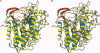Abstract
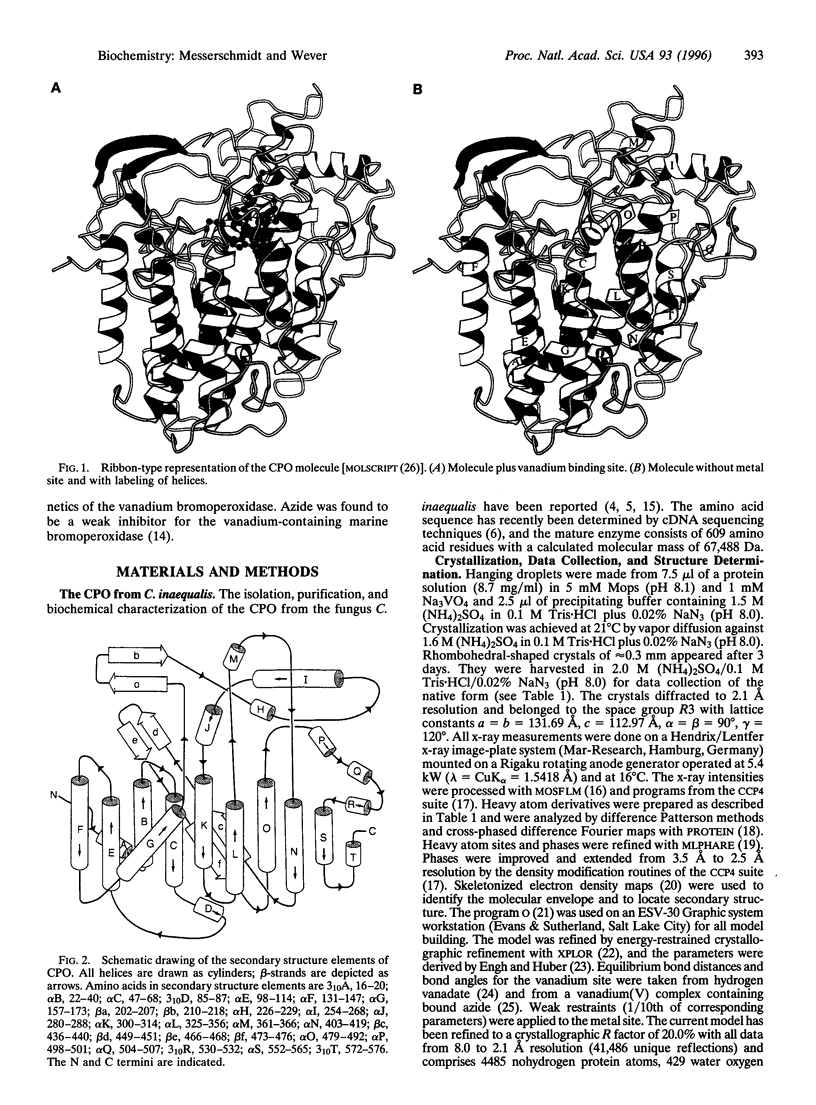
The chloroperoxidase (EC 1.11.1.-) from the fungus Curvularia inaequalis belongs to a class of vanadium enzymes that oxidize halides in the presence of hydrogen peroxide to the corresponding hypohalous acids. The 2.1 A crystal structure (R = 20%) of an azide chloroperoxidase complex reveals the geometry of the catalytic vanadium center. Azide coordinates directly to the metal center, resulting in a structure with azide, three nonprotein oxygens, and a histidine as ligands. In the native state vanadium will be bound as hydrogen vanadate(V) in a trigonal bipyramidal coordination with the metal coordinated to three oxygens in the equatorial plane, to the OH group at one apical position, and to the epsilon 2 nitrogen of a histidine at the other apical position. The protein fold is mainly alpha-helical with two four-helix bundles as main structural motifs and an overall structure different from other structures. The helices pack together to a compact molecule, which explains the high stability of the protein. An amino acid sequence comparison with vanadium-containing bromoperoxidase from the seaweed Ascophyllum nodosum shows high similarities in the regions of the metal binding site, with all hydrogen vanadate(V) interacting residues conserved except for lysine-353, which is an asparagine.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arber J. M., de Boer E., Garner C. D., Hasnain S. S., Wever R. Vanadium K-edge X-ray absorption spectroscopy of bromoperoxidase from Ascophyllum nodosum. Biochemistry. 1989 Sep 19;28(19):7968–7973. doi: 10.1021/bi00445a062. [DOI] [PubMed] [Google Scholar]
- Hecht H. J., Sobek H., Haag T., Pfeifer O., van Pée K. H. The metal-ion-free oxidoreductase from Streptomyces aureofaciens has an alpha/beta hydrolase fold. Nat Struct Biol. 1994 Aug;1(8):532–537. doi: 10.1038/nsb0894-532. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986 Apr;5(4):819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Machius M., Wiegand G., Huber R. Crystal structure of calcium-depleted Bacillus licheniformis alpha-amylase at 2.2 A resolution. J Mol Biol. 1995 Mar 3;246(4):545–559. doi: 10.1006/jmbi.1994.0106. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Structural and genetic analysis of protein stability. Annu Rev Biochem. 1993;62:139–160. doi: 10.1146/annurev.bi.62.070193.001035. [DOI] [PubMed] [Google Scholar]
- Messerschmidt A., Ladenstein R., Huber R., Bolognesi M., Avigliano L., Petruzzelli R., Rossi A., Finazzi-Agró A. Refined crystal structure of ascorbate oxidase at 1.9 A resolution. J Mol Biol. 1992 Mar 5;224(1):179–205. doi: 10.1016/0022-2836(92)90583-6. [DOI] [PubMed] [Google Scholar]
- Nordlund P., Sjöberg B. M., Eklund H. Three-dimensional structure of the free radical protein of ribonucleotide reductase. Nature. 1990 Jun 14;345(6276):593–598. doi: 10.1038/345593a0. [DOI] [PubMed] [Google Scholar]
- Poulos T. L., Kraut J. The stereochemistry of peroxidase catalysis. J Biol Chem. 1980 Sep 10;255(17):8199–8205. [PubMed] [Google Scholar]
- Rosenzweig A. C., Frederick C. A., Lippard S. J., Nordlund P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature. 1993 Dec 9;366(6455):537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- Simons B. H., Barnett P., Vollenbroek E. G., Dekker H. L., Muijsers A. O., Messerschmidt A., Wever R. Primary structure and characterization of the vanadium chloroperoxidase from the fungus Curvularia inaequalis. Eur J Biochem. 1995 Apr 15;229(2):566–574. doi: 10.1111/j.1432-1033.1995.tb20499.x. [DOI] [PubMed] [Google Scholar]
- Van Schijndel J. W., Barnett P., Roelse J., Vollenbroek E. G., Wever R. The stability and steady-state kinetics of vanadium chloroperoxidase from the fungus Curvularia inaequalis. Eur J Biochem. 1994 Oct 1;225(1):151–157. doi: 10.1111/j.1432-1033.1994.00151.x. [DOI] [PubMed] [Google Scholar]
- Verschueren K. H., Kingma J., Rozeboom H. J., Kalk K. H., Janssen D. B., Dijkstra B. W. Crystallographic and fluorescence studies of the interaction of haloalkane dehalogenase with halide ions. Studies with halide compounds reveal a halide binding site in the active site. Biochemistry. 1993 Sep 7;32(35):9031–9037. doi: 10.1021/bi00086a008. [DOI] [PubMed] [Google Scholar]
- van Schijndel J. W., Simons L. H., Vollenbroek E. G., Wever R. The vanadium chloroperoxidase from the fungus, Curvularia inaequalis. Evidence for the involvement of a histidine residue in the binding of vanadate. FEBS Lett. 1993 Dec 27;336(2):239–242. doi: 10.1016/0014-5793(93)80811-8. [DOI] [PubMed] [Google Scholar]
- van Schijndel J. W., Vollenbroek E. G., Wever R. The chloroperoxidase from the fungus Curvularia inaequalis; a novel vanadium enzyme. Biochim Biophys Acta. 1993 Feb 13;1161(2-3):249–256. doi: 10.1016/0167-4838(93)90221-c. [DOI] [PubMed] [Google Scholar]