Abstract
The origin of complex centralized brains is one of the major evolutionary transitions in the history of animals. Monophyly (i.e. presence of a centralized nervous system in urbilateria) vs polyphyly (i.e. multiple origins by parallel centralization of nervous systems within several lineages) are two historically conflicting scenarios to explain such transitions. However, recent phylogenomic and cladistic analysis suggests that complex brains may have independently evolved at least 9 times within different animal lineages. Indeed, even within the phylum Mollusca cephalization might have occurred at least 5 times. Emerging molecular data further suggest that at the genomic level such transitions might have been achieved by changes in expression of just a few transcriptional factors – not surprising since such events might happen multiple times over 700 million years of animal evolution. Both cladistic and genomic analyses also imply that neurons themselves evolved more than once. Ancestral polarized secretory cells were likely involved in coordination of ciliated locomotion in early animals, and these cells can be considered as evolutionary precursors of neurons within different lineages. Under this scenario, the origins of neurons can be linked to adaptations to stress/injury factors in the form of integrated regeneration-type cellular response with secretory signaling peptides as early neurotransmitters. To further reconstruct the parallel evolution of nervous systems genomic approaches are essential to probe enigmatic neurons of basal metazoans, selected lophotrochozoans (e.g. phoronids, brachiopods) and deuterostomes.
Keywords: Brain evolution, Xenoturbella, Deuterostomes, basal Metazoa, origin of neurons
INTRODUCTION AND OUTLINE
Historically, all hypotheses related to the evolution of central nervous systems (CNSs) and neurons can be broadly presented as two classical scenarios: Monophyly vs Polygenesis (see the overview in [54]). Monophyly means that all neurons originated from a single ancestral cell lineage. This explanation predicts that the origin of a complex brain can be traced back to the ancestral CNS or to a centralized “nerve cord” present in a common ancestor of bilaterian animals (Urbilateria) or all animals. In contrast, Polygenesis (or Polyphyly) refers to multiple origins of neurons and complex brains among species in different lineages [54, 59], see Figs 1 and 2. as a result of parallel evolution, different animal lineages were able to develop centralization of their nervous systems independently.
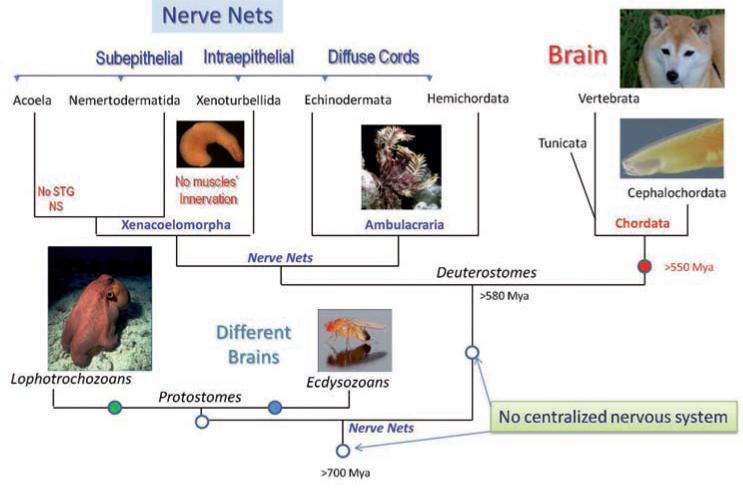
Fig. 1.
Parallel evolution of neuronal centralization in Deuterostomes and Protostomes. The diagram shows types of neural organization among all major Deuterostome lineages. The presented reconstruction of phylogenetic relationships is based upon recent large-scale phylogenomic analysis among all bilaterians [63]. Filled circles indicate possible events of neural centralization from diffuse Nerve Net type of organization in a common ancestor of all Deuterostomes and Protostomes (open circles). Note that nerve nets in Deuterostomes have only superficial similarities and might not be genealogically related to each other. See text for details. Two major superclades of Protostomes (Ecdysozoa and Lophotrochozoa) might also have evolved complex brains independently from Chordates
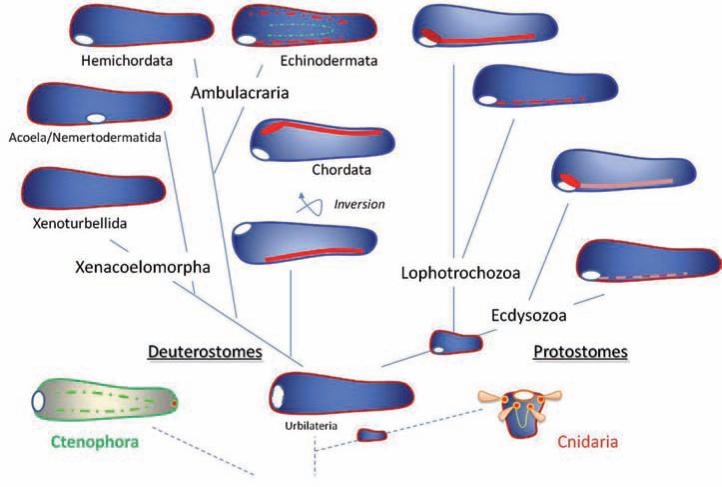
Fig. 2.
The polygenesis of neuronal centralization. The hypothetical scenario outlining multiple origins of neuronal centralization in Bilateria. The ancestral urbilateria had diffuse, possibly only epidermal nerve nets and did not possess central nervous systems. Centralization of neuronal elements and the formation of CNSs and complex brains occurred independently several times in the course of animal evolution (see also Figs 1, 4 and 5). The inversion of the ventral to dorsal axis took place during earlier stages of Chordate evolution. Ctenophore and Cnidaria nervous systems may have evolved in parallel and, at least in part, may not necessarily be related to bilaterian neural organization. Red, orange and green colors schematically illustrate different neuronal structures. Open white circles indicate the position of the mouth. the author supports this hypothesis of neuronal evolution; see text and [54] for details
Early in the 1990s, new molecular data revealed remarkable similarity in spatial expression of genes controlling dorso-ventral and anterio-posterior patterning between vertebrates with a dorsal neural tube and selected invertebrates (insects) with ventral nerve cords (see [10, 17, 18]). These findings revitalized the old hypothesis of Geoffroy Sain-Hilaire about the homology of ventral and dorsal sides of arthropods and vertebrates [27]. In other words, it became widely accepted that during the evolution of chordates dorsoventral body axis inversion took place, and the ventral side of the ancestor of modern arthropods became the dorsal side for chordates [2]. Because similar morphogens and transcription factors control the patterning and formation of the ventral central nervous system in insects and the dorsal nervous system in mammals, it was further suggested that these central nervous systems might be homologous and derived from a common ancestor of all bilaterian animals – the Urbilateria. In its most complete form this hypotheses is summarized by [38, 69]. Under this scenario, an ancestral urbilaterian did possess a relatively complex and centralized nervous system in the form of a ventral cord – a tripartite brain [36, 37]. Such an ancestral state, therefore, implies that in the course of animal evolution some animal lineages might have secondarily lost their centralized nervous systems (Fig. 3).
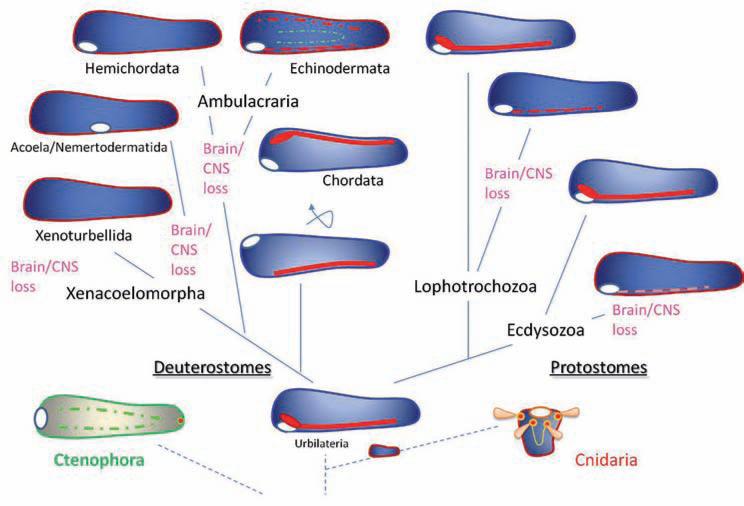
Fig. 3.
The hypothesis of monophyletic origin of the central nervous system in Bilateria. This is an alternative scenario of the neuronal evolution in animals as outlined in Fig. 2. Here, the ancestral urbilateria had a well-defined central nervous system in the form of a ventral cord. However, multiple animal lineages both within Deuterostomes, Ecdysozoa and Lophotrochozoa independently lost their central nervous system (yet many representatives of these groups are still free living organisms, sometimes with complex behavioral repertoires). Ctenophore and Cnidaria nervous systems might be related/homologous to bilaterian neural organization (although at least some parts of them may have evolved independently). Red, orange and green colors schematically illustrate different neuronal structures. Open white circles indicate the position of the mouth. See text and [54] for details
The first problem for this theory is to explain the apparent loss of the centralized brain in many free-living groups of invertebrates such as brachiopods, phoronids, echiurids, priapulids, hemichordates, echinoderms (e.g. in crinoids), Xenoturbella, nemertodermatids, acoels, etc. (Fig. 5). The second difficulty is to accommodate the observed diversity of bilaterian nervous systems which cannot be interpreted in the simpler terms of single dorsal vs ventral cords. Figures 2 and 3 summarize the polyphyletic vs monophyletic hypotheses of the evolution of centralized nervous systems in bilateria.
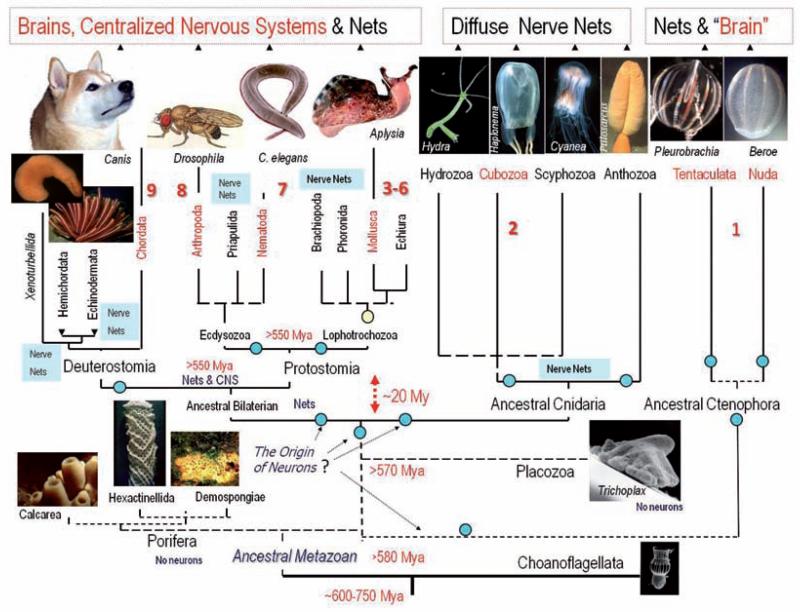
Fig. 5.
Parallel evolution of neuronal centralization in the animal kingdom. The diagram summarizes the current view of evolutionary relationships in the animal kingdom and indicates the presence or absence of a central nervous system (CNS) or brain. From this tree it is possible to see at least 9 possible events of multiple origins of complex brains – shown as red numbers. Circles indicate possible events of multiple origins of neurons. See text for details. This reconstruction of phylogenetic relationships among phyla is a combined view based upon recent large-scale molecular/phylogenomic analyses of several dozen proteins from representatives of more than 15 animal phyla [20, 24, 31, 34, 47, 63, 64, 78, 82, 87]. Only representative groups of the 36 known animal phyla are shown in the diagram. The origin of animals can be traced back to more than 700 Million years ago (Mya) [24]. However, the extant animal phyla might have a more recent evolutionary history and the diversification of the modern bilaterian phyla might be linked to the cambrian explosion. As a result the accurate evolutionary relationships among basal lineages and major bilaterian phyla are not well resolved (dotted lines). Possible timing of the divergence in the diagram is indicated as Mya
In my opinion, multiple origins (Fig. 2) better explain the extant diversity of nervous systems and the enormous plasticity in establishment of complex cell phenotypes, development and differentiation programs, transdifferentiation, and redundancy of molecular components in signal transduction pathways. It offers a possibility to achieve similar neuron-like phenotypes as a result of the combination of evolutionarily conserved molecular regulatory modules (which sometimes reach a convergence in certain cell phenotypes) rather than requiring common ancestry from a single particular cell lineage that will be a predecessor of all neurons and nervous systems in more than 30 phyla [54].
Any interpretation of evolutionary scenarios needs a robust phylogeny for all major animal lineages, especially those without a recognized brain or CNS and with profound nerve net type organization. Indeed, recent reconstructions of animal phylogeny [4, 20, 39] and, in particular, genomics of body patterning in hemichordates [48, 49] challenge the hypothesis of a monophyletic origin of the bilaterian CNS from an ancestral centralized nervous system in the Urbilateria. Here, I will focus on the first insights from the most recent phylogenomic analysis of less investigated groups such as basal metazoans [20, 34] and selected bilaterians and basal deuterostomes including hemichordates, echinoderms, Xenoturbella, nemertodermatids and acoels [4, 63] as well as molluscs [47, 78].
New deuterostome phylogeny and enigmatic nervous systems of basal Deuterostomes
Biology today is the biology of genomes. Advances in sequencing technologies launched a true conceptual revolution in biomedical sciences [51]. Now it is very straightforward and cost-efficient to obtain millions of expressed sequenced tags from virtually any group of organisms on the planet and then use this genome-wide information to unbiasedly reconstruct relationships among these organisms. Although computationally very demanding, these novel approaches, named phylogenomics [6, 11, 19, 22, 23, 40, 62, 86], frequently reveal deep roots of quite complex evolutionary history over more than one billion years.
One of the most surprising insights came from the analysis of deuterostome phylogeny. The story began in 2006 when it was shown that a mysterious marine worm Xenoturbella bocki (westblad, 1949), with no distinct organs except for a ‘statocyst’ containing flagellated statoconia, is not a flat worm or a degenerate mollusc as previously thought [5]. Xenoturbella (and the new phylum of Xenoturbellida) now belongs to a well-defined deuterostome group [4], which also includes three classical phyla: (i) Chordata with a true brain and dorsal nerve tube, (ii) Hemichordata and (iii) Echi-nodermata, forming a clade ambulacraria with diffuse nerve-net like nervous systems and very moderate concentrations of neuronal elements.
The evolutionary relationships between ambulacrarian nervous systems and the chordate CNS are obscure and no reliable homologous regions were convincingly recognized. Burke suggested that the nervous system in Ambulacraria is bipartite with two distinct components: an animal pole nervous system and an axial nervous system [1, 9]. In some echinoderms, these two distinct nervous systems are temporally separated – they are mostly formed in different life history stages. In hemichor-dates, there is temporal co-expression of these two anatomically distinct components. Consequently, in the course of deuterostome evolution, these two components are united in a single central nervous system [9]. Thus, phylogenetically, the chordate nervous system might be a result of an anatomical union of two apparently unrelated neuronal populations where temporal co-expression might be transformed into spatial integration forming what we now know as a single CNS.
Xenoturbella has a nervous system in the form of a uniform intraepidermal nerve plexus [65, 68]. Neuronal elements never cross basal membranes and never anatomically contact with muscles or any mesodermal structures [65]. In fact, it may be the simplest nervous system ever reported in animals – even simpler then the nervous systems in cnidaria and ctenophores. Is this the preservation of the ancestral nerve net organization in a common deuterostome, and possibly an urbilaterian (Fig. 2)? or, did a common deuterostome ancestor have a chordate type nerve tube and then lose this organization in both Xenoturbella and all extant ambulacrarians including crinoids (Fig. 3)? Although the second scenario seems unlikely, future genomic and developmental studies tracing neuronal lineages can clarify the situation.
However, the greatest surprise came from the most recent phylogenomic analysis of all sequenced bilaterians [63]. Using one of the largest sets of comparative genomic data, it was suggested that Acoelomorph flatworms and Nemertodermatida are also deuterostomes related to Xenoturbella (Fig. 1) forming a larger clade Xenacoelomorpha, a sister to Ambulacraria. Thus, the newest Deuterostome phylogeny consists of 6 distinct animal lineages; 5 of them do not have an anatomically recognized CNS but have neural elements that are better described as a nerve net type organization superficially similar to those in basal metazoans and some bilaterians. The most parsimonious cladistic analysis of these results suggests that Chordata is the only extant lineage of deuterostomes that independently from Ecdysozoans and Lopho trochozoans developed a centralized nervous system and complex brain (Fig. 2). The alternative hypothesis is that 5 of 6 lineages of free-living deuterostomes lost their complex brain, which is very unlikely in my view (Fig. 3).
It should be noted however that nervous systems of each of three Xenacoelomorpha lineages (Xenoturbella, Nemertodermatida and Acoela) are very different from each other and also different from the Platyhelminthes. No clear synapomorphies between structures and patterns of transmitter expression have been found so far [65–68]. Thus, the situation can be even more complex with extensive parallel evolution of different nerve net organizations across the entire animal kingdom. Emerging molecular data from acoela also support a simple planula-like urbilaterian and apparently independent origin of many bilaterian features [21, 32, 33] including an ancestral uncentralized nervous system in the Urbilateria.
Further molecular analysis of neurogenesis is highly desirable for all extant deuterostome lineages. For example, by tracing cell lineages in the sea urchin larva Wei demonstrated direct development of some neurons within the foregut endoderm [94]. It would not be a surprise if similar approaches revealed independent origins of multiple neuronal populations in deuterostomes and Xenacoelomorpha in particular.
Multiple origins of centralized brains in molluscs
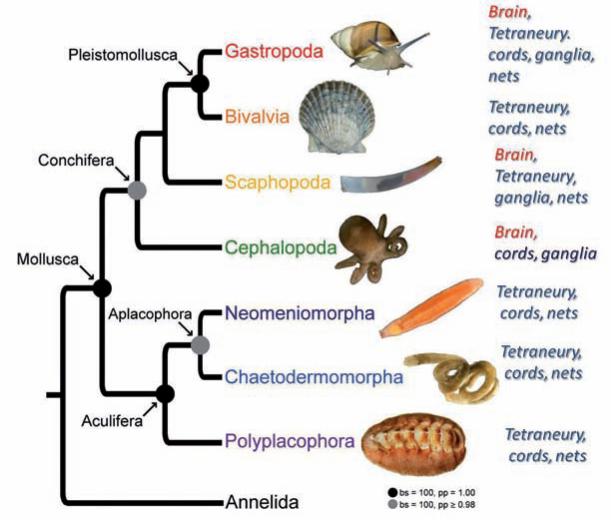
Representatives of the systematically most diverse animal group – Mollusca – include a number of interesting and documented cases where 4–5 independent events of the centralization of nervous systems may have occurred in parallel within different line-ages of the same phylum [54]. Indeed, two very recent independent studies [47, 78] reconstructed relationships within the phylum Mollusca in greater detail and their phylogenomic analyses remain consistent with the hypothesis of multiple origins of the centralized brain, even within the framework of the same phylum (Fig. 4). At least 5 independent events of centralization can be recognized: three in Eutyneura (nudi branchs, some aplysiids, pulmonates), one in Caenogastropoda and one in Cephalopods. The most remarkable example of the novel phylogenomic reconstruction [78] is the sister relationship between Monoplacophora (the earliest known molluscan lineage without a distinct brain and ladder-like diffuse neural organization) and the Cephalopoda – these “primates of sea” that have the most complex brain of any invertebrate and remarkable cognitive abilities. The only possible interpretation of molluscan phylogeny is independent formation of a complex brain in cephalopods rather than secondary loss of a centralized nervous system across many dozen molluscan lineages.
Fig. 4.
The molluscan phylogeny and their neuronal organization. The relationships among major molluscan classes is based on recent phylogenomic analysis [47]. The phylum mollusca has a very broad spectrum of types of neuronal organization. It is based on tetraneury with multiple events of neuronal centralization that are most notable in cephalopods and selected gastropods. See text for details
Parallel evolution of neuronal centralization
In addition to Chordates (Fig. 1) and Molluscs (Fig. 4), the multiple events of nervous system centralization and the formation of a single brain can be found in other animal lineages.
Figure 5 presents the current (yet simplified) view of evolutionary relationships within the animal kingdom. This phylogenetic tree is combined with the data about presence or absence of a central nervous system (CNS) or brain. Only representatives of four (Porifera, Placozoa, Orthonectida and Dicyemida) out of 34–36 animal phyla [87] do not have recognized neurons. Dicyemida and Orthonectida are enigmatic groups of parasitic worm-like animals with uncertain phylogenetic positions and very limited information about their organization [3, 15, 16, 60, 77, 83]; therefore, they will not be reviewed here.
For this discussion a CNS is defined as the concentration of neuronal cells within a distinct organ-type structure (a brain, nerve tube/cords or interconnected ganglia) where neurons and a system of highly concentrated neuronal processes (neuropil) can be supported (surrounded) by other cell types (e.g. glia or connective tissues) to maintain a controllable microenvironment for neuronal functioning. in some extreme cases, a majority or even all neuronal elements can be fused into a single brain such as observed in cephalopods, nudibranchs, vertebrates or some insects. Nevertheless, even these animals with a distinct brain have a well-developed peripheral nervous system.
In Fig. 5 Choanoflagelates (eukaryotic algae-like organisms) are placed at the base of the tree as a sister group for metazoa [46]. Two basal Metazoan phyla (Porifera and Placozoa) do not have recognized neurons [79, 81]. Two other prebilaterian/basal metazoan phyla (Ctenophora and Cnidaria) have well defined neurons and nerves. However, only ctenophores have “true” muscles. Although neuronal organization in basal Metazoa is superficially presented as a nerve net, many species have a prominent concentration of neuronal elements, and numerous and apparently autonomous networks governing surprisingly complex and well-coordinated behaviors [50, 74].
Some representatives of Cnidaria such as Cubozoa have well-developed eyes and a ganglionic organization associated with rhopalia which can be described in terms of a centralized nervous system [25, 26, 28, 56, 74, 76]. Similarly there is a well-defined concentration of neural elements associated with locomotory combs in the phylum Ctenophora [35]. In addition, all comb jellies have a specialized apical organ – the primary, sensory “brain”-type integrative structure located at the aboral pole of the animal and involved in the coordination of locomotion and other animal behaviors [85].
Chordates, Nematodes, Molluscs, Annellids and Arthropods are classical neuro-biological models and all of them have well-defined central nervous systems, while in seven other bilaterian phyla shown in Fig. 5 the gross anatomical organization of their nervous systems can be similar or even simpler than those in selected cnidarians and ctenophores [7, 8].
Using the topology of this tree and standard cladistic analysis, the most parsimonious scenario of the origin of the bilaterial CNS is the hypothesis that the Urbilateria might have an uncentralized, possibly nerve net-like organization (Fig. 2) without the prerequisite well-defined location at the dorsal or ventral surfaces (although some concentration of neuronal elements could be present in locomotory, feeding and sensory regions). Next, current animal phylogeny (Fig. 5) implies that the centralization of nervous systems with the formation of canonical brain-type structures occurred at least 9 times in evolution (possibly more often). For example, it might have happened independently in the lineages leading to (i) chordates, (ii) arthropods, (iii) nematodes, (iv) annelids and (v) at least 5 times in molluscs. Lastly, we should consider the independent origins of rhopalia in Cubozoa and the aboral organ in Ctenophores as true brain analogs in basal Metazoa.
Thus, answering the question posed as the title of this paper, I suggest that complex brains might have evolved at least 9 times in the evolution of bilaterians and at least twice independently in the evolution of basal metazoans. To further reconstruct the parallel evolution of nervous systems genomic approaches are essential to probe enigmatic neurons of basal metazoans, selected lophotrochozoans (e.g. phoronids, brachiopods) and all lineages of basal deuterostomes.
On the multiple origins of neurons
The earlier reference to endodermal genesis of neurons in the sea urchin [94] and the presence of enigmatic neurons in Ctenophores together with their possible position as the earliest branching lineage of all animals [20, 34] implies that neurons themselves might evolved more than once (Figs 5 and 6).
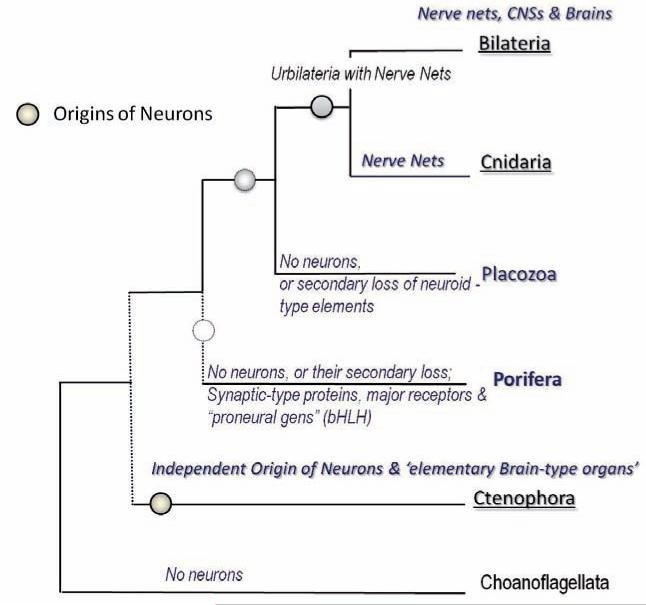
Fig. 6.
Phylogeny of basal metazoa and independent origin of neurons in Ctenophora. The relationships among five basal metazoan groups (Ctenophora, Porifera, Placozoa, Cnidaria and Bilateria) is based upon [20, 34]. Although the exact placement of Ctenophora and Porifera is not currently resolved (dotted lines), these two lineages were branched before Placozoa, Cnidaria and Bilateria (a clade that was named ParaHoxozoa [72]). The topology of this tree and our recent analysis of Ctenophore gene complements suggest that neurons evolved independently in ctenophores. Sponges either primarily lack neurons or they might have lost them secondarily. Placozoa might also be viewed as a secondarily simplified group
Moreover, the emerging data about neurogenesis in molluscs (and other protostomes) suggest that during development neurons might originate from at least three unrelated and distinct histological sources such as the areas (i) close to the apical organ, (ii) around ciliary bands and (iii) even within the body wall [41]. Evidently, different neuronal lineages can be labeled by different molecular markers [12–14, 89–93] indicating that they might not be genealogically related, but instead form a bipartite brain with at least two distinct components [57, 58] or even more complex assembles with multiple distinct cell lineages.
The polygenesis of neurons was originally discussed elsewhere [54, 73]. It was proposed that under this scenario, the origins of neurons can be linked to adaptations to stress/injury factors in the form of integrated regeneration-type cellular response with secretory signaling peptides as early neurotransmitters [54]. Ancestral polarized secretory cells were also likely involved in coordination of ciliated locomotion in early animals [42], and these cells can be viewed as evolutionary precursors of neurons within different lineages.
In the broad comparative context, it would be important to establish universal criteria to be applied to all neurons to analyze the origin and evolution of nervous systems. Are there any such universal criteria equally applied to all neurons across phyla? Is there a universal molecular tool-kit that defines a neuron? What is a neuron from the genomic standpoint? These are long-standing questions to be addressed in the future. Obviously, neither action potentials nor specialized synapses are absolute prerequisites of neurons. Historically, there can be many transition forms within the same cell line-age from a simple secretory cell without defined processes to a highly polarized neuron with hundreds of specialized processes and synapses. In generalized terms, the following definition of a neuron can be considered but needs to be carefully validated in a broad comparative survey which includes the basal Metazoa.
Here, I define neurons as polarized secretory cells [54, 71] specialized for directional active conducting – the features that enable them to transmit signals beyond their immediate neighbors without affecting all intervening cells en route. In the 1950–60s Grundfest suggested that neurons arose from ancestral secretory cells [29, 30] when the secretory activity became confined to the termination of elongated processes (reviewed in [54]). It might not be very ‘difficult’ to develop a neuronal organization in the first place by coupling the formation of a polarized cell and localized secretion of signaling molecules. Emerging molecular data further suggests that at the genomic level such transitions might have been achieved by changes in expression of just 2–4 transcriptional factors [43–45, 52, 61, 84, 88, 95] or miRNAs [96] – not surprising if such events might happen multiple times over 700 million years of animal evolution. Consequently, early neurons/synapses evolved as the next step in development of compartmentalized transmitter secretion, possibly recruiting pre-existing molecular components for polarized transport and signaling (explaining the recruitment of certain RNA binding proteins, secretory machinery, ion channels, etc.) by just a few master regulators either in the form of transcription factors or non-coding RNAS or both.
The first neural circuits evolved to control cilia and coordinate primary (ciliated) locomotion whereas the first muscles controlled hydroskeleton and feeding/defensive movements as in extant Ctenophores. It appears that ctenophores might have evolved their nervous system independently (Fig. 6) from other animal lineages [54, 55]. There is a reasonable possibility that sponges (traditionally viewed as the earliest animal lineage preserving the ancestral organization) might also have been secondarily simplified during evolution; they could have lost some components of the earliest neural systems during evolution, reduced their complement of ancestral molecular toolkits, and even reduced their sensory capabilities (still partially preserved in larvae [70, 71, 80, 81]), as well as their protoneuron-like elements.
However, it would be not an overstatement to say that in spite of the very important position of Ctenophora in the animal tree of life, and the clear presence of the earliest known neurons and muscle cells in these prebilaterian animals, we know virtually nothing about molecular/neuronal organization within this phylum nor about the cellular or transmitter bases of their behaviors. Thus, I consider work on Ctenophores that focuses on identification of their signaling molecules and the genomic bases of neuronal organization in these animals and cnidaria to be most vital steps toward understanding the earliest nervous systems. It would not be surprising that nature used many different molecular toolkits to ‘make neurons’ in the first place.
One of the foreseen applications of these apparently remote theoretical studies would be in synthetic and regenerative medicine [75], opening novel perspectives in the treatment of complex neurodegenerative disorders where trans-differentiation of existing or induced pluripotent cells to form novel or repair injured neural circuits is required.
ACKNOWLEDGEMENTS
We thank Dr. Andrea Kohn and Jim Netherton for their valuable comments. This work is supported by NIH grants 1RO1NS060762, 1R01GM097502, R21RR025699, 5R21DA030118, McKnight Brain Research Foundation, and NSF-0744649.
Footnotes
Presented during the 12th ISIN Symposium on Invertebrate Neurobiology, August 31–september 4, 2011, Tihany, Hungary.
REFERENCES
- 1.Angerer LM, Yaguchi S, Angerer RC, Burke RD. The evolution of nervous system patterning: insights from sea urchin development. Development. 2011;138:3613–3623. doi: 10.1242/dev.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arendt D, Nubler-Jung K. Inversion of dorsoventral axis? Nature. 1994;371:26. doi: 10.1038/371026a0. [DOI] [PubMed] [Google Scholar]
- 3.Aruga J, Odaka YS, Kamiya A, Furuya H. Dicyema Pax6 and Zic: tool-kit genes in a highly simplified bilaterian. BMC Evol. Biol. 2007;7:201. doi: 10.1186/1471-2148-7-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, Kirschner M, lander ES, Thorndyke M, Nakano H, Kohn AB, Heyland A, Moroz LL, Copley RR, Telford MJ. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- 5.Bourlat SJ, Nielsen C, Lockyer AE, Littlewood DT, Telford MJ. Xenoturbella is a deuterostome that eats molluscs. Nature. 2003;424:925–928. doi: 10.1038/nature01851. [DOI] [PubMed] [Google Scholar]
- 6.Boussau B, Daubin V. Genomes as documents of evolutionary history. Trends Ecol. Evol. 2010;25:224–232. doi: 10.1016/j.tree.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Brusca RC, Brusca GJ. Invertebrates. Sinauer Associates, Inc.; Sunderland, Massachusetts: 2003. p. 936. [Google Scholar]
- 8.Bullock TH, Horridge GA. Structure and Function in the Nervous Systems of Invertebrates. Freeman; San Francisco: 1965. [Google Scholar]
- 9.Burke RD. Deuterostome neuroanatomy and the body plan paradox. Evol. Dev. 2011;13:110115. doi: 10.1111/j.1525-142X.2010.00460.x. [DOI] [PubMed] [Google Scholar]
- 10.Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 11.Casci T. Phylogenomics: improving our family tree. Nature Reviews. 2011;12:298. doi: 10.1038/nrg2996. [DOI] [PubMed] [Google Scholar]
- 12.Croll RP. Developing nervous systems in molluscs: navigating the twists and turns of a complex life cycle. Brain, behavior and evolution. 2009;74:164–176. doi: 10.1159/000258664. [DOI] [PubMed] [Google Scholar]
- 13.Croll RP. Development of embryonic and larval cells containing serotonin, catecholamines, and FMRFamide-related peptides in the gastropod mollusc Phestilla sibogae. Biol. Bull. 2006;211:232247. doi: 10.2307/4134546. [DOI] [PubMed] [Google Scholar]
- 14.Croll RP, Voronezhskaya EE, Hiripi L, Elekes K. Development of catecholaminergic neurons in the pond snail, Lymnaea stagnalis: II. Postembryonic development of central and peripheral cells. J. Comp. Neurol. 1999;404:297–309. doi: 10.1002/(sici)1096-9861(19990215)404:3<297::aid-cne2>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 15.Czaker R. Dicyemid's dilemma: structure versus genes. The unorthodox structure of dicyemid reproduction. Cell. Tissue Res. 2011;343:649–658. doi: 10.1007/s00441-010-1124-z. [DOI] [PubMed] [Google Scholar]
- 16.Czaker R. Serotonin immunoreactivity in a highly enigmatic metazoan phylum, the pre-ner-vous Dicyemida. Cell. Tissue Res. 2006;326:843–850. doi: 10.1007/s00441-006-0247-8. [DOI] [PubMed] [Google Scholar]
- 17.De Robertis EM. Evo-devo: variations on ancestral themes. Cell. 2008;132:185–195. doi: 10.1016/j.cell.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Robertis EM, Sasai Y. A common plan for dorsoventral patterning in Bilateria. Nature. 1996;380:37–40. doi: 10.1038/380037a0. [DOI] [PubMed] [Google Scholar]
- 19.Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nature Reviews. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- 20.Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, Sorensen MV, Haddock SH, Schmidt-Rhaesa A, Okusu A, Kristensen RM, Wheeler WC, Martindale MQ, Giribet G. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 21.Egger B, Steinke D, Tarui H, De Mulder K, Arendt D, Borgonie G, Funayama N, Gschwentner R, Hartenstein V, Hobmayer B, Hooge M, Hrouda M, Ishida S, Kobayashi C, Kuales G, Nishimura O, Pfister D, Rieger R, Salvenmoser W, Smith J, Technau U, Tyler S, Agata K, Salzburger W, Ladurner P. To be or not to be a flatworm: the acoel controversy. PLoS ONE. 2009;4:e5502. doi: 10.1371/journal.pone.0005502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisen JA. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- 23.Eisen JA, Fraser CM. Phylogenomics: intersection of evolution and genomics. Science. 2003;300:1706–1707. doi: 10.1126/science.1086292. [DOI] [PubMed] [Google Scholar]
- 24.Erwin DH, Laflamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science. 2011;334:1091–1097. doi: 10.1126/science.1206375. [DOI] [PubMed] [Google Scholar]
- 25.Garm A, Ekstrom P, Boudes M, Nilsson DE. Rhopalia are integrated parts of the central nervous system in box jellyfish. Cell Tissue Res. 2006;325:333–343. doi: 10.1007/s00441-005-0134-8. [DOI] [PubMed] [Google Scholar]
- 26.Garm A, Mori S. Multiple photoreceptor systems control the swim pacemaker activity in box jellyfish. J. Exp. Biol. 2009;212:3951–3960. doi: 10.1242/jeb.031559. [DOI] [PubMed] [Google Scholar]
- 27.Geoffroy Saint-Hilaire, E. Principes de philosophie zoologique. Pichon et Didier; Paris: 1830. [Google Scholar]
- 28.Gray GC, Martin VJ, Satterlie RA. Ultrastructure of the retinal synapses in cubozoans. Biol. Bull. 2009;217:35–49. doi: 10.1086/BBLv217n1p35. [DOI] [PubMed] [Google Scholar]
- 29.Grundfest H. Evolution of conduction in the nervous system. In: Bass AD, editor. Evolution of Nervous Control from Primitive Organisms to Man. American Association for Advancement of Science; Washington DC: 1959. pp. 43–86. [Google Scholar]
- 30.Grundfest H. Evolution of electrophysiological properties among sensory receptor systems. In: Pringle JWS, editor. Essays on Physiological Evolution. Pergamon Press; Oxford: 1965. pp. 107–138. [Google Scholar]
- 31.Halanych KM. The new view of animal phylogeny. Annu. Rev. Ecol. Evol. Syst. 2004;35:229256. [Google Scholar]
- 32.Hejnol A, Martindale MQ. Acoel development indicates the independent evolution of the bilaterian mouth and anus. Nature. 2008;456:382–386. doi: 10.1038/nature07309. [DOI] [PubMed] [Google Scholar]
- 33.Hejnol A, Martindale MQ. Acoel development supports a simple planula-like urbilaterian. Philos. Trans R. Soc. Lond. B Biol. Sci. 2008;363:1493–1501. doi: 10.1098/rstb.2007.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, Martinez P, Baguna J, Bailly X, Jondelius U, Wiens M, Muller WE, Seaver E, Wheeler WC, Martindale MQ, Giribet G, Dunn CW. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. Biol. Sci. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez-Nicaise M-L. Ctenophora. In: Harrison FW, Westfall JA, editors. Microscopic Anatomy of Invertebrates: Placozoa, Porifera, Cnidaria, and Ctenophora. Wiley; New York: 1991. pp. 359–418. [Google Scholar]
- 36.Hirth F. On the origin and evolution of the tripartite brain. Brain, behavior and evolution. 2010;76:3–10. doi: 10.1159/000320218. [DOI] [PubMed] [Google Scholar]
- 37.Hirth F, Kammermeier L, Frei E, Walldorf U, Noll M, Reichert H. An urbilaterian origin of the tripartite brain: developmental genetic insights from Drosophila. Development. 2003;130:23652373. doi: 10.1242/dev.00438. [DOI] [PubMed] [Google Scholar]
- 38.Hirth F, Reichert H. Basic nervous system types: one or many. In: Kaas JH, editor. Evolution of Nervous Systems. Academic Press; Amsterdam: 2007. pp. 55–72. [Google Scholar]
- 39.Holland ND. Early central nervous system evolution: an era of skin brains? Nat. Rev. Neurosci. 2003;4:617–627. doi: 10.1038/nrn1175. [DOI] [PubMed] [Google Scholar]
- 40.House CH. The tree of life viewed through the contents of genomes. Methods Mol. Biol. 2009;532:141–161. doi: 10.1007/978-1-60327-853-9_8. [DOI] [PubMed] [Google Scholar]
- 41.Jacob MH. Neurogenesis in Aplysia californica resembles nervous system formation in vertebrates. J. Neurosci. 1984;4:1225–1239. doi: 10.1523/JNEUROSCI.04-05-01225.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jekely G. Origin and early evolution of neural circuits for the control of ciliary locomotion. Proc. Biol. Sci. 2011;278:914–922. doi: 10.1098/rspb.2010.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim JB, Greber B, Arauzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Scholer HR. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–653. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- 44.Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, Meyer J, Hubner K, Bernemann C, Ortmeier C, Zenke M, Fleischmann BK, Zaehres H, Scholer HR. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 45.Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, Scholer HR. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 46.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kocot KM, Cannon JT, Todt C, Citarella MR, Kohn AB, Meyer A, Santos SR, Schander C, Moroz LL, Lieb B, Halanych KM. Phylogenomics reveals deep molluscan relationships. Nature. 2011;477:452–456. doi: 10.1038/nature10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lowe CJ, Terasaki M, Wu M, Freeman RM, Jr., Runft L, Kwan K, Haigo S, Aronowicz J, Lander E, Gruber C, Smith M, Kirschner M, Gerhart J. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS biology. 2006;4:e291. doi: 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, Gruber CE, Gerhart J, Kirschner M. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113:853–865. doi: 10.1016/s0092-8674(03)00469-0. [DOI] [PubMed] [Google Scholar]
- 50.Mackie GO. The elementary nervous sytems revisited. American Zoologist. 1990;30:907–920. [Google Scholar]
- 51.Mardis ER. A decade's perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 52.Marro S, Pang ZP, Yang N, Tsai MC, Qu K, Chang HY, Sudhof TC, Wernig M. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem. Cell. 2011;9:374–382. doi: 10.1016/j.stem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moroz LL. On the independent origins of complex brains and neurons. Brain, behavior and evolution. 2009;74:177–190. doi: 10.1159/000258665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moroz LL, Kohn AB, Citarella M, Grigorenko A, Kocot K, Halanych K, Rogaev E. The genome of the ctenophore Pleurobrachia bachei: Molecular insights into independent origins of nervous systems. Society for Integrative and Comparative Biology. 2012 [Google Scholar]
- 56.Nakanishi N, Yuan D, Hartenstein V, Jacobs DK. Evolutionary origin of rhopalia: insights from cellular-level analyses of Otx and POU expression patterns in the developing rhopalial nervous system. Evol. Dev. 2010;12:404–415. doi: 10.1111/j.1525-142X.2010.00427.x. [DOI] [PubMed] [Google Scholar]
- 57.Nielsen C. Larval and adult brains. Evol. Dev. 2005;7:483–489. doi: 10.1111/j.1525-142X.2005.05051.x. [DOI] [PubMed] [Google Scholar]
- 58.Nielsen C. Trochophora larvae: cell-lineages, ciliary bands and body regions. 2. Other groups and general discussion. Journal of experimental zoology Part B. 2005;304:401–447. doi: 10.1002/jez.b.21050. [DOI] [PubMed] [Google Scholar]
- 59.Northcutt RG. Cladistic analysis reveals brainless urbilateria. Brain, behavior and evolution. 2010;76:1–2. doi: 10.1159/000316443. [DOI] [PubMed] [Google Scholar]
- 60.Ogino K, Tsuneki K, Furuya H. Distinction of cell types in Dicyemajaponicum (phylum Dicyemida) by expression patterns of 16 genes. J. Parasitol. 2011;97:596–601. doi: 10.1645/GE-2472.1. [DOI] [PubMed] [Google Scholar]
- 61.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peregrin-Alvarez JM, Parkinson J. Phylogenomic analysis of EST datasets. Methods Mol. Biol. 2009;533:257–276. doi: 10.1007/978-1-60327-136-3_12. [DOI] [PubMed] [Google Scholar]
- 63.Philippe H, Brinkmann H, Copley RR, Moroz LL, Nakano H, Poustka AJ, Wallberg A, Peterson KJ, Telford MJ. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature. 2011;470:255–258. doi: 10.1038/nature09676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Philippe H, Derelle R, Lopez P, Pick K, Borchiellini C, Boury-Esnault N, Vacelet J, Renard E, Houliston E, Queinnec E, Da Silva C, Wincker P, Le Guyader H, Leys S, Jackson DJ, Schreiber F, Erpenbeck D, Morgenstern B, Worheide G, Manuel M. Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 65.Raikova OI. In: Department of Biology. Abo Akademi University; Abo, Finland: 2004. Neuroanatomy of basal bilaterians (Xenoturbellida, Nemertodermatida, Acoela) and its phylogenetic applications. p. 86. [Google Scholar]
- 66.Raikova OI, Reuter M, Gustafsson MK, Maule AG, Halton DW, Jondelius U. Basiepidermal nervous system in Nemertoderma westbladi (Nemertodermatida): GYIRFamide immunoreactivity. Zoology (Jena) 2004;107:75–86. doi: 10.1016/j.zool.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 67.Raikova OI, Reuter M, Jondelius U, Gustafsson MK. The brain of the Nemertodermatida (Platyhelminthes) as revealed by anti-5HT and anti-FMRFamide immunostainings. Tissue Cell. 2000;32:358–365. doi: 10.1054/tice.2000.0121. [DOI] [PubMed] [Google Scholar]
- 68.Raikova OI, Reuter M, Jondelius U, Gustafsson MKS. An immunocytochemical and ultrastructural study of the nervous and muscular systems of Xenoturbella westbladi (Bilateria inc. sed.). Zoomorphology. 2000;120:107–118. [Google Scholar]
- 69.Reichert H. A tripartite organization of the urbilaterian brain: developmental genetic evidence from Drosophila. Brain Res. Bull. 2005;66:491–494. doi: 10.1016/j.brainresbull.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 70.Richards GS, Degnan BM. The dawn of developmental signaling in the metazoa. Cold Spring Harb Symp. Quant. Biol. 2009;74:81–90. doi: 10.1101/sqb.2009.74.028. [DOI] [PubMed] [Google Scholar]
- 71.Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr. Biol. 2008;18:1156–1161. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- 72.Ryan JF, Pang K, Mullikin JC, Martindale MQ, Baxevanis AD. The homeodomain complement of the ctenophore Mnemiopsis leidyi suggests that Ctenophora and Porifera diverged prior to the ParaHoxozoa. Evodevo. 2010;1:9. doi: 10.1186/2041-9139-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sakharov DA. Genealogy of Neurons. Nauka; Moscow: 1974. [Google Scholar]
- 74.Satterlie RA. Do jellyfish have central nervous systems? J. Exp. Biol. 2011;214:1215–1223. doi: 10.1242/jeb.043687. [DOI] [PubMed] [Google Scholar]
- 75.Sendtner M. Regenerative medicine: Bespoke cells for the human brain. Nature. 2011;476:158159. doi: 10.1038/476158a. [DOI] [PubMed] [Google Scholar]
- 76.Skogh C, Garm A, Nilsson DE, Ekstrom P. Bilaterally symmetrical rhopalial nervous system of the box jellyfish Tripedalia cystophora. J. Morphol. 2006;267:1391–1405. doi: 10.1002/jmor.10472. [DOI] [PubMed] [Google Scholar]
- 77.Sliusarev GS. [Phylum Orthonectida: morphology, biology, and relationships to other multicellular animals]. Zh. Obshch. Biol. 2008;69:403–427. [PubMed] [Google Scholar]
- 78.Smith SA, Wilson NG, Goetz FE, Feehery C, Andrade SC, Rouse GW, Giribet G, Dunn CW. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature. 2011;480:364–367. doi: 10.1038/nature10526. [DOI] [PubMed] [Google Scholar]
- 79.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, Signorovitch AY, Moreno MA, Kamm K, Grimwood J, Schmutz J, Shapiro H, Grigoriev IV, Buss LW, Schierwater B, Dellaporta SL, Rokhsar DS. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 80.Srivastava M, Larroux C, Lu DR, Mohanty K, Chapman J, Degnan BM, Rokhsar DS. Early evolution of the LIM homeobox gene family. BMC Biol. 2010;8:4. doi: 10.1186/1741-7007-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, Larroux C, Putnam NH, Stanke M, Adamska M, Darling A, Degnan SM, Oakley TH, Plachetzki DC, Zhai Y, Adamski M, Calcino A, Cummins SF, Goodstein DM, Harris C, Jackson DJ, Leys SP, Shu S, Woodcroft BJ, Vervoort M, Kosik KS, Manning G, Degnan BM, Rokhsar DS. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Struck TH, Paul C, Hill N, Hartmann S, Hosel C, Kube M, Lieb B, Meyer A, Tiedemann R, Purschke G, Bleidorn C. Phylogenomic analyses unravel annelid evolution. Nature. 2011;471:95–98. doi: 10.1038/nature09864. [DOI] [PubMed] [Google Scholar]
- 83.Suzuki TG, Ogino K, Tsuneki K, Furuya H. Phylogenetic analysis of dicyemid meso-zoans (phylum Dicyemida) from innexin amino acid sequences: dicyemids are not related to Platyhelminthes. J. Parasitol. 2010;96:614–625. doi: 10.1645/GE-2305.1. [DOI] [PubMed] [Google Scholar]
- 84.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 85.Tamm SL. Ctenophora. In: Electrical conduction and behavior in ‘simple’ invertebrates. Clarendon Press; Oxford: 1982. pp. 266–358. [Google Scholar]
- 86.Telford MJ. Phylogenomics. Curr. Biol. 2007;17:R945–946. doi: 10.1016/j.cub.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 87.Valentine JW. On the origin of phyla. The University of Chicago Press; Chicago: 2004. p. 614. [Google Scholar]
- 88.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Voronezhskaya EE, Elekes K. Expression of FMRFamide gene encoded peptides by identified neurons in embryos and juveniles of the pulmonate snail Lymnaea stagnalis. Cell. Tissue Res. 2003;314:297–313. doi: 10.1007/s00441-003-0800-7. [DOI] [PubMed] [Google Scholar]
- 90.Voronezhskaya EE, Elekes K. Expression of FMRFamide gene neuropeptides is partly different in the embryonic nervous system of the pond snail, Lymnaea stagnalis L. Neurobiology (Bp) 1997;5:91–93. [PubMed] [Google Scholar]
- 91.Voronezhskaya EE, Elekes K. Transient and sustained expression of FMRFamide-like immunoreactivity in the developing nervous system of Lymnaea stagnalis (Mollusca, Pulmonata). Cell. Mol. Neurobiol. 1996;16:661–676. doi: 10.1007/BF02151903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Voronezhskaya EE, Glebov KI, Khabarova MY, Ponimaskin EG, Nezlin LP. Adult-to-embryo chemical signaling in the regulation of larval development in trochophore animals: cellular and molecular mechanisms. Acta Biol. Hung. 2008;59(Suppl.):117–122. doi: 10.1556/ABiol.59.2008.Suppl.19. [DOI] [PubMed] [Google Scholar]
- 93.Voronezhskaya EE, Hiripi L, Elekes K, Croll RP. Development of catecholaminergic neurons in the pond snail, Lymnaea stagnalis: I. Embryonic development of dopamine-containing neurons and dopamine-dependent behaviors. J. Comp. Neurol. 1999;404:285–296. doi: 10.1002/(sici)1096-9861(19990215)404:3<285::aid-cne1>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 94.Wei Z, Angerer RC, Angerer LM. Direct development of neurons within foregut endo-derm of sea urchin embryos. Proc. Natl. Acad. Sci. USA. 2011;108:9143–9147. doi: 10.1073/pnas.1018513108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]