SUMMARY
Molecular analyses of Aplysia, a well-established model organism for cellular and systems neural science, have been seriously handicapped by a lack of adequate genomic information. By sequencing cDNA libraries from the central nervous system (CNS), we have identified over 175,000 expressed sequence tags (ESTs), of which 19,814 are unique neuronal gene products and represent 50%–70% of the total Aplysia neuronal transcriptome. We have characterized the transcriptome at three levels: (1) the central nervous system, (2) the elementary components of a simple behavior: the gill-withdrawal reflex—by analyzing sensory, motor, and serotonergic modulatory neurons, and (3) processes of individual neurons. In addition to increasing the amount of available gene sequences of Aplysia by two orders of magnitude, this collection represents the largest database available for any member of the Lophotrochozoa and therefore provides additional insights into evolutionary strategies used by this highly successful diversified lineage, one of the three proposed super-clades of bilateral animals.
INTRODUCTION
Aplysia and related opisthobranchs are free-living representatives of the Mollusca (class Gastropoda), the second largest phylum in the animal kingdom after the Arthropods. Molluscs have more than 100,000 extant species—marine, freshwater, and terrestrial—and trace their origin to the Cambrian period (Brusca and Brusca, 2003; Pojeta et al., 1987). Molluscs are of further phylogenetic interest because they are an exceedingly diverse and evolutionarily highly successful bilaterian lineage equivalent in their significance to chordates, arthropods, and nematodes. Their success is, however, based upon a different body plan and different adaptive strategies. To understand their strategies better, it is useful to have a better inventory of gene gain and loss in the major bilaterian lineages represented by Aplysia, Drosophila, or C. elegans. Despite their importance, however, the molluscs have not previously been studied on a genome-wide scale. Indeed, while there is now good genomic information on two of the three proposed superclades of bilaterian animals the Deuterostomia (echinoderms, hemichordates, and chordates) and Ecdysozoa (Arthropods, Nematodes, and four to five smaller phyla), evolutionary analysis is seriously limited by lack of genomic information of the third proposed superclade, the Lophotrochozoa (i.e., protostomes with ciliated larvae) to which Mollusca belongs (Aguinaldo et al., 1997; de Rosa et al., 1999; Halanych, 2004).
Although the Lophotrochozoa constitute more than 15 animal phyla (Halanych, 2004), none of the representatives of this group, the largest and systematically most diverse of these three presumed superclades of bilateral animals, has had its genome sequenced. In fact, expressed sequence tag (EST) collections from molluscs are very limited. As a result, the position of molluscs within the Lophotrochozoa and the relationships within the molluscan phylum among the phylogeny of bilateral animals, are all poorly understood and controversial (Brusca and Brusca, 2003; Giribet et al., 2006; Grande et al., 2004a, 2004b; Halanych, 2004; Haszprunar, 2000; Lydeard et al., 2000; Nielsen, 2001; Passamaneck and Halanych, 2006; Valentine, 2004; Vonnemann et al., 2005).
Beyond phylogenetic relationships, our understanding of the basic molecular aspects of nervous system biology, such as genes controlling the maintenance of cellular diversity, the generation of specific neuronal circuitry, and the modifications of neural circuitry by learning and memory, suffers from limited knowledge about the repertoire of genes expressed by individual neurons. For example, we know little about the genes that distinguish the identities of one identifiable neuron from another, or even a motor neuron from a sensory neuron or from an inter-neuron. The anatomical complexity of most nervous systems poses obstacles for the detailed study of cellular identity in relation to behavioral functions.
These obstacles can be overcome in the Aplysia nervous system in part by the accessibility of individual ganglia and specific neurons within these ganglia and the ability to identify individual nerve cells that play roles in specific behaviors (Kandel, 1976, 1979). Aplysia has only about 104 central nerve cells (Cash and Carew, 1989; Kandel, 1976) compared to the 2 × 105 neurons of Drosophila and about 1011 neurons of mammalian brain. Moreover, in mouse, C. elegans, and Drosophila, most of the nerve cells are relatively small. By contrast, many of the neurons located in the Aplysia ganglia are large, and some are gigantic (100–1000 μm in diameter). Because of this, Aplysia has been useful for cell and molecular biological studies of behavior, learning, and memory, complementing the genetic studies in Drosophila and mice. Many of these large identified cells, such as the metacerebral cells (MCC), have well-defined homologs even in distantly related species (Kandel and Tauc, 1966; Sakharov, 1974; Weiss and Kupfermann, 1976), a feature that lends itself to the comparative study of behavior at the cellular level.
As with C. elegans, the identified nerve cells in Aplysia form precise connections with one another. Thus, the connections between identified cells of a neural circuit can be mapped on a cell-to-cell basis for a variety of behaviors ranging in complexity from simple withdrawal reflexes to complex fixed action patterns such as locomotion, feeding, and defense reactions (Hening et al., 1979; Jahan-Parwar and Fredman, 1979; Kupfermann, 1974; Kupfermann and Kandel, 1969). Moreover, these behaviors are modulated by various forms of nonassociative and associative forms of learning, as well as by arousal and motivational states (Cleary and Byrne, 1993; Kupfermann and Weiss, 1982; Rosen et al., 1989; Fitzgerald et al., 1997; Kandel, 2001). Components of these defined neuronal circuits can be reconstructed in cell culture to study both specific cells and cellular compartments at high resolution (Kandel, 2001).
Despite these distinct advantages, research in Aplysia has been seriously handicapped by the deficit in genomic information. As part of an attempt to correct this deficit systematically, we generated more than 175,000 ESTs and characterized the global transcriptome at three levels of neuronal organization: (1) the entire central nervous system (CNS) consisting of nine morphologically distinct ganglia (Figure 1C), (2) individual identified neurons, such as the sensory cells and motor cells of the gill withdrawal reflex and MCC serotonergic modulatory interneurons (Figure 1D), and (3) neuronal processes from the same MCC and sensory neurons (see also Moccia et al., 2003). In this way, we were able to identify general neuronal markers as well as markers for subcellular populations of transcripts, which provide a beginning for exploring genomic bases of neuronal identity and the roles of generalized and localized mRNA translation in synaptic specificity and growth. Gene clusters represented by these ESTs appear to comprise more than half of the protein coding genes of Aplysia, including several hundred novel genes potentially involved in cellular signaling, development, and synaptogenesis.
Figure 1. Construction of an Aplysia Neurotranscriptomics Database.
(A) Table summarizing statistics for the neuronal transcriptome project including collections of ESTs from identified MCC neuron and neuronal processes (neurites) of the same cell.
(B) Aplysia californica, a free moving animal. Scale: 4 cm.
(C) Schematic representation of the Aplysia central nervous system and its nine major ganglia, including paired buccal (BG), pedal (PeG), pleural (PlG), and cerebral (CG) as well as a single abdominal (AG) ganglion; a pair of neurosecretory clusters (Bag cells: Bc) is located in the abdominal ganglion and involved in the control of egg-laying behavior.
(D) Dorsal view of the left and right cerebral ganglia of Aplysia and position of serotonergic modulatory neurons (MCC). All connective tissues were removed to reveal individual neurons located at the surface of the ganglia. Scale: 400 μm.
RESULTS AND DISCUSSION
Neuronal Transcriptome of Aplysia californica: Assembly Statistics and Development of a Neurotranscriptomics Database
We have generated 190,408 ESTs from normalized and nonnormalized cDNA libraries prepared from the Aplysia CNS. Figure 1A summarizes the assembly statistics for the ESTs obtained from the entire collection as well as from individual neurons and their processes. A list of the sources for each cDNA library is listed in Table S1. After removal of ribosomal, mitochondrial (Knudsen et al., 2006), and short sequences (less than 100 bases), the clustering and assembly of 176,232 neuronal ESTs yielded 19,814 total clusters and 65,055 total nonredundant sequences (Figure 1A). Of these, we detected a total of 24,422 open reading frames (ORFs ≥ 100 aa). More than 11,000 predicted Aplysia gene products had recognizable similarity to 3,047 unique protein domains (Pfam, Table S2).
As the next step, we have the characterized transcriptome of individual cells. First, we used microarrays to identify transcripts in the sensory and motor neurons of the gill-withdrawal reflex. Second, we generated libraries from individual identified serotonergic neurons (MCC) and obtained 9,223 unique gene products expressed in these cells. Finally, we generated libraries from pure neuronal processes of the MCCs and obtained about 1,000 nonredundant sequences.
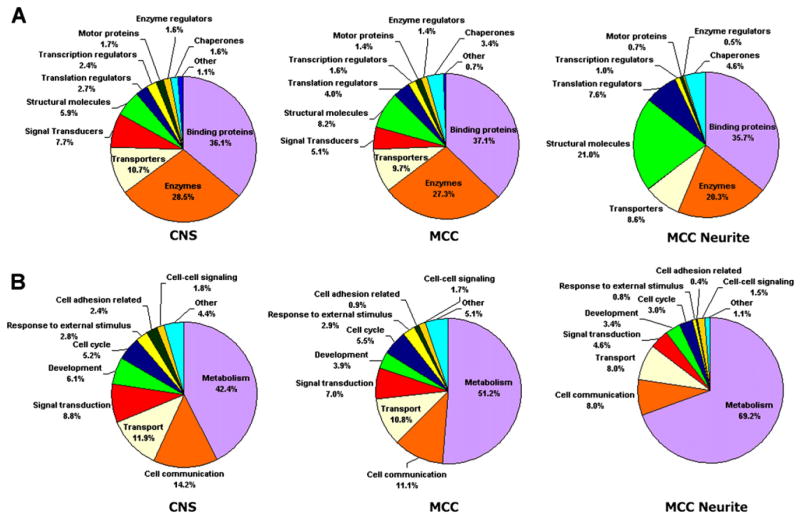
Figure 2 and Table S3 provide a breakdown of the annotated Aplysia sequences using characteristics based on mappings to gene ontology (GO): predicted “molecular function” and “biological process.” The largest functional categories for CNS transcripts were those related to binding proteins (36.1%) and enzymes (28.5%), followed by transporters (10.7%), signal transducers (7.7%), and developmental proteins (6.1%). About 30% of the annotated CNS transcripts in Aplysia were predicted to be membrane-associated (including 6% or 444 transcripts integral to the plasma membrane), and about 5% were putative extracellular secretary proteins and neuropeptides.
Figure 2. Comparison of ESTs Identified from CNS, MCC Somata, and MCC Neurites.
(A and B) Annotated ESTs broken out according to major GO categories by both molecular function (A) and biological process (B) for ESTs obtained from the whole CNS, MCC somata, and MCC neurites. While the majority of categories stay relatively constant in each mRNA source, there are significantly fewer signal transducers and more structural molecules found in MCC neurites.
Sequencing of nearly 10,000 ESTs from nonnormalized libraries from the pedal-pleural ganglia allowed us to estimate the relative abundance of selected transcripts and screen for the presence of potential neuron specific markers that were previously unknown for molluscs and many other members of this clade (see Table S4). The most abundant class of ESTs contains neuropeptides (e.g., pedal peptide, pleurin, achatin, and FMRF-amide), followed by the cytoskeletal proteins (e.g., neuronal isoform of β-tubulin and α-actin), glial secretory proteins (e.g., Aplysia glial protein [Ag] and Acetylcholine Binding Protein [AChBP]), and components related to protein synthesis. With the help of this EST collection, we obtained full-length cDNA sequences for several Aplysia genes relevant for future analysis of interneuronal signaling (Table S5). Web-accessible resources and annotated searchable databases were created and are available from links provided at the end of this paper.
Comparison of our database against National Center for Biotechnology Information’s KOG (eukaryotic clusters of orthologous groups) database (Tatusov et al., 2003) revealed that our ESTs have homologs to about 55% of KOGs shared among all seven represented organisms (Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens, Arabidopsis thaliana, Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Encephalitozoon cuniculi) and 40% of the KOGs when we restrict the analysis to human, Drosophila, and C. elegans. These comparisons imply that the annotated sections of our database cover roughly 50% of commonly expressed genes, including housekeeping genes and other genes expressed in nearly all organisms and about 40% of commonly expressed genes found only in organisms with a nervous system. Table 1 presents a comparison of signal transduction components currently identified in Aplysia with those from the genomes of Drosophila, C. elegans, and humans. Based on the comparison of ion channels and homologs to KEGG metabolic and signaling pathways (Table S6), we estimate that the entire Aplysia EST database represents ~50%–70% of the expressed genes in the nervous system.
Table 1.
Comparison of Gene Orthologs Relevant to Neuronal Functions among Major Model Organisms and Humans
| Gene Orthologs | Aplysia neuronal transcripts | Drosophila genome | C. elegans genome | Human genome |
|---|---|---|---|---|
| Protein Kinases | 452 | 240 | 454 | 518 |
| Protein Phosphatases | 67 | 66 | 106 | 100 |
| Ionotropic Glu Receptors | 15 | 11 | 10 | 16 |
| Metabotropic GluR | 3 | 2 | 3 | 8 |
| Ionotropic ACh receptors | 16 | 12 | 56 | 17 |
| Voltage gated Ca channels | 8 | 9 | 12 | 10 |
| Voltage gated Na channels | 2 | 4 | 4 | 10 |
| K channels | 14–16 | 22 | 64 | 78 |
| Amiloride-sensitive Na channels | 2 | 24 | 27 | 11 |
| Cyclic nucleotide gated channels* | 4 | 7 | 4 | 10 |
| Cadherins/Protocadherins | 12 | 17 | 16 | 113 |
| Synaptotagmins | 8–10 | 3 | 5 | 30 |
| Semaphorins | 4–5 | 6 | 2 | 2 |
| Fragile X MRP (Mental Retardation Protein) | 1 | 1 | 1 | 2 |
| Plexins | 3 | 2 | 2 | 16 |
| Innexins/Pannexins (gap junctions) | 9 | 9 | 29 | 3 |
| Nitric oxide synthase# | 2 | 1 | 0 | 3 |
| Nuclear hormone receptors | 6 | 21 | 270 | 48 |
| ITP/ryanodine receptors | 2–3 | 2 | 4 | 8 |
| Serotonin transporter | 1 | 1 | 1 | 1 |
| 5-HT receptors | 6 | 4 | 5 | 20 |
| DA receptors | 3 | 5 | 3 | 6 |
| GABA A & B receptors | 5/1 | 4/3 | 4/2 | 19/4 |
| Gly Receptors | 2 | 4 | 3 | 6 |
| Histamine receptors | 2 | 0 | 0 | 4 |
| Receptors guanylyl cyclases | 7 | 0 | 31 | 5 |
| Ephrin & EphR | 2 | 1/1 | 3/1 | 16 |
| Importins | 7 | 3 | 10 | 20 |
| Notch/Delta | 3–4/2 | 1/1 | 2/1 | 4/3 |
| Integrins/disintegrin | 29 | 7 | 4 | 67 |
| Laminins | 10 | 3 | 3 | 17 |
| Prominins (stem cell marker) | 2 | 1 | 1 | 11 |
| Microcephalin (MCPH) | 1 | 1 | 1 | 1 |
| P2X purinoreceptors# | 2 | 0 | 0 | 10 |
| Major Vault Complex# | 3 | 0 | 0 | 3 |
| Ependymin# | 1 | 0 | 0 | 1 |
| RAG-1 like (Recombination-Activating Gene)# | 1–5 | 0 | 0 | 1 |
| DNA methyltransferase associating protein 1# | 1 | 0 | 0 | 1 |
| Theromacin (cys-rich antimicrobial peptides)# | 1 | 0 | 0 | 0 |
| Universal Stress Proteins (UspA)# | 7 | 0 | 0 | 0 |
Counts for Drosophila, C. elegans, and human gene orthologs were determined by manually screening Ensembl, Flybase, Worm-base, and NCBI. Genes for which no orthologs were found are counted as 0. P2X-type ATP receptors, Vault proteins, ependymin, and RAG-like transcripts, etc. that were identified in Aplysia and human genomes but were not found in C. elegans or in Drosophila are marked # (see details in the Supplemental Data). One HCN channel, one CNG channel, and two CNG-like ESTs were found in Aplysia (marked *). There are four HCNs in humans and one in Drosophila, but no HCN channels were found in C. elegans.
Aplysia in an Evolutionary Context
The availability of the Aplysia transcriptome allows us to address questions about the phylogenetic position of molluscs among other animal phyla. All bilaterians are thought to be grouped into three major superclades (Aguinaldo et al., 1997; de Rosa et al., 1999; Peterson and Eernisse, 2001; Philippe et al., 2005). The Deuterostomes are proposed to occupy the basal branch of the tree, along with the two more derived sister superclades, the Ecdysozoa and the Lophotrochozoa (containing more than 15 animal phyla including molluscs). This tripartite classification, initially based on analysis of rRNAs and selected HOX genes (de Rosa et al., 1999; Halanych, 2004; Passamaneck et al., 2004), has now been challenged by recent molecular studies (Philip et al., 2005; Rokas et al., 2005; Wolf et al., 2004) and by data from developmental biology (Nielsen, 2001, 2005). As a result of the limited genomic information available from the representatives of many invertebrate phyla, even the existence of two monophyletic protostome superclades is controversial (Passamaneck and Halanych, 2006).
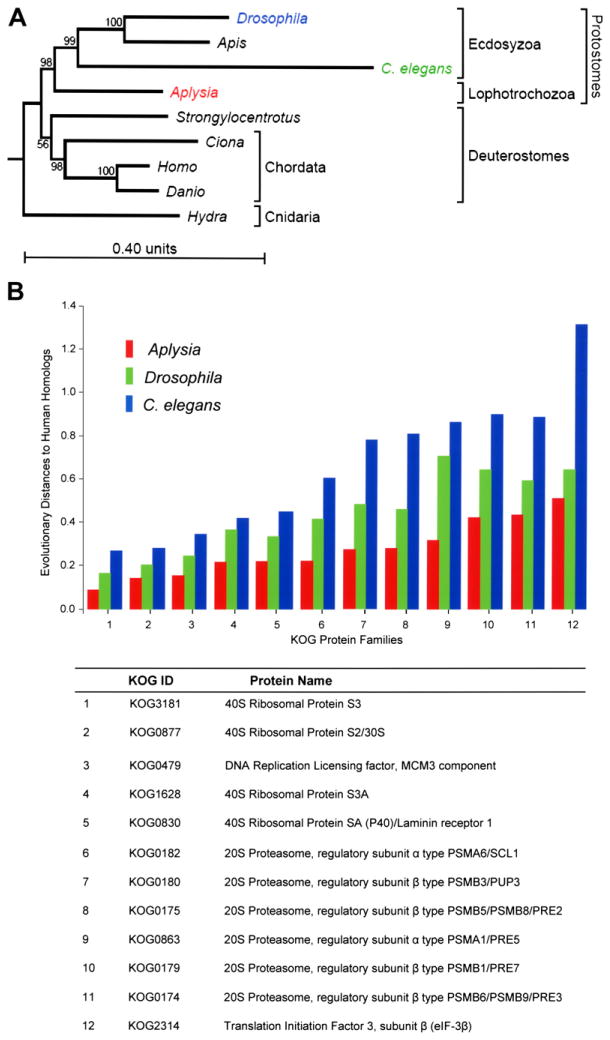
We used the sequence data obtained from Aplysia to re-examine whether molluscs are a sister taxon of Ecdysozoa or whether they are best grouped together with Deuterosomes, as suggested by Davidson and coworkers (Chen et al., 2004) based on their interpretation of new Precambrian fossils. For this phylogenetic analysis, we selected a cnidarian (Hydra) for outgrouping the bilaterian animals, two insect species, two worms, the zebrafish (Danio), the ascidian (Ciona) and an echinoderm, the sea urchin (Strongylocentrotus). The latter two were chosen as members of basal chordates and deuterostomes, respectively. These species represent all major clades, for which genome or large-scale EST projects have been performed. We analyzed 45 protein families (Table S15) based on their known conservative evolution and wide distribution among species (Wolf et al., 2004).
The results of this phylogenetic analysis (Figure 3A) are consistent with a model of three major supeclades of bilaterians and positions Aplysia as a sister to the arthropod/nematode clade. This finding also supports the monophyly of bilaterians (i.e., the same common ancestor for humans, Aplysia, Drosophila, and C. elegans). Furthermore, our analysis supports the idea that the Lophotrochozoans (to which Mollusca belongs) and the Ecdysozoa are sister groups. Thus, the proximity of Hydra to the bi-laterians makes it a significantly better outgroup for the bilateral animals than the yeast/plant outgroup.
Figure 3. Aplysia in Evolutionary Context.
(A) Phylogenetic relationship between Aplysia and other genomic model animals reconstructed based on the KOG (Tatusov et al., 2003) sequence analysis. The mollusc Aplysia californica (a representative of the Lophotrochozoa clade) is placed as a sister to the Arthropod (the fruit fly, Drosophila melanogaster, and the honey bee, Apis mellifera), and nematode (Caenorhabditis elegans) phyla representing the clade Ecdysozoa. The clade of Deuterostomes is represented here by chordates (Homo sapiens, the zebrafish Danio rerio, the ascidian Ciona intestinalis) and the sea urchin (Strongylocentrotus purpuratus). Therefore, this evolutionary analysis shows that Aplysia is more closely related to nematodes and insects than to vertebrates. The evolutionary distance (measured as branch length) from human to Aplysia is shorter, however, than the distance from human to Drosophila and C. elegans, suggesting that the amino acid replacement rate has been lower in the lineage leading to Aplysia than in the lineages leading to Drosophila and C. elegans.
(B) Comparison of relative evolutionary distances for selected protein families between model organisms (C. elegans, Drosophila, Aplysia) and human homologs. The relative distances (the y axis) are measured for each KOG protein family (Tatusov et al., 2003) according to the JTT model of evolution (Jones et al., 1992). The distance units are expected amino acid replacements per site (for further details see Supplemental Data). Note that rates of evolutionary protein changes in the Aplysia lineage (red bars) were slower when compared to C. elegans or Drosophila lineages (blue and green bars).
Figure 3A also illustrates that the evolutionary distance from Aplysia to human is shorter than the distance from Drosophila and C. elegans to human. This indicates that the amino acid replacement rate has been lower in the lineage leading to Aplysia than in the lineages leading to Drosophila and C. elegans. To test this idea further, we measured the corresponding distances among the individual protein families (Table S7) according to the JTT model of evolution. This analysis also showed a significantly shorter distance from humans to Aplysia than to both C. elegans and Drosophila. Figure 3B illustrates the overall comparison between Aplysia ESTs and protein coding genes in the three other organisms. Greater similarity was found between human and Aplysia proteins, further confirming that the amino acid replacement rate has been lower in the molluscan lineage than in the lineages leading to Drosophila and C. elegans. Similarly, annelid and human proteins were also found to be more closely related to each other than to their ecdysozoan orthologs (Raible et al., 2005).
Predicted Gene Loss in Bilaterians
The topology of the bilaterian phylogenetic tree (Figure 3A) and novel Aplysia sequences make possible an analysis of gene loss within each of the four bilaterian lineages leading to humans, Aplysia, flies, and C. elegans. Indeed, the presence of a shared gene ortholog between Aplysia and humans as well as its absence in the sequenced genomes of Drosophila or C. elegans suggest the loss of these orthologs in the lineages of Ecdysozoa rather than its improbable independent origin in molluscs and deuterostomes. As an illustrative example of such an analysis, ORFs from each of the Aplysia nonredundant sequences were compared against protein databases from the other model organisms using blastx. Selected genes identified in this analysis can also be found in Tables S8 and S9.
We focused in particular on proteins from about 40 annotated transcripts relevant to brain development and functions that are only present in humans and Aplysia and are not present in C. elegans or Drosophila. For example, we found an Aplysia homolog of Churchill, a zinc finger transcription activator that in vertebrates acts as a switch between different roles of the fibroblast growth factors and regulates the transition between gastrulation and neurulation (Sheng et al., 2003). Interestingly, the Churchill homolog, though not found in nematodes, arthropods, or Ciona, was recently found in two cnidarians, the coral Acropora (Kortschak et al., 2003) and the freshwater polyp Hydra. The origin of this gene can be traced to the most basal metazoan predecessors, and it has apparently been lost in more derived ecdysozoans, such as nematodes and arthropods.
A similar fate can be seen for the P2X receptor genes that encode ATP-gated cationic channels known to be key elements in purinergic transmission involved in pain and certain forms of long-term plasticity. We found a homolog of this gene in Aplysia and Hydra, yet homologs are apparently absent from sequenced ecdysozoan genomes. Other genes found in Aplysia but lost in insects and nematodes include Cystatin B encoding a specific cysteine protease inhibitor in humans (Brannvall et al., 2003), which is associated with a progressive myoclonic type of epilepsy that leads to mental deterioration and dementia (Lehesjoki, 2003). Neuronal transcripts shared by Aplysia and humans also include a unique group of Selenoproteins (e.g., the predicted selenoprotein N homolog of Aplysia has no known homologs in other invertebrates), Ependymin-like neurotrophic factor, Major Vault proteins with RNA binding capacities but unknown neurological functions, and proteins related to innate immunity (e.g., complement components).
Some newly identified Aplysia transcripts have only been reported in basal deuterostomes and were apparently lost in the entire vertebrate lineage of deuterostomes as well as ecdysozoans with sequenced genomes. One such example is the apextrin-like secretory protein found in the Aplysia CNS. In the sea urchin Heliocidaris erythrogramma, apextrin is involved in establishing cell polarity, ectoderm development, and metamorphosis (Haag et al., 1999). The fact that apextrin is found in such divergent lineages as echinoderms, cephalochordates, and molluscs implies it had a role in the predecessor of all bilaterians.
We conservatively estimate that there are at least 20 genes expressed in the CNS of Aplysia that are shared by Aplysia and Hydra (prebilaterian organism, phylum Cnidaria) that have no obvious homologs in vertebrates, arthropods, or nematodes (Table S9), suggesting that these 20 genes were a part of the most basal gene pool present in early metazoans even before the origin of bilaterian animals. Some of these 20 transcripts from this list have homologs only within the basal or unicellular eukaryotes (e.g., the apicomplexans (Cryptosporidium parvum) or the slime molds (Dictyostelium discoideum).
In Aplysia we identified seven nonredundant sequences (UspA) of one ancient superfamily of “Universal Stress Proteins” (Usp, Table S9), a well-known and conserved group of proteins that are found in Archaea, Eubacteria, and plants. However, the functions of these stress proteins in eukaryotes are currently not known. In bacteria, these proteins act as global regulators of gene expression triggered by a large variety of environmental insults. In Escherichia coli, for example, an integrated adaptive response has been described resulting in changes of cell motility, adhesion, and oxidative stress resistance (Nachin et al., 2005). Recently, Usp-like ESTs were also found in the cnidarians Nematostella and Acropora (Technau et al., 2005) as well as in the parasitic flat worm Schistosoma japonicum. This implies greater conservation of UspA proteins in basal metazoans and lophotrochozoans but a noticeable loss of this class of proteins during the evolution of echinoderms, chordates, nematodes, and some arthropods.
In summary, information from the Aplysia transcriptome together with recent comparative data (Kusserow et al., 2005; Miller et al., 2005; Technau et al., 2005) support the idea that there was a common bilateran ancestor (Urbilateria) that had a complex genome. As a result, more derived genomes of insects and nematodes represent a substantial level of gene loss from an ancestral state as compared to slower evolved genomes within selected lophotrochozoan (e.g., molluscs, annelids) and deuterostome (vertebrates) lineages. Furthermore, the fact that many ancient regulatory systems are expressed in neurons is important in the re-evaluation of novel hypotheses about the evolution of neural systems (Holland, 2003).
The Transcriptome of the Aplysia Nervous System: Overview of Predicted Neuronal Genes
As expected, the Aplysia neuronal transcriptome is enriched for many developmentally related genes. The transcriptome reflects nearly every aspect of neuronal signaling and includes genes such as Microcephalin and Abnormal Spindle-like Microcephaly associated (ASPM) that determine brain size and glial markers such as AChBP (AAL37251, Figure 4D), Aplysia glial protein (Ag), and glia maturation factor β. Perhaps most surprising is the finding of markers for various human disorders (Table S10). We illustrate the diversity of new information contained within this database with several selected examples.
Figure 4. Expression Patterns of Selected Neuronal Genes in the CNS of Aplysia californica (In Situ Hybridization).
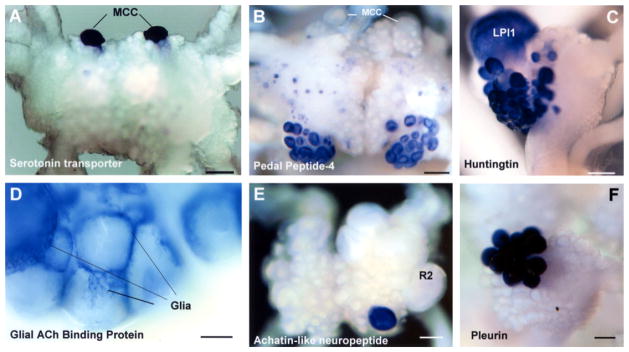
RNA probes were designed based on cloned sequences to examine their expression in individual Aplysia ganglia.
(A) Expression of serotonin transporter (AAK94482) in paired MCC neurons, in a dorsal view of the cerebral ganglion.
(B) Novel isoform of pedal-type neuropeptide (DQ479396) is predominantly expressed in two symmetrical clusters of motoneurons (A/B groups) in the cerebral ganglion.
(C) Expression of Huntingtin in the left pleural ganglion (LPl1 is the giant cholinergic neuron involved in control of mucus release from the body wall).
(D) Acetylcholine Binding Protein (AAL37251) is exclusively expressed in glial type cell of Aplysia (the caudal part of the abdominal ganglion).
(E) Expression of Achatin-like neuropeptide precursor (AY842441) in the abdominal ganglion.
(F) Highly localized expression of Pleurin neuropeptide precursor (AY833131) in the right pleural ganglion. Scales: 300 μm (A and B); 200 μm (C); 100 μm (D); 350 μm (E); 120 μm (F).
Neurotransmitter Systems
The Aplysia neuronal transcriptome contains transcripts encoding genes for the synthesis of most major transmitters, their receptors, and essential components of their signal transduction pathways (Table 1) and provides the first insight into the existence of purinergic transmission in molluscs. The availability of many of these markers allowed us to map cholinergic and octopaminergic neurons in Aplysia (Figure S2).
Disease-Relevant Gene Products
A comparative analysis of 146 human genes implicated in 168 neurological diseases (Cravchik et al., 2001) allowed us to identify 104 orthologs (71%) of these genes in Aplysia (Table S10), including genes relevant to Parkinsons and Alzheimer’s disease, thus affording a unique opportunity to define their functions in simpler networks. Eleven Aplysia homologs of human genes, relevant to different neurodegenerative diseases, are differentially lost either in Drosophila (e.g., Cochilin, Fukutin, CLN8, GM2 activator) or C. elegans (e.g., Huntingtin [Figure 4C], Fragile X Mental Retardation Protein, Hyperpolarization-activated cyclic nucleotide-modulated ionic channel, and Wolframin) lineages. These comparisons suggest that the distinctive features of the CNS of Aplysia present opportunities to study molecular and cellular functions of these genes, as well as to develop relevant models for these diseases in Aplysia. Interestingly, Aplysia has an ortholog of the mammalian antiaging hormone, Klotho. In mice, a defect in Klotho gene expression accelerates the aging process, while its overexpression extends the life span (Kurosu et al., 2005).
Immunity and Posttranslational Gene Silencing in CNS
Orthologs of 25 predicted antimicrobial, antiviral, and immune-related proteins have been found in the Aplysia EST database, including lysozymes, transcripts similar to blood coagulation factors, Ig heavy chain precursors, and complement precursors. Until now, complement genes were only found in vertebrates (as an illustrative example of vertebrate lineage-specific innovations related to adaptive immunity), and their discovery in Aplysia suggests that orthologs of these genes were lost in Ecdysozoa (nematode and arthropod) lineages.
Although RNAi is broadly found in different organisms, there previously has been no evidence in molluscs for the presence of proteins involved in RNAi. We have identified the main components of RNAi in Aplysia, including homologs to RNase III, the Dicer & Argonaute gene family.
Identification of New Signaling Pathways in Invertebrate Nervous System
Our analysis has identified two previously undescribed signaling pathways in the invertebrate CNS: thyroid hormone (TH) related signaling (Heyland et al., 2006 and Table S11) and CpG methylation. TH hormones are involved in the development of the nervous system in a variety of vertebrate taxa. We find a peroxidasin ortholog, potentially involved in TH metabolism, expressed in the cerebral ganglion. The presence of this gene in the CNS of Aplysia californica, along with several other putative transcripts from the TH-like signaling pathway, suggests the presence of TH-like signaling in molluscs as well (Heyland et al., 2006).
DNMT1 and DNA methyltransferases in general have been found to be lacking in C. elegans, Schistosoma (Rosado Fantappie et al. 2001), and dipteran insects (Goll et al. 2006), which all are known to have only trace levels of 5-methyl cytosine at CpG sites if any (Simpson et al. 1986, Rae and Steele 1979). We find in the Aplysia EST database homologs for the DNA methyltransferase DNMT1, DMAP1 (DNA methyltransferase associating protein), and the transcriptional repressor MBD2 (Methyl-CpG binding Domain Protein 2). These findings demonstrate that Aplysia has many of the fundamental components for transcriptional regulation by CpG methylation.
Portrait of the Transcriptome from Individual Neurons and Processes
How does the mRNA expression profile of one neuron differ from that of different neuronal types? How does the mRNA population in the cell body of a neuron differ from the mRNAs present in the neuronal processes? As a first step to address these questions, we have examined the composition of the components of the gill-withdrawal reflex: the sensory neuron and the motor neuron. In addition, we examined a model serotonergic cell capable of modulating the strength of the synaptic connections.
Identification of Circuit-Specific Transcripts: Transcriptomic Profiling of Sensory and Motor Neurons Using Representative Aplysia Microarrays
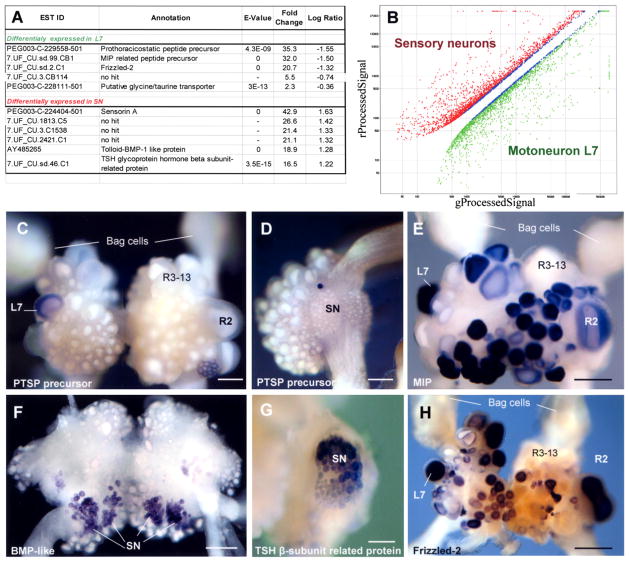
Using custom-made highly representative Aplysia-specific oligonucleotide (60-mer) microarrays, we examined a critical monosynaptic component of the neural circuit of the gill-withdrawal reflex, the motor neuron L7, and the mechanosensory neurons. After performing significance analysis with a 5% false discovery rate and 2-fold change cut-off, we identified 853 differentially expressed transcripts: 362 were enriched in L7 and 491 were enriched in sensory neurons (see illustrative examples in Figure 5 and Tables S12 and S13). This approach has provided new ideas about the specificity of neuro-transmitter action and identification of candidate cell-specific signaling molecules.
Figure 5. Identification of L7 and Sensory Neuron-Specific Transcripts Using Microarrays.
(A) Selected differentially expressed transcripts in L7 and sensory neurons.
(B) Expression results from oligoarray experiments comparing L7 and sensory neurons. Spots representing transcripts common to the two neuronal types have been removed for clarity.
(C–H) Neuron-specific expression (in situ hybridization) of selected transcripts identified in the microarray experiments (C, E, and H: abdominal ganglion; D and G: pleural ganglia; F: cerebral ganglia). Transcripts encoding Aplysia prothoraracicostatic peptide (PTSP) homolog (DQ375548, C and D), MIP-like neuropeptide (AAF80382, E) and Frizzled-2 (AY535406, H) are expressed in L7 (but not in sensory neurons, e.g., D), while BMP-like (AY485265, F) and TSH glycoprotein hormone β subunit-related protein (AAX35673), G) genes are expressed in sensory neurons. Note that MIP-like peptide prohormone and Frizzled-2 are also expressed in a subset of abdominal neurons, including some gill motoneurons and R2 neuron. Scales: 300 μm (C); 250 μm (D); 400 μm (E and H); 200 μm (F); 150 μm (G).
Our initial analysis pointed to three L7 specific proteins as candidate transmitter molecules. The first is the Mytilus inhibitory peptide (MIP) related precursor (Figure 5E), initially cloned from bivalves and later from Aplysia (Fujisawa et al., 1999). This precursor can produce opioid-like peptides known to be specific modulators in molluscan neurons. The second candidate is a novel Aplysia neuro-peptide precursor (PTSP, Figures 5C and 5D), distantly related to a prothoracicostatic hormone in the silkworm Bombyx mori that controls insect development via cAMP/Ca2+ signaling cascades. Using whole-mount in situ hybridization (Figure 5), coupled with Lucifer yellow injection of L7 (not shown), we demonstrated that the MIP related peptide precursor, the PTSP, and a third putative neurotransmitter transporter are differentially expressed in L7 but not sensory neurons, confirming the microarray data.
In situ hybridization with choline acetyltransferase (ChAT, a marker for cholinergic neurons) revealed that L7 is negative for ChAT and thus not likely a cholinergic motor neuron (Giller and Schwartz, 1971). No GABA immunoreactive neurons have been observed in the abdominal ganglion (Diaz-Rios et al., 1999), and this essentially rules out the possibility that L7 is GABAergic. This leaves both taurine-/ glycine-type transporters as candidates for L7 specific uptake mechanisms. In addition, the micro-array data suggested L7-specific expression of several molecules known to be key regulators in morphogenesis and cell-cell signaling. Of these, we cloned Frizzled-2 (AY535406) and confirmed its high and specific expression in L7 but not sensory neurons (Figure 5H).
Microarray data for the presynaptic (sensory) neuron confirmed the presence of bone morphogenic (BMP) type/tolloid protein (confirmed by in situ hybridization, Figure 5F), a candidate involved in structural changes associated with cell-cell communication. A second major cell-specific transcript in the sensory neurons is the sensorin prohormone. Other predicted gene products differentially expressed in sensory neurons also include genes known to be involved in the pathways leading to short-and long-term plasticity (Tables S12 and S13).
Serotonergic Modulatory Cells (MCC)
Normalized and nonnormalized cDNA libraries from five to eight individual MCC neurons were analyzed to obtain good representation of both common and rare cell-specific transcripts. The assembly of ESTs obtained from MCC neurons resulted in 9,945 nonredundant sequences, suggesting that roughly 10,000 unique gene products are expressed in a single Aplysia neuron. Of these, 1,263 genes were assigned to GO categories based on their predicted molecular function or biological process (Figure 2A and Table S3). Among the most abundant transcripts in MCC neurons are those for serotonin transporter and predicted pheromone receptors. The transcriptome of MCC also contains putative photoreceptors, suggesting that these interneurons might also be involved in light sensitivity or regulation of circadian clocks (supported by the expression of Nocturnin homologs).
Subcellular Transcriptome of a Neuron: EST Collection from MCC Neurites and Comparison to the Neurites of Sensory Neurons
Since these peripheral processes sustain the requirement of local protein synthesis at the synaptic sites, a cohort of mRNAs must be transported to the peripheral processes. We addressed this issue by sequencing normalized and nonnormalized cDNA libraries from neuronal processes (neurites) of MCC. The assembly of 4515 ESTs resulted in 198 different clusters of transcripts and 830 singlets. We annotated 268 unique nonoverlapping sequences in MCC processes and assigned them to predicted molecular functions using GO categories (Figure 2A and Table S3).
Table 2 shows several transcripts, including actin, ubiquitin, and several ribosomal proteins that are enriched several-fold in the MCC processes in comparison to the MCC somata. The enrichment of several ribosomal proteins in the MCC processes agrees with recent information on the processes of the sensory neuron and suggests that in the processes the proteins present on the ribosome surface may be replenished directly and have the capability for local protein synthesis (Moccia et al., 2003). Interestingly, the subcellular transcriptome of MCC also contains several cell communication and signal transduction components, including mRNAs encoding four kinases (diglyceride kinase, delta; calcium independent protein kinase C; and two serine-threonine protein kinases similar to PAK-1 and CDC2L), inositol-1,4,5-triphosphate receptor, NMDA receptor, presenilin, hedgehog homologs, Slit homolog 2, RNA binding ELAV-like protein, cadherin, and FMRP.
Table 2.
Abundance of Transcripts in MCC Somata (MCC_N) and MCC Neuronal Processes (MCC_P)
| EST Annotation | Blast Hit Accession | Blast E-value | MCC_N% | MCC_P% |
|---|---|---|---|---|
| Tubulin B-alpha-1 | Q71U36 | 0 | 1.3 | 1 |
| Soma Ferritin | P42577 | 1E-76 | 1.1 | 1.5 |
| Similar to putative pheromone receptor | XP_578826 | 6E-16 | 1.1 | 1.1 |
| Thymosin beta | Q9W7M8 | 4E-12 | 0.5 | 0.9 |
| Sodium-dependent serotonin transporter * | P51143 | 3E-51 | 0.5 | 0.07 |
| Synaptotagmin-1 (p65) | P41823 | 1E-88 | 0.4 | 0.07 |
| 7B2 [Lymnaea stagnalis] | AAB41699 | 1E-107 | 0.3 | 0.6 |
| Receptor of activated protein kinase C | O42248 | 1E-160 | 0.3 | 0.4 |
| Calmodulin | P62153 | 1E-79 | 0.3 | 0 |
| Eukaryotic translation initiation factor 1A | P47813 | 1E-41 | 0.3 | 0 |
| Major vault protein (MVP100) | Q90405 | 3E-48 | 0.3 | 0 |
| 60S ribosomal protein L6 | Q02878 | 2E-66 | 0.2 | 0.07 |
| 40S ribosomal protein S15A | P48149 | 3E-62 | 0.2 | 0 |
| Spectrin beta chain, brain 1 | Q01082 | 1E-174 | 0.2 | 0 |
| Aromatic-L-amino-acid decarboxylase (AADC) * | P05031 | 1E-51 | 0.2 | 0 |
| 60S ribosomal protein L3 | O16797 | 1E-115 | 0.1 | 0.4 |
| 40S ribosomal protein S2 | Q90YS3 | 8E-85 | 0.1 | 0.3 |
| Translationally controled tumor protein (TCTP) | O18477 | 4E-11 | 0.1 | 0.1 |
| 60S ribosomal protein L7 | P18124 | 1E-75 | 0.1 | 0.07 |
| Amelogenin | Q28462 | 2.00E-07 | 0.1 | 0 |
| 60S ribosomal protein L5 | Q26481 | 3E-66 | 0.05 | 0.6 |
| Nucleoside diphosphate kinase NBR-A | P52174 | 5E-58 | 0.05 | 0.4 |
| 60S ribosomal protein L7a | O57592 | 7E-99 | 0.05 | 0.3 |
| 40S ribosomal protein S8 | Q90YR6 | 7E-86 | 0.02 | 0.3 |
| Unknown transcript | - | - | 0.02 | 0.4 |
| 60S ribosomal protein L9 | P50882 | 3E-67 | 0.02 | 0.3 |
| Hypothetical protein Bd1086 | NP_968019 | 1.00E-08 | 0.02 | 0.3 |
| Cytochrome c oxidase polypeptide Vb | P19536 | 9E-15 | 0.02 | 0.3 |
| Ubiquitin | P68201 | 6E-30 | 0.02 | 0.3 |
| Actin, cytoplasmic | Q964E0 | 1E-166 | 0.02 | 0.2 |
| 60S ribosomal protein L10 | O96647 | 1E-103 | 0.02 | 0.1 |
| 60S ribosomal protein L13a | P40429 | 1E-68 | 0.02 | 0.1 |
| 40S ribosomal protein S3B | P47835 | 1E-116 | 0.02 | 0.1 |
| Tax_Id = 9606 47 kDa protein | ENSP00000348114 | 2.00E-07 | 0.02 | 0.07 |
| 60S ribosomal protein L13 | Q90Z10 | 3E-64 | 0.02 | 0.07 |
| 40S ribosomal protein S27 | Q6ZWU9 | 2E-34 | 0.02 | 0.05 |
| 40S ribosomal protein S5 | P46782 | 3E-96 | 0.02 | 0.05 |
| 60S ribosomal protein L18B (L14B) | P02412 | 1E-79 | 0.02 | 0.04 |
| Non-neuronal cytoplasmic intermediate filament protein (IF) | P22488 | 2E-24 | 0.02 | 0.02 |
Transcripts enriched in MCC neuronal processes are marked in bold. Markers of serotonin containing cells are labeled with asterisk (*).
We took advantage of our Aplysia transcriptome database to further annotate a set of 668 ESTs, independently collected from pure neuronal processes of pleural sensory neurons (Moccia et al., 2003) and to compare them to the subset of extrasomatic transcripts identified in MCC. 564 ESTs derived from sensory neuronal processes matched Aplysia transcripts in the present EST database (495 of them were identical or nearly identical and 169 had significant similarity). As a result, we annotated 66 novel transcripts, including ribosomal proteins, RNA binding proteins, and signaling proteins.
Direct comparison of the MCC neurite transcriptome to the transcriptome of the neurites of the sensory neuron suggests that a subset of extrasomatic transcripts is neuron-specific, including transcripts encoding signaling molecules and neuropeptides characteristic of a single cell type. Several identified transcripts that are associated with synaptic plasticity were found in sensory neuron processes, but not in MCC. These are the frizzled related protein, TGF β-inducible protein, ephrin type receptor, receptor for activated protein kinase C, the apoptosis inhibitor protein, CREB, and fasciclin. In addition, sensory neuron neurites are enriched for mRNAs that encode caveolin and the neuropeptide sensorin. By contrast, MCC neurites contain mRNAs encoding the serotonin transporter and serotonin synthetic enzyme tryptophan 5-monooxygenase that were not present in sensory neuron processes.
Conclusions
As a result of large-scale sequencing of the Aplysia neuronal transcriptome, we have established a database for future gene discoveries, expression profiling, and characterization of signaling pathways. Our evolutionary analysis has suggested the Lophotrochozoa are closer to nematodes and arthropods than to chordates and other deuterostomes. Indeed, we have been able to clarify some of the evolutionary relationships and have identified genes that have been lost in the three major lineages of bilaterian animals. Thus, our neural transcriptome data have helped delineate a set of genes derived from a common bilaterian ancestor. Genes found in this database include many formerly unknown in molluscs, including many associated with human neurological diseases that are not found in other classic model organisms. Further, the initial gene identification and expression analyses conducted on individual neurons and their processes in Aplysia provide a bridge between genomics at the level of identified neurons and physiological analysis within a defined cellular network and its appropriate linked behaviors. As the emphasis in neural science shifts from cellular to circuit studies, Aplysia can provide a number of interesting behaviors accessible for detailed investigation. The Aplysia transcriptome thus reveals many potential targets for investigation, particularly in terms of the more complex neural circuitry of behaviour, such as the fixed action potential patterns of feeding and locomotion.
EXPERIMENTAL PROCEDURES
cDNA Library Construction
Specimens (5–80 g) of Aplysia californica (Opisthobranchia: Anaspidea) were obtained from the NIH/University of Miami National Resource for Aplysia. Prior to dissection, animals were anesthetized by injecting a volume of isotonic MgCl2 (337 mM) equivalent to 50%–60% of their weight. Normalized and nonnormalized cDNA libraries were constructed from four types of tissues: (1) the whole CNS, (2) pedal-pleural ganglia, (3) individual MCC neurons, and (4) neuronal processes from MCC neurons. All details of cell isolation and culture protocols have been described elsewhere (Moccia et al., 2003; Lovell and Moroz, 2006). Normalization of the CNS, MCC, and MCC process libraries was performed using both a novel cDNA normalization method (duplex-specific nuclease or DSN normalization [Zhulidov et al., 2004]) and standard techniques (Fu et al., 2002; Soares et al., 1994). See additional details, including a complete list of libraries (Table S1), in the Supplemental Data.
DNA Sequencing and Sequence Analysis
After cloning cDNA libraries into pBluescript, individual clones were amplified using either PCR or TempliPhi rolling circle amplification (GE Healthcare) and analyzed on either an ABI3730 (Applied Biosystems) or a MegaBACE 1000 (GE Healthcare) DNA sequencer.
All sequences were checked for quality prior to downstream analysis using custom PERL scripts. Using Paracel TranscriptAssembler (PTA) version 2.7.0 (Paracel Inc, Pasadena, CA), ribosomal RNA sequences, E. coli contamination, and mitochondrial genes were removed, and all vector and adaptor sequences were masked. Resulting sequences were then clustered and sequence clusters were assembled to yield a unique set of sequences (800 bases average length). Unique sequences were annotated using Paracel Blast through blastx searches against the SwissProt/Trembl and NCBI nr databases using an E-value cutoff of 10−10. Further details concerning EST annotation and analysis can be found in the Supplemental Data.
All annotation information, clustering analysis, and sequence information are stored in a MySQL database. Public access is available through two project websites at http://aplysia.uf-genome.org and http://aplysia.cu-genome.org. Data can be accessed through annotations that are broken down into GO categories for easy browsing or through blast searches with user-input sequences. Databases are also linked to graphical representations of metabolic pathways and relevant references in PubMed. Further detailed descriptions of the databases are available at the indicated websites.
Phylogenetic Analysis
Reference sequences from the latest release of the KOG database (Tatusov et al., 2003) were obtained from NCBI. Orthologs for each KOG were extracted from publicly available protein and EST databases as well as the Aplysia EST database for each of the species analyzed. EST sequences were translated into all reading frames, and the protein translation with the highest homology was extracted. This yielded a total of 45 protein families (see Table S15) with at least 11 orthologous proteins in each. Alignments of orthologous sequences were performed with T-Coffee and manually verified to ensure accuracy. Phylogenetic trees were determined from the alignments (excluding regions with more than two gaps) with the PHYML program using the JTT model and γ distributed rates (Guindon and Gascuel, 2003; Jones et al., 1992; Yang, 1994). Bootstrap analyses were performed to evaluate the reliability of the phylogenies. Further details can be found in the Supplemental Data.
In Situ Hybridization
Sense and antisense probes were generated with the DIG RNA Labeling Kit (Roche Diagnostics); all details for in situ hybridization protocols have been described elsewhere (Jezzini et al., 2005; Jezzini and Moroz, 2004).
Microarray Experiments
Two custom 44,000 oligonucleotide arrays were constructed in collaboration with Agilent Technologies using 60-mer oligonucleotide sequences designed from each nonredundant sequence in the Aplysia EST database. The data discussed in this publication have been deposited in NCBIs Gene Expression Omnibus (GEO) and are accessible through the GEO Series accession number GSE4628. Additional details about the protocols used including a list of features on each array can be found at the GEO web site and are summarized in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Tom Carew, Kelsey Martin, Kausik Si, Tom Jessel, Wayne Sossin, John Byrne, and Larry Zipursky for their comments on earlier versions of this paper. Our work is supported by HHMI, NSF, and National Institutes of Health Center of Excellence in Genomic Science Grants P50 HG002806 and R01 MH075026, NS39103, and in part by the McKnight Brain Research Foundations, UF Opportunity Funds, and RFBR-05-04-48401. We also would like to thank Mrs. E. Bobkova, T. Brough, E. Meleshkevitch, J. Netherton, and Drs. M. Matz, N. Alieva, and R. Sadreyev for technical help and comments at earlier stages of this project. We thank MOgene (LC), and in particular Shaukat Rangwala, for the microarray analysis, and Drs. E. Koonin and Y. Wolf for KOGs discussions. Computational work for this project was supported by The AMDeC Bioinformatics Core Facility at the Columbia Genome Center, Columbia University, and the ICBR Core facility at the University of Florida. Currently, Y. Panchin is at Moscow State University, Moscow, Russia; and B. Knudsen is at CLC bio A/S Aarhus, Denmark.
Footnotes
Supplemental Data include fifteen tables, two figures, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.cell.com/cgi/content/full/127/7/1453/DC1/.
Accession Numbers
The Aplysia EST sequences have been deposited in GenBank with accession numbers EB187504-EB359573, BF707524-BF708380, BF713631-BF713632, BI273615-BI273627, CK327631-CK329175, and CK320902-CK325388 (see Table S5 for full-length genes).
References
- Aguinaldo AM, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- Brannvall K, Hjelm H, Korhonen L, Lahtinen U, Lehesjoki AE, Lindholm D. Cystatin-B is expressed by neural stem cells and by differentiated neurons and astrocytes. Biochem Biophys Res Commun. 2003;308:369–374. doi: 10.1016/s0006-291x(03)01386-x. [DOI] [PubMed] [Google Scholar]
- Brusca RC, Brusca GJ. Invertebrates. 2. Sunderland, MA: Sinauer Associates, Inc; 2003. [Google Scholar]
- Cash D, Carew TJ. A quantitative analysis of the development of the central nervous system in juvenile Aplysia californica. J Neurobiol. 1989;20:25–47. doi: 10.1002/neu.480200104. [DOI] [PubMed] [Google Scholar]
- Chen JY, Bottjer DJ, Oliveri P, Dornbos SQ, Gao F, Ruffins S, Chi H, Li CW, Davidson EH. Small bilaterian fossils from 40 to 55 million years before the Cambrian. Science. 2004;305:218–222. doi: 10.1126/science.1099213. [DOI] [PubMed] [Google Scholar]
- Cleary LJ, Byrne JH. Identification and characterization of a multifunction neuron contributing to defensive arousal in Aplysia. J Neurophysiol. 1993;70:1767–1776. doi: 10.1152/jn.1993.70.5.1767. [DOI] [PubMed] [Google Scholar]
- Cravchik A, Subramanian G, Broder S, Venter JC. Sequence analysis of the human genome: implications for the understanding of nervous system function and disease. Arch Neurol. 2001;58:1772–1778. doi: 10.1001/archneur.58.11.1772. [DOI] [PubMed] [Google Scholar]
- de Rosa R, Grenier JK, Andreeva T, Cook CE, Adoutte A, Akam M, Carroll SB, Balavoine G. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- Diaz-Rios M, Suess E, Miller MW. Localization of GABA-like immunoreactivity in the central nervous system of Aplysia californica. J Comp Neurol. 1999;413:255–270. doi: 10.1002/(sici)1096-9861(19991018)413:2<255::aid-cne7>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KK, Takacs CA, Carew TJ. Nonassociative and associative modification of head-waving produced by aversive tentacular stimuli in Aplysia. Learn Mem. 1997;3:366–375. doi: 10.1101/lm.3.5.366. [DOI] [PubMed] [Google Scholar]
- Fu GK, Starnes S, Stuve LL. Construction of uni-directionally cloned cDNA libraries from messenger RNA for improved 3′ end DNA sequencing. 6, 387,624 US patent. 2002 May;
- Fujisawa Y, Furukawa Y, Ohta S, Ellis TA, Dembrow NC, Li L, Floyd PD, Sweedler JV, Minakata H, Nakamaru K, et al. The Aplysia mytilus inhibitory peptide-related peptides: identification, cloning, processing, distribution, and action. J Neurosci. 1999;19:9618–9634. doi: 10.1523/JNEUROSCI.19-21-09618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giller E, Jr, Schwartz JH. Choline acetyltransferase in identified neurons of abdominal ganglion of Aplysia californica. J Neurophysiol. 1971;34:93–107. doi: 10.1152/jn.1971.34.1.93. [DOI] [PubMed] [Google Scholar]
- Giribet G, Okusu A, Lindgren AR, Huff SW, Schrodl M, Nishiguchi MK. Evidence for a clade composed of molluscs with serially repeated structures: Monoplacophorans are related to chitons. Proc Natl Acad Sci USA. 2006;103:7723–7728. doi: 10.1073/pnas.0602578103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA Methyltransferase Homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- Grande C, Templado J, Cervera JL, Zardoya R. Molecular phylogeny of Euthyneura (Mollusca: Gastropoda) Mol Biol Evol. 2004a;21:303–313. doi: 10.1093/molbev/msh016. [DOI] [PubMed] [Google Scholar]
- Grande C, Templado J, Cervera JL, Zardoya R. Phylogenetic relationships among Opisthobranchia (Mollusca: Gastropoda) based on mitochondrial cox 1, trnV, and rrnL genes. Mol Phylogenet Evol. 2004b;33:378–388. doi: 10.1016/j.ympev.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haag ES, Sly BJ, Andrews ME, Raff RA. Apextrin, a novel extracellular protein associated with larval ectoderm evolution in Heliocidaris erythrogramma. Dev Biol. 1999;211:77–87. doi: 10.1006/dbio.1999.9283. [DOI] [PubMed] [Google Scholar]
- Halanych KM. The new view of animal phylogeny. Annu Rev Ecol Syst. 2004;35:229–256. [Google Scholar]
- Haszprunar G. Is the Aplacophora monophyletic? A cladistic point of view. Amer Malacol Bull. 2000;15:115–130. [Google Scholar]
- Hening WA, Walters ET, Carew TJ, Kandel ER. Motorneuronal control of locomotion in Aplysia. Brain Res. 1979;179:231–253. doi: 10.1016/0006-8993(79)90441-4. [DOI] [PubMed] [Google Scholar]
- Heyland A, Price AD, Bodnarova-Buganova M, Moroz LL. Thyroid hormone metabolism and peroxidase function in two non-chordate animals. J Exp Zoolog B Mol Dev Evol. 2006;306:551–566. doi: 10.1002/jez.b.21113. [DOI] [PubMed] [Google Scholar]
- Holland ND. Early central nervous system evolution: an era of skin brains? Nat Rev Neurosci. 2003;4:617–627. doi: 10.1038/nrn1175. [DOI] [PubMed] [Google Scholar]
- Jahan-Parwar B, Fredman SM. Neural control of locomotion in Aplysia: role of the central ganglia. Behav Neural Biol. 1979;27:39–58. doi: 10.1016/s0163-1047(79)92744-4. [DOI] [PubMed] [Google Scholar]
- Jezzini SH, Moroz LL. Identification and distribution of a two-pore domain potassium channel in the CNS of Aplysia californica. Brain Res Mol Brain Res. 2004;127:27–38. doi: 10.1016/j.molbrainres.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Jezzini SH, Bodnarova M, Moroz LL. Two-color in situ hybridization in the CNS of Aplysia californica. J Neurosci Methods. 2005;149:15–25. doi: 10.1016/j.jneumeth.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kandel ER. Cellular Basis of Behavior. San Francisco: W.H. Freeman and Company; 1976. [Google Scholar]
- Kandel ER. Behavioral Biology of Aplysia. San Francisco: W.H. Freeman and Company; 1979. [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kandel ER, Tauc L. Anomalous rectification in the meta-cerebral giant cells and its consequences for synaptic transmission. J Physiol. 1966;183:287–304. doi: 10.1113/jphysiol.1966.sp007867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen B, Kohn AB, Nahir B, McFadden CS, Moroz LL. Complete DNA sequence of the mitochondrial genome of the sea-slug, Aplysia californica: conservation of the gene order in Euthyneura. Mol Phylogenet Evol. 2006;38:459–469. doi: 10.1016/j.ympev.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Kortschak RD, Samuel G, Saint R, Miller DJ. EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr Biol. 2003;13:2190–2195. doi: 10.1016/j.cub.2003.11.030. [DOI] [PubMed] [Google Scholar]
- Kupfermann I. Feeding behavior in Aplysia: a simple system for the study of motivation. Behav Biol. 1974;10:1–26. doi: 10.1016/s0091-6773(74)91644-7. [DOI] [PubMed] [Google Scholar]
- Kupfermann I, Kandel ER. Neuronal controls of a behavioral response mediated by the abdominal ganglion of Aplysia. Science. 1969;164:847–850. doi: 10.1126/science.164.3881.847. [DOI] [PubMed] [Google Scholar]
- Kupfermann I, Weiss KR. Activity of an identified serotonergic neuron in free moving Aplysia correlates with behavioral arousal. Brain Res. 1982;241:334–337. doi: 10.1016/0006-8993(82)91072-1. [DOI] [PubMed] [Google Scholar]
- Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- Lehesjoki AE. Molecular background of progressive myoclonus epilepsy. EMBO J. 2003;22:3473–3478. doi: 10.1093/emboj/cdg338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell P, Moroz LL. The largest growth cones in the animal kingdom and dynamics of neuronal growth in cell culture of Aplysia. Integr Comp Biol. 2006;46:847–870. doi: 10.1093/icb/icl042. [DOI] [PubMed] [Google Scholar]
- Lydeard C, Holznagel WE, Schnare MN, Gutell RR. Phylogenetic analysis of molluscan mitochondrial LSU rDNA sequences and secondary structures. Mol Phylogenet Evol. 2000;15:83–102. doi: 10.1006/mpev.1999.0719. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Ball EE, Technau U. Cnidarians and ancestral genetic complexity in the animal kingdom. Trends Genet. 2005;21:536–539. doi: 10.1016/j.tig.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Moccia R, Chen D, Lyles V, Kapuya EEY, Kalachikov S, Spahn CM, Frank J, Kandel ER, Barad M, Martin KC. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23:9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachin L, Nannmark U, Nystrom T. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J Bacteriol. 2005;187:6265–6272. doi: 10.1128/JB.187.18.6265-6272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C. Animal Evolution: Interrelationships of the Living Phyla. 2. Oxford: Oxford Univ. Press; 2001. [Google Scholar]
- Nielsen C. Larval and adult brains. Evol Dev. 2005;7:483–489. doi: 10.1111/j.1525-142X.2005.05051.x. [DOI] [PubMed] [Google Scholar]
- Passamaneck Y, Halanych KM. Lophotrochozoan phylogeny assessed with LSU and SSU data: Evidence of lophophorate polyphyly. Mol Phylogenet Evol. 2006;40:20–28. doi: 10.1016/j.ympev.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Passamaneck YJ, Schander C, Halanych KM. Investigation of molluscan phylogeny using large-subunit and small-subunit nuclear rRNA sequences. Mol Phylogenet Evol. 2004;32:25–38. doi: 10.1016/j.ympev.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Peterson KJ, Eernisse DJ. Animal phylogeny and the ancestry of bilaterians: inferences from morphology and 18S rDNA gene sequences. Evol Dev. 2001;3:170–205. doi: 10.1046/j.1525-142x.2001.003003170.x. [DOI] [PubMed] [Google Scholar]
- Philip GK, Creevey CJ, McInerney JO. The Opisthokonta and the Ecdysozoa may not be clades: stronger support for the grouping of plant and animal than for animal and fungi and stronger support for the Coelomata than Ecdysozoa. Mol Biol Evol. 2005;22:1175–1184. doi: 10.1093/molbev/msi102. [DOI] [PubMed] [Google Scholar]
- Philippe H, Lartillot N, Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of ecdysozoa, lophotrochozoa, and protostomia. Mol Biol Evol. 2005;22:1246–1253. doi: 10.1093/molbev/msi111. [DOI] [PubMed] [Google Scholar]
- Pojeta J, Runnegar B, Peel JS, Gordon MJ. Phylum Mollusca. In: Boardman RS, Chetham AH, Rowell AJ, editors. Fossil Invertebrates. Cambridge, MA: Blackwell Science; 1987. pp. 270–435. [Google Scholar]
- Rae PM, Steele RE. Absence of cytosine methylation at C-C-G-G and G-C-G-C sites in the rDNA coding regions and intervening sequences of Drosophila and the rDNA of other insects. Nucleic Acids Res. 1979;6:2987–2995. doi: 10.1093/nar/6.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible F, Tessmar-Raible K, Osoegawa K, Wincker P, Jubin C, Balavoine G, Ferrier D, Benes V, de Jong P, Weissenbach J, et al. Vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii. Science. 2005;310:1325–1326. doi: 10.1126/science.1119089. [DOI] [PubMed] [Google Scholar]
- Rokas A, Kruger D, Carroll SB. Animal evolution and the molecular signature of radiations compressed in time. Science. 2005;310:1933–1938. doi: 10.1126/science.1116759. [DOI] [PubMed] [Google Scholar]
- Rosen SC, Weiss KR, Goldstein RS, Kupfermann I. The role of a modulatory neuron in feeding and satiation in Aplysia: effects of lesioning of the serotonergic metacerebral cells. J Neurosci. 1989;9:1562–1578. doi: 10.1523/JNEUROSCI.09-05-01562.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado Fantappie M, Rodrigues Pereira Gimba E, Rumjanek FD. Lack of DNA Methylation in Schistosoma mansoni. Exp Parasitol. 2001;98:162–166. doi: 10.1006/expr.2001.4630. [DOI] [PubMed] [Google Scholar]
- Sakharov DA. Evolutionary aspects of transmitter heterogeneity. J Neural Transm Suppl. 1974;11:43–59. [PubMed] [Google Scholar]
- Sheng G, dos Reis M, Stern CD. Churchill, a zinc finger transcriptional activator, regulates the transition between gastrulation and neurulation. Cell. 2003;115:603–613. doi: 10.1016/s0092-8674(03)00927-9. [DOI] [PubMed] [Google Scholar]
- Simpson VJ, Johnson TE, Hammen RF. Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res. 1986;14:6711–6719. doi: 10.1093/nar/14.16.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares MB, Bonaldo MF, Jelene P, Su L, Lawton L, Efstratiadis A. Construction and characterization of a normalized cDNA library. Proc Natl Acad Sci USA. 1994;91:9228–9232. doi: 10.1073/pnas.91.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U, Rudd S, Maxwell P, Gordon PM, Saina M, Grasso LC, Hayward DC, Sensen CW, Saint R, Holstein TW, et al. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 2005;21:633–639. doi: 10.1016/j.tig.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Valentine JW. On the origin of phyla. Chicago: The University of Chicago Press; 2004. [Google Scholar]
- Vonnemann V, Schrodl M, Klushmann-Kolb A, Wagele H. Reconstruction of the phylogeny of Opisthobranchia (Mollusca, Gastropoda) by means of 18S and 28S rRNA gene sequences. J Molluscan Stud. 2005;71:113–125. [Google Scholar]
- Weiss KR, Kupfermann I. Homology of the giant serotonergic neurons (metacerebral cells) in Aplysia and pulmonate molluscs. Brain Res. 1976;117:33–49. doi: 10.1016/0006-8993(76)90554-0. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Koonin EV. Coelomata and not Ecdysozoa: evidence from genome-wide phylogenetic analysis. Genome Res. 2004;14:29–36. doi: 10.1101/gr.1347404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Estimating the pattern of nucleotide substitution. J Mol Evol. 1994;39:105–111. doi: 10.1007/BF00178256. [DOI] [PubMed] [Google Scholar]
- Zhulidov PA, Bogdanova EA, Shcheglov AS, Vagner LL, Khaspekov GL, Kozhemyako VB, Matz MV, Meleshkevitch E, Moroz LL, Lukyanov SA, Shagin DA. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 2004;32:e37. doi: 10.1093/nar/gnh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.