially rescues the H19 knockout phenotype. Thus, H19 promotes HSC quiescence by regulating the Igf2-Igfr1 pathway at the transcriptional and posttranscriptional levels.
Interestingly, several other imprinted lncRNAs are upregulated in HSCs or in other adult stem cells relative to their differentiated progeny [30, 36]. These include Air, which acts in cis to silence the paternal allele of Igfr2 via transcriptional interference and also Slc22a3 and Slc22a2 via recruitment of the G9a histone methyltransferase [37–39], as well as the small RNA hosts from the Dlk1-Dio3 imprinted region Rian and Gtl2, thought to act in stem cell maintenance [28, 40].
lncRNAs in the myeloid and erythroid lineages
The first lncRNA with a role in hematopoiesis to be described was EGO, a conserved gene transcribed anti-sense to ITPR1 that modulates the development of eosinophils [41]. EGO is normally expressed in human CD34 + HSCs and becomes upregulated during their differentiation into eosinophils. The EGO transcript is non-coding, as it does not associate with ribosomes. Knockdown of EGO by siRNAs in cultured CD34 + progenitors impaired the expression of genes critical for eosinophil development, including major basic protein and eosinophil-derived neurotoxin. Thus, EGO can contribute to eosinophilopoiesis by enhancing the expression of genes needed for this process.
lncRNAs are also implicated in the regulation of myelopoiesis, the formation of granulocytes and monocytes. Zhang et al. [42] identified HOTAIRM1, a gene transcribed antisense to the HOXA1/2 intergenic region that is strongly upregulated during retinoic acid-induced granulocytic differentiation of myeloid progenitors. The HOTAIRM1 transcript is ~500 nt and does not associate with ribosomes. Its expression is coordinated along the body plan with that of HoxA1 and HoxA2, suggesting that it may be involved in keeping these genes active. Indeed, knockdown of HOTAIRM1 inhibits HoxA1 and HoxA4 activation during granulocytic differentiation in the NB4 acute promyelocytic leukemia cell line, resulting in impaired expression of myeloid differentiation markers such as CD11b and CD18. This effect may be mediated through the interaction of HOTAIRM1 with various chromatin modifiers [28], suggesting that this lncRNA modulates myelopoiesis by regulating the epigenetic state of neighboring genes at the HoxA locus.
Our group has extensively profiled the lncRNA transcriptome of the erythroid lineage, uncovering numerous erythroid-specific lncRNAs that become dramatically induced during terminal differentiation of mouse fetal liver red blood cells in vivo [43, 44]. Many of these RNAs—including intergenic, antisense, and enhancer transcripts—are targets of erythroid-important transcription factors GATA1, TAL1 or KLF1 and are needed for proper red cell development. We characterized 13 such lncRNAs and showed that they are nuclear-localized and exhibit complex developmental expression patterns. Depleting any of them severely impaired erythrocyte maturation, inhibiting cell size reduction and subsequent enucleation. One antisense lncRNA, alncRNA-EC7, is transcribed from an enhancer and is specifically needed for activation of the neighboring gene encoding BAND3. One intergenic lncRNA, lincRNA-EPS, produces a ~2.5 kb capped and polyadenylated transcript that is alternatively spliced and resides in the nucleus. Depleting lincRNA-EPS by shRNAs severely compromised terminal differentiation of erythroid progenitors and resulted in elevated apoptosis. Conversely, ectopic expression of lincRNA-EPS protected erythroid progenitors from apoptosis triggered by erythropoietin starvation. These effects persisted after disruption of putative short ORFs within the transcript, and rather are mediated by a conserved 500 bp 3′ terminal domain that is sufficient for anti-apoptotic activity. Functional studies indicated that lincRNA-EPS acts by repressing a number of pro-apoptotic proteins, most prominently the caspase activating adaptor protein Pycard. Thus, lincRNA-EPS acts to promote red blood cell maturation by downregulating pro-apoptotic pathways. These examples show that lncRNAs can modulate the development of myeloid and erythroid cells through their capacity to regulate gene expression.
lncRNAs in lymphoid lineages
Roles for lncRNAs in lymphoid cells were first proposed by early observations of lymphoid-specific lncRNAs that are dynamically regulated during T cell differentiation and activation [45–47]. Recent studies have now provided evidence for the importance of several lncRNAs in immune cell function. The lncRNA NeST (Tmevpg1) modulates the ability of mice to respond to viral and bacterial infections [48, 49]. The NeST locus was identified through a forward genetic screen as a susceptibility locus for sensitivity to pathogenesis of Thelier’s virus in mice [50–52]. The NeST locus encodes an lncRNA specifically expressed by the TH1 subset of helper T cells. Endogenous or ectopic expression of NeST regulates the degree of inflammation induced by infecting pathogens, such as Thelier’s virus or Salmonella. Mechanistically, NeST regulates expression of the cytokine IFN-γ, critical for innate and adaptive immunity, by specifically interacting with WDR5, a component of the H3K4 methyltransferase complex, in CD8+ T cells. Thus, NeST acts in immune effector cells to regulate the outcome of viral or bacterial infections by epigenetically activating expression of the IFN-γ locus via direct interactions with chromatin modifiers.
Fitzgerald and colleagues have characterized lincRNA-Cox2, which becomes dramatically upregulated by Toll-like receptor (TLR) signaling in mouse bone marrow-derived dendritic cells and macrophages [1, 53]. The lin-cRNA-Cox2 locus encodes several RNA isoforms that do not associate with ribosomes. Upon silencing them with shRNAs, Ccl5, Ccrl, and several IFN-stimulated genes were particularly upregulated. Overexpression of lincRNA-Cox2 in turn resulted in severe attenuation of Ccl5 and overexpression of TLR-induced interleukin 6. lincRNA-Cox2 is found in both nucleus and cytoplasm and for inhibitory activity interacts with heterogeneous nuclear ribonucleoproteins A/B and A2/B2. Thus, lincRNA-Cox2 also acts during inflammatory signaling by modulating the expression of several immune response genes via interactions with regulatory complexes.
In another recent study, Hu et al. globally profiled lncRNA gene expression during the differentiation of naïve CD8+ T cells into various helper T cell subsets, and characterized LincR-Ccr2-5′AS, a TH2-specific gene activated by the transcription factor GATA3 [54]. LincR-Ccr2-5′AS is located between the genes encoding the chemokine receptors Ccr3 and Ccr2 and is coregulated with them. Depleting LincR-Ccr2-5′AS with shRNAs in TH2 cells downregulated expression of the nearby Ccr1, Ccr2, Ccr3 and Ccr5 genes, without affecting their chromatin architecture, and impaired the ability of the cells to migrate to the lungs in vivo. The global gene expression response of TH2 cells depleted for LincR-Ccr2-5′AS significantly overlaps with that following GATA3 depletion, suggesting a functional link between the two during T cell differentiation and immunological function. These examples document critical roles for lncRNAs in modulating immune responses by associating with regulatory complexes to specify immune gene expression programs.
lncRNAs during malignant hematopoiesis
Defects in the proper coordination of hematopoiesis can result in the uncontrolled proliferation of blood cells at different stages of development, resulting in cancer. Because of their roles in regulating blood cell fates, including differentiation, proliferation and survival, lncR-NAs are likely suspects in the pathogenesis of hematopoietic malignancies. Thus far, a handful of lncRNAs have been conclusively linked to cancers of the blood (Table 2; Fig. 3). We discuss these in the next sections, with an emphasis on their mechanism of action.
Table 2.
Examples of lncRNAs with roles in malignant hematopoiesis
| Name | Cell type | Loss of function molecular phenotype | Loss of function malignant phenotype | Functional assays | Reference |
|---|---|---|---|---|---|
| DLEU2 | B cells | Dysregulated cell cycle progression and apoptosis | MBL, CLL, DLBCL | Mouse knockout models; ectopioc expression | [58] |
| Xist | HSC | Compromised X inactivation; impaired maturation; loss of long-term HSCs | Mixed MPN/MDS | Mouse knockout models; transplantation assays | [72] |
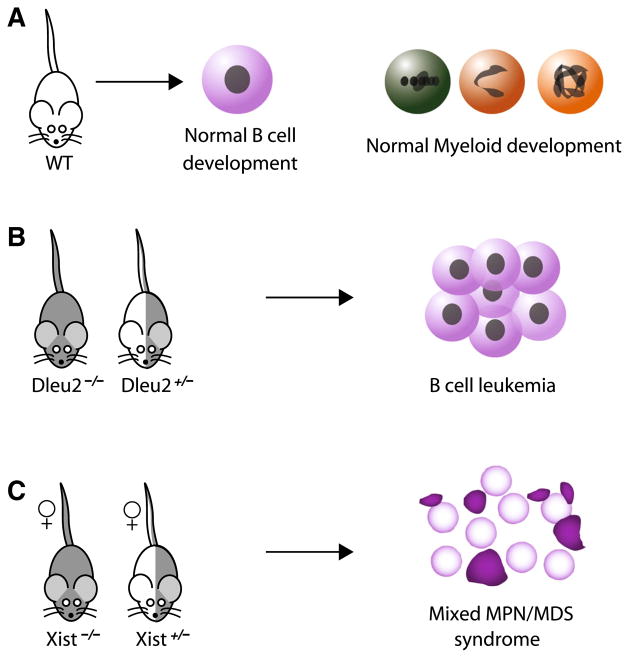
Fig. 3.
Mouse knockout models link lncRNAs to blood cancer pathogenesis. Proper expression of Dleu2 and Xist is required for normal hematopoiesis. a Wild-type mice develop B cells and myeloid cells normally. b Dleu2 −/− and Dleu2∓ mice fail to properly regulate cell cycle progression and apoptosis of B cells, developing a B cell chronic lymphocytic leukemia reminiscent of human CLL that significantly reduces lifespan. c Xist −/− and Xist∓ female mice fail to maintain proper X dosage, developing a deficiency in HSC maturation that leads to a lethal mixed myeloproliferative neoplasm and myelodysplastic (MPN/MDS) syndrome
lncRNAs and leukemia
B cell chronic lymphocytic leukemia (CLL), the most common adult B cell-derived malignancy, has been linked to deletion of 13q14, which is observed at >50 % frequency in both CLL and CD5+ monoclonal B cell lymphocytosis (MBL), and at lower frequencies in CD5− B cell-derived malignancies and T cell lymphomas [55–58]. The minimal deleted region (MDR) within 13q14 consists of a ~110 kb stretch comprising the DLEU2 lncRNA, which hosts microRNAs 15a and 16-1 and is spliced into a mature RNA that can be found in the nucleus and cytoplasm [44]. Mice deleted for the entire MDR or only for miR-15a/16-1 in all cells (or only in B cells) displayed MBL, developed CLL at moderate penetrance, and in a fraction of cases progressed into diffuse large B cell lymphoma, thus recapitulating the spectrum of human CLL phenotypes [59]. Re-expression of miR-15a/16-1 in a human CLL cell line reduced proliferation and downregulated multiple genes involved in the G0/G1-S phase transition but did not affect expression of antiapoptotic ones such as BCL2 or NF-kB, which are normally dysregulated in CLL. Moreover, disease penetrance in MDR−/− mice was 42 % as opposed to 26 % in miR-15a/16-1−/− mice, which translated in shortened lifespan in the former but not in the latter. These findings indicated that loss of DLEU2 leads to CLL at least in part through the absence of the cell cycle inhibitory microRNAs 15a and 16-1, but additional roles of DLEU2 that contribute to a more aggressive disease course remained unexplained. Indeed, recent studies have now linked DLEU2 to the in cis repression of protein-coding neighboring genes that positively regulate NF-kB activity, consistent with the NF-kB downregulation and resistance to apoptosis seen in DLEU2-deficient CLL cells [44, 60, 61]. Thus, DLEU2 acts as a tumor suppressor by regulating both cell cycle progression and the NF-kB signaling pathway in B cells, although the precise mechanisms for the latter role remain elusive. This lncRNA is therefore not only a useful prognostic maker in CLL, but also a potential target for novel CLL therapies.
Interestingly, other lncRNAs are dysregulated in other types of leukemia. MEG3 lncRNA is commonly down-regulated in acute myeloid leukemia (AML) and hypermethylation of its promoter is a marker of poor prognosis [62, 63]. Indeed, MEG3 has been implicated in regulation of the Rb and p16INK4a pathway and thus of cell proliferation in a variety of cancers [64, 65]. Similarly, the lncRNA ANRIL is upregulated in acute lymphoblastic leukemia (ALL) and modulates cell proliferation via regulation of the Rb and p16INK4a pathway in multiple cancers [66, 67]. Although no clear role in the etiology of AML or ALL has emerged for MEG3 and ANRIL, it is likely that they contribute to the disease course via their known effects on cell cycle regulation. Thus, dysregulation of these or of other lncRNAs in specific blood cancer subtypes may present at the very least useful markers of disease progression.
lncRNAs in myeloproliferation and myelodysplasia
Perhaps the most well-understood lncRNA to date, Xist, has recently been found to act as a potent suppressor of hematological cancer in vivo. The Xist locus is transcribed into multiple polyadenylated, nuclear transcripts that can reach ~18–19 kb in length in mouse and human and are required for X chromosome inactivation during embryogenesis [68]. Xist is expressed from a region within the future inactive X chromosome called the X-inactivation center (Xic). Xist recruits the chromatin repressive complex PRC2 to its transcription site through a structured RNA domain, and the two co-migrate to spatially proximal gene-dense regions of active chromatin within the X chromosome, leading to their PRC2-mediated silencing [69–72]. These regions are repositioned into a growing heterochromatin compartment, effectively bringing new sites into close contact with the Xic for further proximity transfer of the Xist-PRC2 complex. By this mechanism, Xist spreads over a ~ 150 Mb scale to silence most genes in the inactive X chromosome. Attesting to its importance during development, paternally inherited loss of Xist is lethal due to lack of X-inactivation in extra-embryonic tissues [16].
X chromosome dosage has long been associated with cancer, since selective loss of the inactive X and duplication of the active one is frequently seen in breast and ovarian malignancies. However, it was not until a recent report by Lee and colleagues that regulation by the Xist lncRNA was causally linked to blood cancer [73]. Conditional deletion of Xist in HSCs after the occurrence of X-inactivation was lethal for both Xist−/− and Xist−/+ females (at 100 % penetrance) but not for their male counterparts. Deceased mutant females exhibited massive splenomegaly and extramedullary hematopoiesis, associated with hyperproliferation of all hematopoietic lineages, with myeloid cells out proliferating those of the other lineages. Bone marrow dysfunction was also observed, including myelofibrosis, myeloproliferation and myelodysplasia, leading to chronic myelomonocytic leukemia and erythroleukemia and thus recapitulating human myeloproliferative neoplasm and myelodysplastic syndrome (MPN/MDS). Wild-type mice transplanted with Xist−/− bone marrow developed MPN/MDS, whereas Xist−/− mice transplanted with wild-type bone marrow did not, indicating a cell autonomous HSC defect. Indeed, Xist−/− mice displayed impaired HSC maturation and loss of long-term HSCs. Molecularly, these effects were mediated by widespread X reactivation leading to genome-wide changes including upregulation or downregulation of oncogenes or tumor suppressors implicated in MPN and MDS, respectively. Thus, the Xist lncRNA is required for both establishment and long-term maintenance of proper X dosage in vivo, and its loss of function leads to blood cancer.
Interestingly, several other lncRNAs modulate Xist expression and thus X chromosome dosage. These include Tsix, which silences Xist in cis via recruitment of the DNMT3A methyltransferase to its promoter, and Jpx, which acts to upregulate Xist in trans at the Xic. Importantly, loss of Tsix or of Jpx is female-lethal [17, 19]. Since X chromosome dosage is so frequently associated with human cancer, the lncRNAs that regulate it present a unique opportunity for the study of functional lncRNAs in the context of hematological cancer.
Conclusions and future directions
The number of human lncRNAs described has exploded over the past 10 years, owing to the rapid development of high throughput transcriptome profiling techniques and their application to a variety of developmental and pathological conditions. Interestingly, many of these genes are highly cell/tissue-specific. Functional characterization of lncRNAs has revealed and likely will continue to reveal roles in various physiological processes, including hematopoiesis. Clearly, lncRNAs are a group of novel regulatory genes that contribute to animal development and pathogenesis. Here, we discuss from our own perspectives some of the outstanding challenges and opportunities for the study of lncRNAs in the hematology field.
Investigating the molecular mechanisms by which lncRNAs regulate gene expression
lncRNAs regulate gene expression during hematopoiesis via multiple mechanisms. One common theme for nuclear-localized lncRNAs is to recruit protein partners, such as chromatin modifying complexes, to target regions in the genome to modulate gene expression. Thus, it is important to identify both the protein partners of lncRNAs and the target genomic regions with which they associate. To this end, an expanding toolbox of biochemical approaches for RNA purification coupled with high throughput sequencing or with mass spectrometry is rapidly becoming available. These include CHART [74], ChIRP-Seq [75], RIA-Seq [76] and RAP [71], which share the same principle of using biotinylated antisense probes that hybridize to a target RNA to purify it together with any associated proteins, RNAs or genomic DNA segments from cross-linked cell lysates. In addition to these biochemical approaches, simple cell biology approaches, such as direct visualization of lncRNAs in situ by single-molecule RNA-FISH can provide important insights into their molecular mechanisms. For example, RNA-FISH can evidence localization to functional subcellular compartments, such as paraspeckles or the nucleolus [38, 77–79], or even to specific chromosomal regions [80, 81], and can also be used to examine multimerization potential and co-localization with RNA or protein partners [82–84]. Thus, when used in combination, these biochemical and cell biology approaches can greatly facilitate the exploration of lncRNA mechanisms during hematopoietic cell differentiation.
Integrating lncRNAs into regulatory networks in control of hematopoiesis
Hematopoiesis is regulated at multiple levels by both intracellular factors and cytokines to ensure the proper generation of multiple cell lineages under both normal and pathophysiological conditions [85, 86]. As more and more functions for lncRNAs in the hematopoietic system are being described, it is increasingly clear that lncRNAs are important components of these regulatory circuitries. Thus, it is of great interest to determine just how these noncoding transcripts fit into known regulatory networks in control of hematopoiesis.
The expression of many lncRNAs that modulate differentiation into a specific cell type is indeed controlled by the key TFs needed for differentiation of that cell type. For example, during erythroid terminal differentiation, the induction of many lncRNAs that are required for proper red cell maturation is mediated by the master erythroid TFs GATA1 and TAL1 [44]. Since the genomic binding sites of many TFs with roles during hematopoiesis have been already assayed by ChIP-seq studies, it is now feasible to intersect these binding sites with active lncRNA loci. This should help integrate lncRNAs into the transcriptional networks established by these TFs, and can additionally help researches prioritize which lncRNA candidates to choose for functional studies. Interestingly, some lncRNAs may physically bind TFs [29], suggesting that lncRNAs might also regulate TF activity. Further progress in identifying the global binding sites of key TFs during hematopoiesis, as well as the protein interactome of lncRNAs, should be of great help in deciphering potential lncRNA and TF regulatory networks.
MicroRNAs also control the differentiation of many blood cell lineages [87, 88]. Interestingly, certain lncRNAs and microRNAs can regulate each other [89–91]. However, functional validation of lncRNA targets of microRNAs in the hematopoietic system remains largely unexplored. Identifying miRNA and lncRNA genes with complementary expression patterns during blood cell differentiation may thus generate candidate lncRNA-microRNA pairs to be tested for inference of lncRNA-microRNA regulatory relationships. These studies should not only lead to the identification of ncRNA regulatory networks in the hematopoietic system, but may also enhance our understanding of how these RNA genes contribute to its development.
Testing lncRNA functions in vivo
The effects of inhibiting some lncRNAs in vitro are relatively small, usually only a ~2 fold change in the expression of target loci, placing them closer to the effects of miRNAs than of TFs. This may be in part due to limitations in achieving efficient depletion of lncRNAs by current si/shRNA approaches, or it may indicate that many lncRNAs, like microRNAs, mainly act to fine tune target gene expression. In addition, any effects of lncRNA perturbation observed in vitro are not guaranteed to elicit phenotypic changes in vivo. Thus, complete deletion of lncRNA loci is required to properly assess the relative contribution of lncRNAs to target gene expression and to establish roles for lncRNAs during hematopoiesis in vivo.
Only a few lncRNA knockout mouse models have been generated to date to address these questions. Encouragingly, in both Xist−/− and Xist−/+ female mice, compromised maintenance of X-inactivation causes a critical deficiency in HSC maturation that leads to a highly aggressive myeloproliferative neoplasm and myelodysplastic syndrome with full penetrance [73]. Similarly, Dleu2−/− and Dleu2−/+ mice fail to properly regulate cell cycle progression and apoptosis of B cells, developing a chronic lymphocytic leukemia reminiscent of human CLL with moderate penetrance [59]. Thus, proper expression of the Xist and Dleu2 lncRNAs is clearly required for normal hematopoiesis, and dysregulation of these genes leads to lethal blood cancer phenotypes.
Despite the examples of Xist and Dleu2, whether other lncRNAs, particularly those that are lineage specific and affect blood cell differentiation in vitro, contribute to hematopoiesis in vivo remains largely unanswered. Examining the phenotypic effects of animal models deleted for these RNAs will require careful evaluation of the specific tissue, developmental stage and environmental condition in which the gene is active. Valuable lessons have been learned in this respect through recent studies. For example, despite the fact that Xist is continuously expressed throughout female life, it had been widely believed to be indispensable for maintenance of X chromosome inactivation, since deleting Xist post-inactivation does not cause immediate reactivation in fibroblasts, in somatic cell hybrids or in ES models of autosomal silencing by an autosomal Xist transgene [92–95]. However, deleting Xist post-inactivation in the blood compartment in vivo clearly leads to X reactivation and potent hematologic cancer. Analogously, the lncRNA MALAT1, first identified as a marker for metastasis development of lung adenocarcinoma [96], is dispensable for life and development [97–99], but targeted deletion of MALAT1 in human lung tumor cells indeed impaired tumor metastasis in a mouse xenograft model [21]. These examples highlight the need for studying lncRNAs under informative physiological conditions.
The strategy for deleting an lncRNA gene is also a crucial factor for elucidating its function in vivo. This is exemplified by the case of the lncRNA HOTAIR, transcribed from the HoxC gene cluster and originally found to act in trans to epigenetically repress the HoxD cluster in human cells with anterior and distal positional identities [100]. Deleting a large chromosomal region including eight HoxC genes, two microRNAs, HOTAIR and several other lncRNAs in a mouse model did not elicit obvious developmental defects and only resulted in modest de-repression of HoxD genes. However, targeted deletion of only the HOTAIR locus indeed affected HoxD gene expression in forming anterior and distal skeletal structures, leading to homeotic transformations of the spine and malformation of metacarpal-carpal bones. Thus, deletions that specifically target lncRNAs may be necessary for uncovering their specific roles in vivo, especially in the case of lncRNAs that overlap other loci. Recently developed techniques for efficient targeted genome engineering, such as the CRISPR-Cas9 system, should aid researchers in this respect. Such targeted deletion studies in vivo will be necessary to uncover the true functions of the many uncharacterized lncRNAs that are active during normal and malignant hematopoiesis.
Acknowledgments
W.H. is a Merck Fellow of the Life Sciences Research Foundation and is supported by a Pathway to Independence Award (1K99HL118157-01) from the National Institutes of Health, National Heart, Lung, and Blood Institute. This research was supported by grants from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases DK068348, and from the National Institutes of Health, National Heart, Lung, and Blood Institute, 5P01 HL066105.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Contributor Information
Juan R. Alvarez-Dominguez, Whitehead Institute for Biomedical Research, Cambridge, MA 02142, USA. Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02142, USA
Wenqian Hu, Whitehead Institute for Biomedical Research, Cambridge, MA 02142, USA.
Austin A. Gromatzky, Whitehead Institute for Biomedical Research, Cambridge, MA 02142, USA. Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02142, USA
Harvey F. Lodish, Email: lodish@wi.mit.edu, Whitehead Institute for Biomedical Research, Cambridge, MA 02142, USA. Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02142, USA. Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02142, USA
References
- 1.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banfai B, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22(9):1646–57. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman M, et al. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154 (1):240–51. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Kretz M, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26(4):338–43. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dethoff EA, et al. Functional complexity and regulation through RNA dynamics. Nature. 2012;482(7385):322–30. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10(11):R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulitsky I, et al. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147(7):1537–50. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–61. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Gibb EA, et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6(10):e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marahrens Y, et al. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11(2):156–66. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 17.Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103(1):17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 18.Grote P, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–14. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143(3):390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauvageau M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutschner T, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5(1):3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anguera MC, et al. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PloS Genet. 2011;7(9):e1002248. doi: 10.1371/journal.pgen.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng JC, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20(11):1470–84. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blank U, Karlsson G, Karlsson S. Signaling pathways governing stem-cell fate. Blood. 2008;111(2):492–503. doi: 10.1182/blood-2007-07-075168. [DOI] [PubMed] [Google Scholar]
- 26.Rossi L, et al. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 2012;11(3):302–17. doi: 10.1016/j.stem.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheik Mohamed J, et al. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16(2):324–37. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31(3):522–33. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venkatraman A, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500(7462):345–9. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pachnis V, Brannan CI, Tilghman SM. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J. 1988;7(3):673–81. doi: 10.1002/j.1460-2075.1988.tb02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brannan CI, et al. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai XZ, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13(3):313–6. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keniry A, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and lgf1r. Nat Cell Biol. 2012;14(7):659–65. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32(6):473–80. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 36.Berg JS, et al. Imprinted genes that regulate early mammalian growth are coexpressed in somatic stem cells. PLoS One. 2011;6(10):e26410. doi: 10.1371/journal.pone.0026410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415(6873):810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 38.Nagano T, et al. The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 39.Latos PA, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338 (6113):1469–72. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 40.Stadtfeld M, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465(7295):175–85. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner LA, et al. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood. 2007;109(12):5191–8. doi: 10.1182/blood-2006-06-027987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113(11):2526–34. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu W, et al. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25(24):2573–8. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alvarez-Dominguez JR, et al. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123(4):570–81. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu AY, et al. The human NTT gene: identification of a novel 17-kb noncoding nuclear RNA expressed in activated CD4(+) T cells. Genomics. 1997;39(2):171–84. doi: 10.1006/geno.1996.4463. [DOI] [PubMed] [Google Scholar]
- 46.Pang KC, et al. Genome-Wide Identification of Long Noncoding RNAs in CD8(+) T Cells. J Immunol. 2009;182(12):7738–48. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 47.Haasch D, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene. BIC Cell Immunol. 2002;217(1–2):78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 48.Collier SP, et al. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol. 2012;189(5):2084–8. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gomez JA, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152(4):743–54. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bureau JF, et al. The interaction of two groups of murine genes determines the persistence of Theiler’s virus in the central nervous system. J Virol. 1992;66(8):4698–704. doi: 10.1128/jvi.66.8.4698-4704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vigneau S, et al. Homology between a 173-kb region from mouse chromosome 10, telomeric to the Ifng locus, and human chromosome 12q15. Genomics. 2001;78(3):206–13. doi: 10.1006/geno.2001.6656. [DOI] [PubMed] [Google Scholar]
- 52.Vigneau S, et al. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J Virol. 2003;77(10):5632–8. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carpenter S, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–92. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu G, et al. Expression and regulation of intergenic long non-coding RNAs during T cell development and differentiation. Nat Immunol. 2013;14(11):1190–8. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dohner H, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 56.Rawstron AC, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008;359(6):575–83. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 57.Liu Y, et al. 13q deletions in lymphoid malignancies. Blood. 1995;86(5):1911–5. [PubMed] [Google Scholar]
- 58.Rosenwald A, et al. A biological role for deletions in chromosomal band 13q14 in mantle cell and peripheral t-cell lymphomas? Genes Chromosomes Cancer. 1999;26(3):210–4. doi: 10.1002/(sici)1098-2264(199911)26:3<210::aid-gcc4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 59.Klein U, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17(1):28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 60.Mertens D, Stilgenbauer S. CLL and deletion 13q14: merely the miRs? Blood. 2012;119(13):2974–5. doi: 10.1182/blood-2012-01-400747. [DOI] [PubMed] [Google Scholar]
- 61.Garding A, et al. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the In Cis downregulation of a gene cluster that targets NF-kB. PLoS Genet. 2013;9(4):e1003373. doi: 10.1371/journal.pgen.1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benetatos L, et al. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res. 2010;34(2):148–53. doi: 10.1016/j.leukres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 63.Khoury H, et al. An upstream insulator regulates DLK1 imprinting in AML. Blood. 2010;115(11):2260–3. doi: 10.1182/blood-2009-03-212746. [DOI] [PubMed] [Google Scholar]
- 64.Zhang X, et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010;70(6):2350–8. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129(4):773–9. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- 66.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451(7175):202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–74. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol. 2011;12(12):815–26. doi: 10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- 69.Zhao J, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300(5616):131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 71.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):U233–767. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simon MD, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504(7480):465–9. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yildirim E, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152(4):727–42. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Simon MD, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USA. 2011;108(51):20497–502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chu C, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493(7431):231–5. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sasaki YT, et al. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106(8):2525–30. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19(3):347–59. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clemson CM, et al. An architectural role for a nuclear non-coding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–26. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Redrup L, et al. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development. 2009;136(4):525–30. doi: 10.1242/dev.031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reinius B, et al. Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics. 2010;11:614. doi: 10.1186/1471-2164-11-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106(28):11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chakraborty D, et al. Combined RNAi and localization for functionally dissecting long noncoding RNAs. Nat Methods. 2012;9(4):360–2. doi: 10.1038/nmeth.1894. [DOI] [PubMed] [Google Scholar]
- 84.Fanucchi S, et al. Chromosomal contact permits transcription between coregulated genes. Cell. 2013;155(3):606–20. doi: 10.1016/j.cell.2013.09.051. [DOI] [PubMed] [Google Scholar]
- 85.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–44. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Novershtern N, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen CZ, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 88.Bissels U, Bosio A, Wagner W. MicroRNAs are shaping the hematopoietic landscape. Haematologica. 2012;97(2):160–7. doi: 10.3324/haematol.2011.051730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salmena L, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karreth FA, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147(2):382–95. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brown CJ, Willard HF. The human X-inactivation center is not required for maintenance of X-chromosome inactivation. Nature. 1994;368(6467):154–6. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- 93.Rack KA, et al. Absence of the XIST gene from late-replicating isodicentric X chromosomes in leukaemia. Hum Mol Genet. 1994;3(7):1053–9. doi: 10.1093/hmg/3.7.1053. [DOI] [PubMed] [Google Scholar]
- 94.Csankovszki G, et al. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22(4):323–4. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- 95.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5(4):695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 96.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 97.Zhang B, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2(1):111–23. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eissmann M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–87. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nakagawa S, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487–99. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]