Abstract
RNA-seq or transcriptome analysis of individual cells and small-cell populations is essential for virtually any biomedical field. It is especially critical for developmental, aging, and cancer biology as well as neuroscience where the enormous heterogeneity of cells present a significant methodological and conceptual challenge. Here we present two methods that allow for fast and cost-efficient transcriptome sequencing from ultra-small amounts of tissue or even from individual cells using semiconductor sequencing technology (Ion Torrent, Life Technologies). The first method is a reduced representation sequencing which maximizes capture of RNAs and preserves transcripts’ directionality. The second, a template-switch protocol, is designed for small mammalian neurons. Both protocols, from cell/tissue isolation to final sequence data, take up to 4 days. The efficiency of these protocols has been validated with single hippocampal neurons and various invertebrate tissues including individually identified neurons within a simpler memory-forming circuit of Aplysia californica and early (1-, 2-, 4-, 8-cells) embryonic and developmental stages from basal metazoans.
Keywords: Single-cell RNA-seq, CA1 neurons, Transcriptome, Aplysia, Ctenophores, Hippocampus, Ion Torrent, Ion proton
1 Introduction
DNA and RNA sequencing has and will continue to make a profound impact on medicine, clinical, and basic research. It has become indispensable for virtually any biomedical field, providing an unbiased view of the entire molecular machinery within a biological system of interest. The current trends are to adapt high-throughput sequencing technologies to the level of small-cell populations and even individual cells [1-3]. It is especially critical for developmental and aging biology and neuroscience where the enormous heterogeneity of cells present a significant methodological and conceptual challenge [4, 5]. Equally important is to achieve fast and cost-efficient sequencing targeting true realtime genomics where direct physiological measurements from defined cells or cell populations are coupled with genome-wide sequencing and analysis from the very same cells with feedback for investigators within 3–4 days or even 1 day. The recently introduced semiconductor sequencers (Ion Torrent Personal Genome Machine (PGM) and Ion Torrent Proton) provide novel opportunities toward coupling transcriptional changes and physiological measurements [6, 7]. These platforms, tested in our laboratory, allow us to perform multiple experiments, even single-cell RNA-seq, at low cost with 3–4 day’s turnaround time from cell sampling to sequencing and initial annotation. Here we briefly describe the principle of semiconductor sequencing and provide a practical protocol for both invertebrate and vertebrate preparations.
Beginning with Frederic Sanger’s Nobel prize winning classical methodology and followed by the majority of commercial next generation sequencing platforms (such as 454/Roche, Solexa/Illumina, SOLiD and Helicos and Pacific Bioscience), detection schemes have relied upon optical measurements which, although precise, have a number of limitations with regard to sample and data processing, as well as manufacturing, sample preparation cost, and starting amount. In 2011 Life Technologies reported an electrochemical (light-independent) detection scheme integrated with a semiconductor platform; as the proof-of-the-concept for the technology, they resequenced the human genome [7]. The principle of semiconductor sequencing is illustrated in Fig. 1. It is based on the simple, natural chemistry of DNA synthesis where addition of a new base in the growing DNA strand leads to release of hydrogen ions (H+, Fig. 1a). As a result, the detector is a modified ultra-small pH sensor (Fig. 1b) on a silicon substrate—it is a semiconductor in which each well lies above an ion-sensitive metallic oxide layer coupled to an electronic sensor that registers miniscule (0.02 pH unit) and transient (with a half-life <1 s) pH changes [7]. The process can be performed in parallel on millions of ultra-small wells within electronic semiconductor chips (Fig. 1c, d). The density of these nano-wells is growing with the accumulated advances of more than 30 years of silicon/computer technologies. Today, there are four currently available sequencing chips with different densities of wells per chip; their sequencing parameters and yield are summarized in Fig. 2. Here, the data reported for the three series of chips (314, 316, and 318) and P1 were obtained in our laboratory at the University of Florida in 2012, while the parameters for novel Ion Proton (IP) chips are provided by Life Technologies.
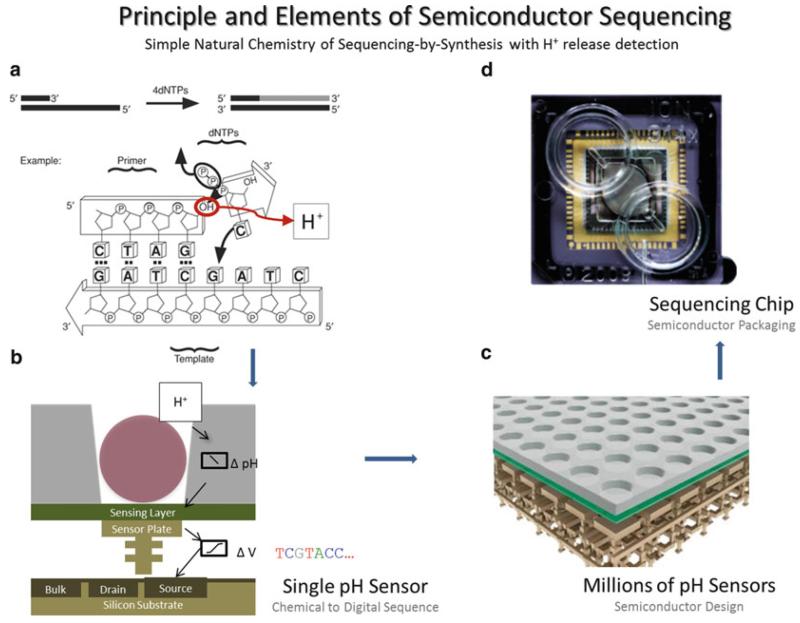
Fig. 1.
Principles and elements of semiconductor sequencing. Simpler natural chemistry of sequencing by synthesis is implemented in the Ion Torrent platform. As the second strand of DNA is synthesized, the addition of every new nucleotide leads to a release of H+ (a) which is detected by a silicon pH sensor (b). Several million pH sensors (c) are arranged within a sequencing chip (d). Cross-sectional view (c) shows the ion-sensitive layer in green with the microwells on the top surface (3 μm) and the transistor stack underneath. The present design reduces the complexity of light detection schemes such as the use of modified and fluorescent bases and optical/laser detection. PGM functions by delivering natural unmodified nucleotides one at a time over the surface of the chip. Because nucleotides are unmodified and detection does not require any additional enzyme/amplification cascades, it also eliminates many sources of error, thereby delivering long accurate reads with highly uniform genome coverage. Modified from ref. 7
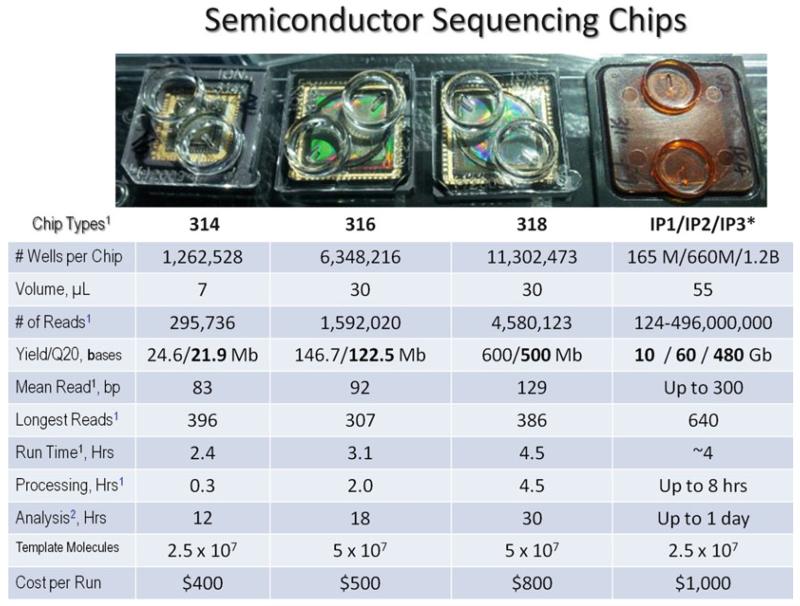
Fig. 2.
Semiconductor sequencing chips. The table summarizes statistics for various sequencing runs and their assessments for each chip type: 314, 316, 318 Ion Torrent PGM, and Ion Proton (IP1/IP2/IP3). All runs were based on 200 bp chemistry and OneTouch™ template preparation with Torrent Suite server version 2.2. Q20 refers to 99 % accuracy of a base call. 1Data were obtained in our laboratory at the University of Florida. 2Analysis includes assembly and initial annotation of a given sequencing run using a High Performance Computer Cluster (64 Intel(R) Xeon(R) X7550 2.00GHz CPUs, 512GB of RAM, and 6TB of storage). *All Proton data were provided by Ion Torrent, Life Technologies
The overall experimental workflow for the semiconductor sequencing is outlined in Fig. 3. After cell and RNA isolation and construction of sequencing libraries from a target cDNA sample (Figs. 6 and 7), each individual DNA fragment is immobilized on an Ion Sphere Particle and clonally amplified (Fig. 4). The process is automatic with a supplementary OneTouch System (Fig. 4). The resulting beads with amplified (emulsion PCR), individually cloned DNA fragments are then enriched to eliminate “empty” beads— this process is also performed by a robotic enrichment system (ES) (Fig. 4). Finally, after being loaded on a selected sequencing chip, the beads containing clonal populations of the DNA from an experimental sample are arrayed in wells and incubated serially with pure nucleotides of DNA. Incorporation of a nucleotide is continuously detected by measuring changes in the hydrogen ion concentration during the sequencing process on the PGM machine and simultaneously processed on a server for further analysis and assembly (Fig. 3).
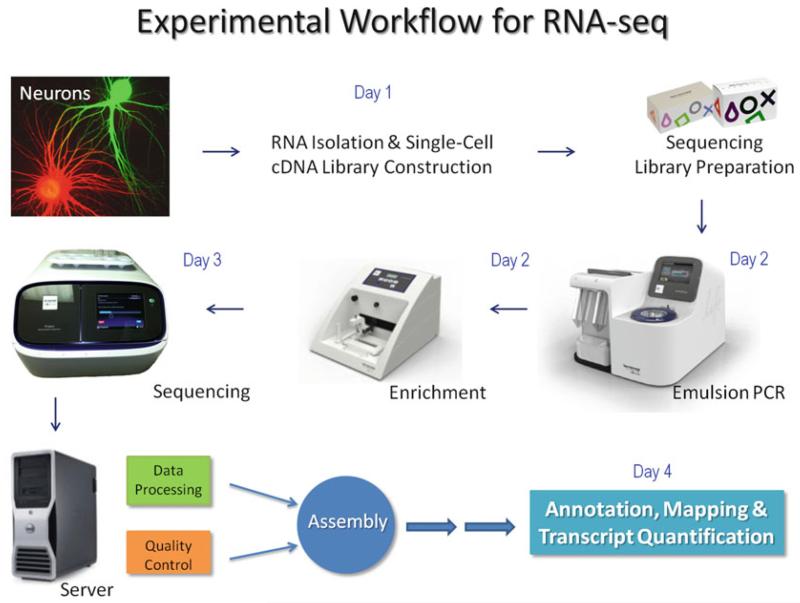
Fig. 3.
Experimental workflow for RNA-seq using the Ion Torrent (see details in the text). The equipment set includes OneTouch™ Instrument for templated bead preparation (Subheadings 3.5.1, 3.5.2, and 3.5.3), an Ion OneTouch™ ES robotic system for sample enrichment (Subheadings 3.5.4, 3.5.5, and 3.5.6), Personalized Genome Sequencer™ (Subheading 3.6), and an Ion Torrent server containing Torrent Software Suite for base calling and mapping and web portal access for data review and sharing for sequencing and primary data analysis within hours. Combined, this innovative approach allows us to perform multiple, even single-cell RNA-seq experiments at the lowest possible cost today within 3–4 day’s turnaround time: from cell sampling to sequencing and initial annotation
Fig. 6.
Preparation of sequencing libraries using a reduced representation protocol. The diagram presents a workflow for library construction and sequencing on Ion PGM. The number of days and corresponding sections are listed in the far left. Tested samples (e.g., hippocampal or Aplysia neurons or developmental/aging cell populations) are prepared for RNA isolation (see Subheadings 3.1 and 3.2) with quality of the RNA checked by a Bioanalyzer (quality control). The insert electropherograms are illustrative examples of RNA isolated from a single Aplysia neuron, 1 day embryos of Pleurobrachia, and rat hippocampal neuronal cluster from the CA3 region (ng amount of total RNA is listed for the entire extraction from each corresponding sample). The library construction process is summarized in Subheading 3.3.1 (see text). Illustrative examples of an E-gel for two libraries are shown in the second insert (here two different markers were run between the samples, and the bright bands are labeled with appropriate sizes). The library quality control assessment is summarized in Subheading 3.4 (see text). The template preparation and sequencing is followed as in Subheadings 3.5 and 3.6 (see text)
Fig. 7.
Preparation of sequencing libraries using template switch from single neurons. The diagram presents a workflow for library construction and sequencing on Ion PGM or Proton. The number of days and corresponding sections are listed in the far left. Tested samples are single mouse hippocampal neurons. The library construction process is summarized in Subheading 3.3.2 (see text). Illustrative example of a TapeStation 2200 analysis from a single CA1 neuron is in the left side insert. The library quality control assessment is summarized in Subheading 3.4 (see text). The template preparation and sequencing is followed as in Subheadings 3.5 and 3.6 (see text)
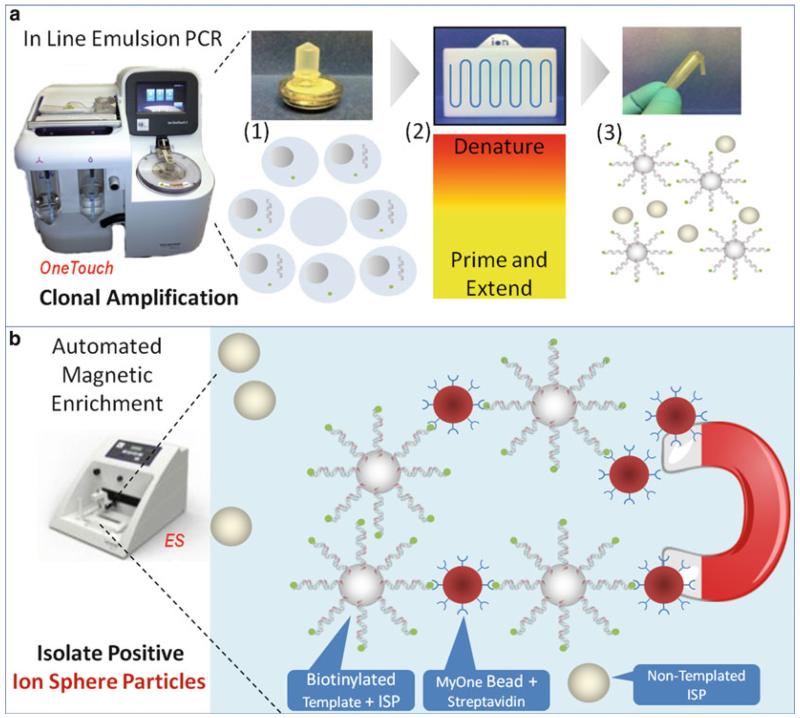
Fig. 4.
In-line emulsion PCR technology. The schematic diagram illustrates key steps in the process using OneTouch™ instrumentation. (a) The Ion OneTouch™ Instrument has three key technologies that enable automated delivery of templated Ion Sphere™ particles. The first is a reaction filter (A1) that creates millions of microreactors in which clonal amplification occurs. The second is the in-line PCR amplification plate (A2) that enables thermal cycling of the microreactors. The third is the integrated centrifuge (A3), which recovers the templated Ion Sphere™ particles. The green dots represent Biotin that has been incorporated on the primer 5′-end of the template or DNA molecule during the emPCR process. The Biotin is used to isolate only the template-positive ISP by binding to Streptavidin-linked C1 Magnetic Beads (large red dots) during the enrichment step on the Ion OneTouch™ ES. (b) The Ion OneTouch™ ES uses magnetic bead (large red dots) technology to isolate template-positive Ion Sphere™ particles that can be loaded directly onto the Ion semiconductor chip, thus delivering automated, highly reproducible enrichment with every run (Color figure online)
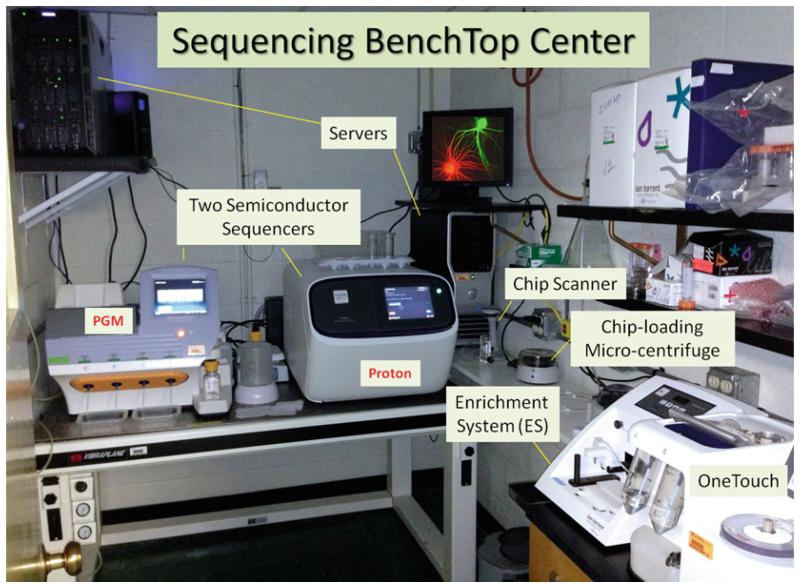
The Ion Torrent PGM and Proton are benchtop sequencers that can be put in any individual laboratory and not necessarily a sequencing center or core facility. Figure 5 illustrates the setup in our laboratory. Initial training and practical experience can be obtained within a couple of weeks for most laboratories. As such, the Ion PGM and Proton can facilitate the genome-wide sequencing components for physiological, developmental, cell, and aging biology-focused laboratories in a simple and scalable way.
Fig. 5.
Sequencing BenchTop Center. The photo shows the arrangement of key instruments required for semiconductor sequencing in an individual laboratory
Here we summarize two protocols that allow fast and cost-efficient transcriptome sequencing from ultra-small amounts of tissues or even from individual cells. We show that as little as 40 pg of total RNA can be used to produce reliable sequencing libraries (we tested it using single mechanosensory neurons from Aplysia with 40–50 μm cell body diameter—see ref. 8). Specifically, we implemented reduced representation sequencing for the current Ion Torrent platform and developed an unbiased method of RNA-seq library construction using commercially available kits to ensure consistency and reproducibility. Second, we implemented a template-switch method, where single cells or neurons are lysed in the buffer for the library construction and then sequenced.
Both protocols, from cell/RNA isolation to the final sequencing data, can be completed within a week. After sequencing libraries are prepared, it takes about 7 h for PGM instrument and about 1 day for Ion Proton to complete the RNA-seq run, including templated bead preparation and sequencing. For example, this combination of library construction and semiconductor sequencing enables us to measure changes in small-cell populations following physiological tests associated with learning and memory formation or as a function of age-related memory loss.
The efficiency of both protocols has been validated using individually identified neurons of Aplysia californica, one of the well-recognized models in learning and memory research [9-11] and cell biology of development and aging [4, 9-17]. We also successfully tested the protocol on single hippocampal CA1 neurons and more than three dozen different invertebrate tissues and cells including early embryonic stages of the ctenophore Pleurobrachia bachei. It can also be applied to a broad spectrum of experimental preparations in the biology of aging.
2 Materials
Reagents
RNAqueous-Micro™ (Cat # 1931, Ambion, Life Technologies).
RNAqueous™ (Cat # 1912, Ambion, Life Technologies).
RiboMinus™ Eukaryote Kit (Cat # A10837-08, Invitrogen, Life Technologies).
RiboMinus™ Concentration Module (Cat # K1550-05, Invitrogen, Life Technologies).
Marathon cDNA Amplification Kit (Cat # 634913, Clontech, Takara Bio).
All enzymes for first and second strand synthesis are inclusive
- 5× First-Strand Buffer:
- 250 mM Tris–HCl (pH 8.5).
- 40 mM MgCl2.
- 150 mM KCl.
- 5 mM Dithiothreitol (DTT).
- 20× Second Strand Enzyme Cocktail
- E. coli DNA polymerase I (6 U/μL).
- E. coli DNA ligase (1.2 U/μL).
- E. coli RNaseH (0.25 U/μL).
SMARTer® PCR cDNA Synthesis Kit (Clontech Cat # 634926).
Alu I and NEB buffer 4 (Cat # R0137S, New England BioLabs).
Rsa I (Cat # R0167S, New England BioLabs).
Ligate-IT Rapid Ligation Kit (Cat # 78400, Affymetrix).
- 5× Quick Ligation Reaction Buffer:
- 132 mM Tris–HCl.
- 20 mM MgCl2.
- 2 mM Dithiothreitol.
- 2 mM ATP.
- 15 % Polyethylene glycol (PEG 6000) pH 7.6 at 25 °C.
AMPure XP Reagent (Cat # A63880, Agencourt, Beckman Coulter, Inc.).
ArrayControl™ RNA Spike-in control RNAs (Catalog # 1780, Ambion, Applied Biosystems).
Advantage® UltraPure PCR Deoxynucleotide Mix (10 mM each dNTP) (Clontech cat # 639125).
Ion Plus Fragment Library Kit (Cat. no. 4471252, Life Technologies).
- Platinum® PCR SuperMix High Fidelity (Cat # 12532-016, Invitrogen, Life Technologies)
- 22 U/ml complexed recombinant Taq DNA polymerase.
- Pyrococcus species GB-D thermostable polymerase.
- Platinum® Taq Antibody.
- 66.0 mM Tris–SO4 (pH 8.9).
- 19.8 mM (NH4)2SO4.
- 2.4 mM MgSO4.
- 220.0 μM dNTPs stabilizers.
- Advantage® 2 Polymerase Mix and the buffer (Cat # 639201, Clontech).
- 50 × Advantage® 2 Polymerase Mix.
- Includes TITANIUM Taq DNA Polymerase and a small amount of proofreading polymerase and TaqStart Antibody (1.1 μg/μl).
- 50 % Glycerol.
- 15 mM Tris–HCl (pH 8.0) 0.3 mM.
- 75 mM KCl 1.5 mM.
- 0.05 mM EDTA 1.0 μM.
- 10 × Advantage® 2 PCR Buffer.
- 400 mM Tricine–KOH (pH 8.7 at 25 °C).
- 150 mM KOAc.
- 35 mM Mg(OAc)2.
- 37.5 μg/ml BSA. 0.05 % Tween 20.
- 0.05 % Nonidet-P40.
Ion PGM™ 200 Xpress Template Kit (Cat # 4474280 Ion Torrent, Life Technologies).
Ion OneTouch™ 200 Template Kit v2 (Cat # 4478316 Ion Torrent, Life Technologies).
Ion PGM™ 200 Sequencing Kit (Cat # 4474004, Ion Torrent, Life Technologies).
Ion 314™ Chip (Cat # 4462923; 8 pack), 316™ Chip (Cat # 4466616; 4 pack), 318™ Chip (Cat # 4466617; 4 pack) (Ion Torrent, Life Technologies).
Ion Sphere™ Quality Control Kit (Cat # 4468802, Ion Torrent, Life Technologies).
Dynabeads® MyOne™ Streptavidin C1 Magnetic Beads (Cat # 65001, Invitrogen, Life Technologies).
Covaris microTUBE AFA Fiber Screw-Cap 6 × 16 mm (Cat # SKU:520096, Covaris).
Agilent Bioanalyzer™ High Sensitivity DNA Kit (Cat # 5067-4626, Agilent Technologies).
Agilent Bioanalyzer™ RNA 6000 Pico Kit (Cat # 5067-1513, Agilent Technologies).
Agilent Bioanalyzer™ RNA 6000 Nano Kit (Cat # 5067-1511, Agilent Technologies).
E-Gel® SizeSelect™ 2 % Agarose (Cat # G6610-02, Invitrogen, Life Technologies).
Primers 0.2 μM scale HPLC purified, IDT, Integrated DNA Technologies, Inc (see Table 1 for all primer and adaptor sequences).
Table 1. Adaptors and primers for Ion Torrent libraries.
| Primer name | Primer sequence |
|---|---|
| Trsa | 5′-CGCAGTCGGTAC (T)13-3′ |
| Adaptor A | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′ and 5′-CTGAGTCGGAGACACGCAGG-3′ |
| P1 adaptor | 5′-CCACTACGCCTCCGCTTTCCTCTCTATGGGCAG TCGGTGAT-3′ and 5′-ATCACCGACTGCCCATAGAGA GGAAAGCGGA-3′ |
| 3′ SMART CDS Primer IIA | 5′-AAGCAGTGGTATCAACGCAGAGTACT(30)N-1N-3′ |
| SMARTer II A Oligonucleotide | 5′-AAGCAGTGGTATCAACGCAGAGTACXXXXX–3′ |
| 5′ PCR Primer IIA | 5′-AAGCAGTGGTATCAACGCAGAGT-3′ |
| A PCR Primer | 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′ |
| P1 PCR Primer | 5′-CCACTACGCCTCCGCTTTCCTCTCTATG-3′ |
Gray shading is primers for reduced representation library, and yellow shading is primers for template-switch single cell/neuron library. The same primers are used for the sequencing library PCR amplification
Equipment
LoBind tubes 0.5 and 1.5 μL (Cat # 80077-236, 80077-230 Eppendorf, VWR International, LLC).
DynaMag™-2 magnet (microcentrifuge tube magnet) (Cat # 123-21D, Invitrogen, Life Technologies).
Covaris M220 Focused-ultrasonicator (Covaris).
Agilent Bioanalyzer™ 2100 (Cat # G2947CA, Agilent Technologies).
Agilent TapeStation 220 (Cat # G2965AA, Agilent Technologies).
Galaxy MiniStar Centrifuge (120 V, 50/60 Hz) (Cat # 93000-196,VWR International).
onicating bath (2- to 3-L tank; 80 W, 40 kHz transducer) (Cat # 15-335-20, Fisher).
Thermolyne Labquake Tube Shaker/Rotator (Cat # 56264-302, VWR International).
Qubit® 2.0 Fluorometer (Cat # Q32866, Invitrogen, Life Technologies).
MJ Research Thermo Cycler (Cat # PTC-100, MJ Research).
E-Gel® iBase™ and E-Gel® Safe Imager™ Combo Kit (Cat # G6465, Invitrogen, Life Technologies).
Personal Genome Machine (PGM™) Sequencer (Cat # 4462917, Ion Torrent Life Technologies).
Ion PGM™ OneTouch™ System which is comprised of two modules: the Ion OneTouch™Instrument and the Ion One-Touch™ ES (enrichment system) (Cat # 4470001, Ion Torrent Life Technologies).
Torrent Server (Cat # 4462918, Ion Torrent, Life Technologies).
3 Methods
3.1 Sample Preparation
Sample preparation is dependent on species, tissue, and even the type of cell from which RNA will be isolated. The efficiency of the protocols below has been validated using both small numbers (20–200) of cells and even single hippocampal neurons (CA1, CA3, and DG areas in collaboration with the Dr. Thomas Foster and C. Jason Frazier laboratories, University of Florida), various invertebrate tissues including early embryonic (1-, 2-, 4-, 8-, 16-, 32, 64-cells) and later developmental stages from basal metazoans (Pleurobrachia bachei), and individually identified neurons within a simpler memory-forming circuit of Aplysia californica [11, 14]. Typically animals are anesthetized appropriately followed by removal of tissue such as regions of the central nervous system or ganglia. For example, if identified neurons are to be isolated from the mollusc Aplysia, ganglia are dissected, connection tissues are removed, and single cells are isolated as described in [1, 13] (see Note 1 for tissue/single-cell storage).
For single-cell library construction from small mammalian neurons, RNA is not isolated because the amount of RNA is too low to detect by conventional instruments such as a Bioanalyzer. Single CA1 neurons from mice were placed directly into a buffer for library construction, briefly sonicated (5–15 s) in a sonicating bath, then snap frozen at −80 °C. Critical to this process is performance of cell isolations under controllable or sterile conditions. The rig, experimental chamber, microelectrodes, solutions, and anything that comes in contact with the tissue/cells can be potential sources for bacterial, fungal, or human contamination.
3.2 RNA Isolation
Obtaining high-quality RNA is the first and often the most important step in performing many molecular techniques such as high-throughput sequencing. We find the best RNA isolation method is quite often species-dependent and needs to be experimentally determined (see Note 2 for a comment on RNA isolation kits). We choose the RNA isolation kit or method that has been experimentally tested based on the quality and quantity of RNA for a specific animal, tissue, or even single cell (see additional details about single neuron isolation in ref. 1). RNA quality is analyzed using an Agilent 2100 Bioanalyzer™ on a 6000 Nano LabChip or the new Agilent TapeStation. For very small quantities, the 6000 Pico LabChip can be used. We may also use the Qubit to determine quantity of RNA for a sample but only if there is a sufficient amount of material. Understanding the properties of a species RNA is important. For example, many molluscs tested have a hidden break in their 28s ribosomal subunit causing the 28s subunit rRNA to migrate with the 18s subunit, thus generating one peak; see Fig. 6 for an electropherogram from an Aplysia single neuron (see Note 3 on invertebrate rRNA [18]). Once the quality and quantity is satisfactory, this material is used to construct a sequencing library. This can also be a pause point and samples should be stored at −80 °C.
3.3 Library Construction: Timing 6 h
Quality sequencing such as RNA-seq depends on a good library. We present here two RNA-seq library protocols based on the amount of the planned sequencing output. First, we present a reduced representation RNA-seq library protocol for maximum capture of transcriptional output which requires relatively lower sequence coverage compared to the complementary protocol. This second protocol is designed for small single mammalian neurons which are lysed directly in the buffer for library construction, then sequenced.
The first protocol, Subheading 3.3.1 describes an unbiased method of library construction. Specifically, it is designed for both quantitative and qualitative analysis of mRNA from ultra-small amounts of material and can be applicable for small neuronal populations and even large single neurons. The distinct feature of this protocol is preservation of the direction of transcripts which allows equal identification of both sense and antisense RNA. It was validated for both individual identified Aplysia neurons as well as for small clusters of 20–200 CA1, CA3 hippocampal neurons from rat, and 100–200 μm invertebrate embryos (e.g., Pleurobrachia).
We can use commercially available kits such as the Marathon® cDNA Amplification Kit from Clontech to ensure consistency and reproducibility but suggest cheaper alternatives to these kits. Although we focused on the use of limited amounts of starting material (starting from 20 to 40 pg), our protocol is also applicable to larger quantities of material (up to 1 μg).
Library construction starts with total RNA isolation (see Sub-heading 3.2), then RNA is reverse transcribed to cDNA with an oligo dT primer and a second strand generated (see Fig. 6). The double stranded cDNA is fragmented with a combination of restriction enzymes. Sequential ligation of adaptors is used to generate a reduced representation library. Fragmented DNA with ligated adaptors is processed through an emulsion-based clonal amplification (template bead preparation see Subheading 3.5 and Fig. 4) and captured onto Ion Sphere Particles (from Ion Torrent) as required for subsequent sequencing steps. DNA-captured beads are placed onto a semiconductor chip for sequencing. Because our libraries are constructed in a reduced representation process, smaller sequencing coverage is needed to capture a representative transcriptional output (e.g., using 314 or 318 sequencing chips).
The second library construction protocol (Subheading 3.3.2) uses a template-switch method and is designed for single mammalian neurons. Here, freshly isolated neurons are lysed directly in the buffer for library construction (see Fig. 7). We take advantage of the unique properties of the Moloney murine leukemia virus (MMLV) reverse transcriptase for the first and second strand synthesis (see Note 6). The MMLV reverse transcriptase supports a template-switch method to synthesize both the first and second strand cDNA from an RNA template using an oligo (dT) primer. This enzyme synthesizes additional nucleotides at the 3′ end of the first-strand cDNA, then with a template-switch oligo, the direction is switched and the enzyme makes the complementary second strand (see Fig. 7). Commercial kits can be used such as SMARTer® PCR cDNA Synthesis Kit (Clontech), or some components of the procedure can be made or combined separately. Once cDNA is constructed, it is amplified, fragmented, ligated with sequencing adaptors, and the final sequencing library amplified. This method is not a reduced representation of the transcriptome and is best paired with either a 318 PGM chip or a new Proton (P1) chip providing greater sequencing yield. This method is also not directional. It should be noted that a similar protocol using the template-switch method for sequencing single cells has been independently suggested [19]. However, Ramskold et al. [19] used the Nextera method of fragmentation and adaptor insertion, called “tagmentation,” that is specialized for human and mouse DNA, then followed with Illumina sequencing which takes weeks to obtain data. Below, we present a protocol that can be used for single-neuron RNA-seq.
3.3.1 Library Construction Protocol: Reduced Representation
(Optional) If there is a sufficient amount of material (i.e., > 100 ng), RiboMinus™ is employed, and the protocol is followed according to manufacturer’s recommendations (see Note 4). If not, the total RNA is processed to cDNA.
Whenever possible, use a master mix of reagents. For X number of samples, multiply all reagent volumes by X.5 so as to accommodate pipetting errors. Use of master mixes will save time, produce fewer errors, and generate more reproducible results.
cDNA is generated using the Marathon cDNA Amplification Kit to promote consistency between libraries (Fig. 6) (see Note 5 for other options and Note 6).
To begin first-strand synthesis, 5 μL of total RNA or RiboMinus RNA and 1 μL of Trsa (sequence in Table 1) cDNA synthesis primer (10 mM) are incubated at 70 °C for 2 min then briefly spun (see Note 7). Ribosomal-depleted starting material will ensure no ribosomal “contamination” in the library.
- The following is added to the sample RNA containing tube:
2.0 μL 5× First Stand Buffer 1.0 μL dNTP Mix (10 mM) 1.0 μL AMV Reverse Transcriptase (20 U/μL) 10 μL Total volume This mixture is triturated gently then incubated at 42 °C for 1 h.
(Optional) Eight individual bacterial RNA transcripts ranging in size from 750 to 2,000 bases with a 30-base 3′ poly (A) tail (ArrayControl™ RNA Spike-in control RNAs) at different concentrations have been added at the beginning of the library construction for previous sequencing libraries to test the quality of the library (see Note 8).
Reaction is terminated by placing on ice.
- Second strand synthesis is performed by adding the following to the first-strand tube:
48.4 μL RNase-free water 16.0 μL 5× Second Strand Buffer 1.6 μL dNTP Mix (10 mM) 4.0 μL 20× Second Strand Enzyme Cocktail 80.0 μL Total volume -
This mixture is triturated gently then incubated at 16 °C for 2h.
Then 2 μL (6 U) of T4 DNA polymerase is added and incubated at 16 °C for an additional 30 min.
Comment 1: The presented protocol is relatively short compared to other protocols [3, 20-22]. For low amounts of starting material, we recommend cDNA production before fragmentation, because the alternative ribosomal depleted RNA fragmentation methods require much larger quantities of starting material. Efficient removing of ribosomal RNA is not possible when working with very low amounts of RNA such as that from small cells and their compartments.
- The library is purified with AMPure XP Reagent.
- 1.8 volumes of AMPure XP Reagent (144 μL) is added to the dsDNA tube and set 5 min.
- The mixture is placed on the Magnetic Particle Separator for 1 min or until the solution is cleared and the supernatant removed.
- The mixture is washed two times with freshly prepared 70 % ethanol and allowed to dry for 5 min.
- The dsDNA is then eluted with 13 μL of RNase-free water.
-
This mixture is incubated for 10 min at room temperature.
Comment 2: The ligation of Adaptor A at this point also allows for reduced representation of the library. Only one fragment per transcript can be ligated with the Adaptor A and later the P1. As a result, such reduced representation libraries allow observation of more distinct transcripts given the same amount of sequencing coverage. Our quantification of 1 transcript per 1 read produces higher coverage of the transcriptome than alternative protocols, when all fragments from a single transcript are ligated with library adaptors for the same amount of sequencing.
The ligation mixture is purified with 1.8 volumes of AMPure XP Reagent (36.0 μL) as described above and eluted with 17.0 μL of RNase-free water.
- The purified A Adaptor-ligated dsDNA is fragmented by digestion with two 4 base blunt-end cutting restriction enzymes by adding:
1.0 μL AluI (10 U) 1.0 μL RsaI (10 U) 2.0 μL NEB buffer 4 17.0 μL dsDNA 21.0 μL Total volume Mixture is incubated at 37 °C for 2 h (see Note 11).
The digested mixture is then ligated with the P1 adaptor.
- 3′ Adaptor ligation is performed by adding the following to the purified dsDNA:
6.0 μL 5× Quick Ligation Reaction Buffer 1.0 μL Quick T4 DNA Ligase 2.0 μL Adaptor P1 (see Table 1) 21.0 μL dsDNA 30.0 μL Total volume This mixture is incubated for 10 min at room temperature.
The ligation mixture is purified with 1.8 volumes of AMPure XP Reagent (54 μL) as described above and eluted with 40 μL of Nuclease-free water.
- The above mixture is amplified with the following conditions (see Note 13):
- 95 °C for 30 s 1 cycle
- X cycles: (typical cycles are 16–18 for >100 μg total RNA and 18–23 for single cells)
- 95 °C for 30 s
- 58 °C for 30 s
- 72 °C for 1 min
- Hold 4 °C ∞
This is can be a pause point and samples can be stored at −20 °C (see Note 14).
3.3.2 Single-Neuron Library Construction Using a Template-Switch Method
Library construction with SMARTer® PCR cDNA Synthesis Kit (Fig. 7) (see Note 15).
Thaw the single cell/neuron in 4.5 μL of 1× First-Strand Buffer (see Subheading 3.1).
- First and second strand synthesis with the template-switch method. Add to the single cell sample.
1 μL 3′ SMART CDS Primer IIA (12 μM) (Table 1) 1 μL 5× First-Strand Buffer 0.25 μL DTT (100 mM) 1 μL dNTP Mix (10 mM ) 1 μL SMARTer II A Oligonucleotide (12 μM) (see Note 16) 0.25 μL RNase Inhibitor 1 μL SMARTScribe Reverse Transcriptase (100 U)* 10 μL Total *Add the reverse transcriptase just prior to use. Mix well by pipetting and spin the tube briefly in a microcentrifuge Incubate the tubes at 42 °C for 1½ h.
- PCR amplification is performed on the cDNA. Prepare a PCR Master Mix for all reactions. Combine the following reagents in the order shown then add to cDNA:
74 μL Deionized H2O 10 μL 10× Advantage 2 PCR Buffer 2 μL 50× dNTP Mix (10 mM) 2 μL 5′ PCR Primer II A (12 μM) (Table 1) 2 μL 50× Advantage 2 Polymerase Mix 10 μL cDNA 100 μL Total - The above mixture is amplified with the following conditions:
- 95 °C 1 min 1 cycle
- X cycles (16–23):
- 95 °C 15 s
- 65 °C 30 s
- 68 °C 3 min
- Hold 4 °C ∞*
Check amplification on an agarose gel with 10 μL (see Note 17).
Purify the amplified cDNA with 1.8 volumes of AMPure XP Reagent (180 μL) as described in Subheading 3.3.1, step 10 and eluted with 50 μL of Nuclease-free water. Transfer the supernatant containing the eluted DNA to a Covaris microTUBE AFA Fiber Screw-Cap tube. It is optional to Qubit the sample to obtain a DNA concentration. Run a gel with 5 μL.
-
Covaris shearing: DNA Shearing with M220 Focused-ultrasonicator.
Sonicate to 200–300 bp depending on the sequencing protocol (300 for PGM and 200 for Proton).
- End repair and purify the DNA with components from an Ion Plus Fragment Library Kit (see Note 18). Add 29 μL Nuclease-free water to the fragmented DNA to bring the total volume to 79 μL.
79 μL Fragmented gDNA 20 μL 5× End Repair Buffer 1 μL End Repair Enzyme 100 μL Total Incubate the end repair reaction for 20 min at room temperature.
Purify the amplified cDNA with 1.8 volumes of AMPure XP Reagent (180 μL) as described in Subheading 3.3.1, step 10 and eluted with 25 μL of Nuclease-free water.
- Ligate and nick repair the end repair DNA.
- 25 °C 15 min
- 72 °C 5 min
- Hold 4 °C ∞
25 μL DNA 10 μL 10× Ligase Buffer 2 μL Adapters* 2 μL dNTP Mix 51 μL Nuclease-free water 2 μL DNA Ligase 8 μL Nick Repair Polymerase 100 μL Total *If barcoded libraries are needed, P1 Adapter and the desired barcode adapters can be substituted - Place the tube in a thermal cycler and run the following program:
Purify the amplified cDNA with 1.8 volumes of AMPure XP Reagent (180 μL) as described in Subheading 3.3.1, step 10 and eluted with 40 μL of Nuclease-free water.
- The above mixture is amplified with the following conditions (see Note 13):
- 95 °C for 30 s 1 cycle
- X cycles: (16–18)
- 95 °C for 30 s
- 58 °C for 30 s
- 72 °C for 1 min
- Hold 4 °C ∞
Purify the amplified cDNA with 1.8 volumes of AMPure XP Reagent (180 μL) as described in Subheading 3.3.1, step 10 and eluted with 25 μL of Nuclease-free water. Proceed to size selection.
This can be a pause point and samples can be stored at −20 °C (see Note 14).
3.4 Library Size Selection Quality/ Quantity Assessment: Timing 2 h
There are two criteria for a quality library: the library size and DNA amount. Ideally when a library is visualized on an agarose gel, there will be one very sharp band at the appropriate size. However, there is usually a smear. There are several options to obtain the perfect size. One way is to size select with the AMPure XP or SPRIselect reagent kit (Cat # B23317, Beckman Coulter, Inc.). Here, a double extraction is performed to remove the larger then smaller fragments of dsDNA. The new SPRIselect reagent kit is reported to give a tighter selection but was not demonstrated in our lab. If the bead size selection is not precise enough, we use the E-Gel® Size-Select™ system run with several markers to obtain the appropriate size. With very low amounts of starting RNA, we can adjust the number of cycles and/or increase the volume in the PCR to increase the final amount of product. The size and quantity are optimized by analyzing the collected samples on a Bioanalyzer/TapeStation and Qubit. However, the disadvantage to the E-Gel® system is a larger amount of material lost.
- The amplified PCR product is visualized on a 2 % agarose gel for adequate concentration.
- The amplified PCR product is purified with 1.8 volumes of AMPure XP Reagent (450 μL) as described above and eluted with 20 μL of Nuclease-free water.
- If sample concentration is adequate, the sample is ready for emulsion PCR.
In general, 2–3 libraries can be constructed in parallel by one person and, including the quality assignment step, we recommend reserving a separate day for this part of the protocol.
Fig. 8.
Quality controls for Ion PGM sequencing. (a) Electropherograms of high-quality Bioanalyzer runs of sequencing libraries showing a sharp peak at 200 bp on left. The second electropherogram is an example of overloading that will not give accurate concentrations. (b) Examples of different loading density on an Ion chip. The higher density, indicated by red, gives more sequences. (c) Examples of sequencing read distributions from two different Ion Torrent sequencing runs. The plot on left shows a higher-quality distribution compared to a low-quality distribution of the sequencing reads in the plot on the right. The high-quality plot was from ~30 cells in the CA1 area of the rat hippocampus and the lower-quality plot was from Aplysia californica (Color figure online)
3.5 Template Preparation: Day 2, Timing 5 h Total
3.5.1 Clonal Amplification and Enrichment
Resultant cDNA produced from a library cannot be sequenced directly due to a limited sensitivity of the Ion Torrent sequencing platform. Thus, amplification steps are required using emulsion PCR (emPCR, Fig. 4). Key to the success of this procedure is obtaining one DNA molecule per Ion Sphere™ particle (single primer-coated bead) in an oil emulsion. During this amplification, each droplet contains a single DNA template attached to an Ion Sphere™ particle forming a clonal colony; this is the final template (Note 21). Using the described number of molecules in Ion Torrent, sample preparations are critical for an optimal sequencing run. The amount of DNA in the library (e.g., in picomoles) can be measured using a typical Bioanalyzer assay step; the number of molecules can be determined by multiplying with Avogadro’s number and then performing a unit conversion. For the 200 bp templating and sequencing kits, the optimal concentration is 15.5 × 106 molecules per μL. An alternative would be qRT-PCR, but this is a more time-consuming step. The final determination of the optimal number of molecules in the sequencing sample has been determined by Ion Torrent; thus performing titration steps might not be necessary.
There are two ways to achieve the template bead preparation: (a) the automated OneTouch™ system coupled with the enrichment Ion OneTouch™ ES instrument (Fig. 4) or (b) a manual method to set up clonal amplification and enrichment.
Here, we describe the automated OneTouch™ System requiring as little as 5 min of hands-on time and less than 5 h for the total duration of the procedure.
3.5.2 Preparation of Template-Positive Particles (ISPs) on the Ion OneTouch™ Instrument
The Ion OneTouch™ Instrument has three key technologies that enable automated delivery of templated Ion Sphere™ particles. First is a reaction filter that creates millions of microreactors in which clonal amplification occurs. Second is the in-line PCR amplification plate that enables thermal cycling of the microreactors. Third is the integrated centrifuge, which recovers the templated Ion Sphere™ particles. Combined with the Ion OneTouch™ ES, this is the first automated massively parallel clonal amplification and recovery process that permits minimal hands-on time on the market today.
- OneTouch setup:
- Install the amplification plate from the kit.
- Ion OneTouch™ Oil is installed on the left front port.
- Ion OneTouch™ Recovery Solution is installed on the right front port.
- 2 Ion OneTouch™ Recovery Tubes and the Ion One Touch™ Recovery Router are installed in the centrifuge.
- Prepare the amplification solution in order listed:
280.0 μL Nuclease-free water 500.0 μL Ion OneTouch™ 2× Reagent Mix (see Note 17) 100.0 μL Ion OneTouch™ Enzyme Mix 100.0 μL Ion OneTouch 200 Reagent P 20.0 μL Diluted library (15.5× 106 molecules per pL) 100.0 μL Ion OneTouch™ 200 Ion Sphere™ Particles (vortexed for
1 min)1,000.0 μL Total volume Mix this amplification solution by pipetting up and down, then transfer the entire volume to the Ion OneTouch™ Reaction Tube/filter assembly.
Gently layer and fill the tube with 1,500 μL Ion OneTouch™ Reaction Oil over the amplification solution.
Secure an Ion OneTouch™ Reaction Filter to the Reaction Tube.
Slowly invert the Reaction Filter/Reaction Tube assembly.
Firmly insert the three prongs of the inverted Reaction Filter into the three ports on the top stage of the Ion OneTouch™ Instrument.
Run the Ion OneTouch™ Instrument (we typically do two samples a day and run the second sample overnight). This can be a pause point.
3.5.3 Ion Sphere™ Particles Recovery
This step removes template-positive ISPs from the OneTouch™ Recovery Solution and prepares them for enrichment.
Remove both Recovery Tubes from the centrifuge, then remove all but ~50 μL of Ion OneTouch™ Recovery Solution from each Ion OneTouch™ Recovery Tube (see Note 22).
Resuspend the template-positive ISPs in the remaining Recovery Solution by pipetting the pellet up and down.
Add 1 mL of Ion OneTouch™ Wash Solution to the pooled template-positive ISPs.
Centrifuge for 2.5 min at 15,500 × g, and then remove all but 100 μL of supernatant, and vortex the pellet for 10 s to completely resuspend.
Transfer a 2.0 μL aliquot of the recovered material to a 0.2 mL PCR Tube; then assess the quality using a Qubit® 2.0 Fluorometer.
Sample is ready for enrichment.
3.5.4 Enriching the Template-Positive ISPs with the Ion OneTouch™ ES
After clonal amplification, the non-templated ISPs are removed from the templated ISPs. The Ion OneTouch™ ES uses magnetic bead technology to isolate template-positive Ion Sphere™ particles that can be loaded directly onto the Ion semiconductor chip thus delivering automated, highly reproducible enrichment with every run.
- Prepare fresh Melt-Off Solution daily.
865.0 μL Nuclease-free water 125.0 μL 1 M NaOH 10.0 μL 10 % Tween 20 in Nuclease-free water 1,000.0 μL Total volume Add 130.0 μL of MyOne™ Beads Wash Solution and 13.0 μL of Dynabeads MyOne™ Streptavidin C1 magnetic beads to a 1.5 mL LoBind Tube and mix.
Place the tube on a magnet such as a DynaMag™-2 magnet for 2 min, then remove and discard the supernatant. Repeat two times.
Resuspend MyOne™ Beads in 130.0 μL of MyOne™ Beads Wash Solution.
Obtain an 8-well strip from the Ion OneTouch™ kit. Ensure that the square-shaped tab of the 8-well strip is on the left.
Transfer the entire volume (100.0 μL) of the sample to Well 1.
Add the resuspended MyOne™ Beads (130.0 μL) to Well 2.
In Wells 3, 4, and 5 add Ion OneTouch™ Wash Solution.
Leave Wells 6 and 8 empty and fill Well 7 with 300.0 μL of fresh melt-off solution.
3.5.5 Prepare the Ion OneTouch™ ES
Place a new tip on the Tip Arm.
Place a new 0.2 mL PCR Tube in the base of the Tip Loader with cap open.
Place the eight-well strip in right side of slot.
Run the enrichment.
3.5.6 Remove and Wash the Template-Positive Ion Sphere™ Particles
Spin the enriched sample at 15,500 × g for 1.5 min. Check that there is no carryover of the brown MyOne™ Beads in the pellet (see Note 23).
Remove all but ~10 μL of supernatant without disturbing the pellet, then add 200 μL of Ion OneTouch™ Wash Solution, resuspend, and repeat centrifugation.
Remove all but ~10 μL of supernatant without disturbing the pellet and add up to 100 μL of Ion OneTouch™ Wash Solution and resuspend. Sample is ready to sequence.
Transfer a 10 μL aliquot of the recovered material to a 0.2 mL PCR Tube; then assess the quality using a Qubit® 2.0 Fluorometer (see Note 24 for the Ion Sphere™ Quality Control Kit process).
3.6 Sequencing on the Ion Torrent: Timing 4 h (Depending on Chip)
All reagents are from the Ion PGM™200 Sequencing Kit (Cat # 4474004, Ion Torrent, Life Technologies) and Ion 314™ Chip Cat # 4462923 (8 pack), 316™ Chip Cat # 4466616 (4 pack), 318™ Chip Cat # 4466617 (4 pack) Life Technologies. The protocol is basically the same with variation in the volumes of some reagents for the three different chips. We will focus on the 314 chip because this is the chip we have used the most.
Clean the Ion Torrent as recommended by following on-screen prompts.
Wash all bottles with MilliQ water (or an equivalent 18 MΩ water system) three times.
Prepare fresh solutions of 1 M and 100 mM NaOH.
Add buffers to Wash 1 and Wash 3 bottles.
Prepare Wash 2 solution.
Add dNTPs to conical tubes.
Initialize the Ion Torrent and follow on-screen prompts.
3.6.1 Sample Preparation
Prepare enriched sample (template-positive ISPs) for sequencing.
Transfer half of the volume of enriched ISPs to a 200.0 μL PCR Tube.
Vortex the Control Ion Sphere™ particles and add 5.0 μL to the sample.
Add 100.0 μL of Annealing Buffer, mix by pipetting, and centrifuge for 1.5 min at 15,500 × g.
Carefully remove the supernatant leaving approximately 3.0 μL in the tube, add 3.0 μL of Sequencing Primer, and pipette up and down thoroughly to disrupt the pellet.
- PCR:
- 94 °C for 2 min
- 37 °C for 2 min
3.6.2 Chip Check and Preparation
Place the chip on the PGM™ System grounding plate.
Press Experiment on the main menu of the PGM.
When prompted, lift the chip clamp and replace the old chip with your new chip, then close the chip clamp.
Press Chip Check and look for leaks.
Add 50.0 μL of 100 % isopropanol to the loading port of the chip, and then remove excess liquid from the other port. Then repeat twice with 50.0 μL of Annealing Buffer. When pipetting liquid into the chip, keep the pipette tip at a 90° angle to the chip, press the tip firmly into the circular loading port, and apply gentle pressure between the pipette tip and chip.
3.6.3 Load the Chip
Remove the template-positive ISPs from the thermal cycler and add 1.0 μL of PGM™ 200 Sequencing Polymerase to a final volume of 7.0 μL, mix, and let set at room temperature for 5 min.
Remove residual Annealing Buffer from the chip by tilting the chip at 45 °C, then inserting the pipette tip into the loading port, and removing all liquid.
Centrifuge the chip upside-down with tab pointing in for 5 s.
Following polymerase incubation, load the entire sample (~7 μL) into the loading port by dialing down the pipette to gently and slowly deposit the ISPs at a rate of ~1 μL per second.
Remove any displaced liquid from the other port of the chip.
Centrifuge for 30 s.
- Mix the sample on the chip.
- Set the pipette volume to 5.0 μL.
- Tilt the chip at a 45° angle so that the loading port is the lower port.
- Insert the pipette tip into the loading port and slowly pipette the sample in and out three times. Avoid creating small bubbles.
- Centrifuge for 30 s with tab pointing out.
- Repeat step (iii).
- Centrifuge for 30 s with tab pointing in.
Tilt the chip at a 45° angle and slowly remove as much liquid as possible from the loading port by dialing the pipette. Discard the liquid (see Note 25).
Spin or tap the chip to remove excess liquid.
Perform Run.
Upload Run Plan with all information and follow on-screen prompts.
Press Next to begin the run.
When the run is complete, the touch screen will return to the Main Menu. You can then proceed with another run or perform a cleaning/initializing step if required (see Note 26).
3.7 Assessing the Sequencing Run
- The quality of a run is dependent on many factors:
- Homopolymers are problematic with flow-based sequencing technology. With Torrent Suite Software version 3.2’s Variant Caller, sensitivity has increased more than two times, allowing indels to be called accurately within homopolymer regions 2–8 bp in length.
3.7.1 Summary Statistics for a Sequencing Run and Assessment
The number of reads and bases generated are dependent on the chip type, sequencing kit, and server software used in the process (see Fig. 2 table). In the last year we have experienced a 100-fold increase in sequence output with the cost remaining the same. The existing four different chips offer substantial flexibility to design different projects from pilot and time-course studies to deep sequencing of selected samples. For example, samples with less confidence can quickly be sequenced on a 314 chip for $350.00 (see Note 27). Once data are generated, a decision can be made whether to proceed with greater sequencing coverage as the next step—which can still be performed in less than 2 days. The speed of the described pipeline allows for a plethora of experiments following development, neuroplasticity, or aging steps in a rapid and cost-efficient manner that can be rapidly adjusted for emerging experimental needs.
3.7.2 Bioinformatic Pipeline “Zero-Click”
Although it is not described in the current protocol, we developed and implemented an automated transcriptome analysis pipeline [23] that assembles Ion Torrent output directly from the server without human intervention, following its automatic annotation using NCBI and Swiss-Prot databases. This fast turnaround-time pipeline is also integrated with GO ontology, KEGG pathways, and Pfam Domains. Other prediction systems can be incorporated such as secretory/signaling peptide predictions. With the “Zero-click pipeline,” once sequencing begins, the data are removed from the server, automatically processed, and uploaded to our database for viewing with all features accessible.
3.7.3 Update and Future Directions
For all practical purposes, most of the current 314 and 316 series chips optimally target the smRNA, amplicon, and smaller microbial genome sequencing applications. The current 318 chips for semiconductor sequencing are also well suited for deep de novo transcriptome analysis, small genome resequencing, and even enriched methylome sequencing [1]. However, the Ion PGM is not the best choice for sequencing larger genomes (>0.1 Gb) because of its limited throughput. To accommodate the demand for applications requiring higher throughput, such as sequencing of more complex/animal genomes, Life Technologies has released a new sequencer called the Ion Proton [6] that we successfully tested in our laboratory (IP1 chip), collecting about 9–10 Gb of sequencing data for the same applications indicated above, as well as for human exomes. Although it uses the same principle, the second version of the IPII chip is expected to produce 33 Gb, and the third iteration IP2 chip might produce about 124 Gb of sequencing data (Fig. 2). This throughput will be sufficient to sequence a human-size genome within a day or multiple transcriptomes/exomes using barcoding options. Along with the growth of the sequence output, the length of reads is also improving, with sequencing lengths of 400 bp currently achievable (Fig. 2) and longer reads of up to 640 bp reported at the latest AGBT-2013 meeting (http://ioncommunity.lifetechnologies.com/community/programs_and_events/agbt2013). Further technology improvements [6] include introduction of the Ion Chef™ as an integrated liquid handling and robotic station, as well as development of alternatives to emulsion PCR (e.g., Avalanche™). Critical to genome sequencing is the paired end and mate-pair library options which are also available. In addition, the upgrade of the Ion server software suite to version 2.2 and now 3.2 greatly improved accuracy compared to the 2.0 version, as has been reported [24].
For practical purposes, a new OneTouch kit was recently released, Ion OneTouch™ 200 Template Kit v2 DL (Cat # 4480285 Ion Torrent, Life Technologies), together with new Ion server software suite version 3.2. The new OneTouch™ 200 Template Kit v2 DL kit helps prevent clogging issues with the One-Touch instrument, and the new software has new user interfaces for planning. Additional resources, descriptions, and demonstrations can be found on the Ion community at http://ioncommunity.iontorrent.com. The webpage provides both the latest updates and allows download of novel protocols. The site also includes a discussion page where users can submit problems and discuss potential problems with both other users and consultants.
In summary, the combination of the Ion PGM and the Ion Proton instruments can make a single laboratory as a task-oriented complementary sequencing center able to sequence transcriptomes, genomes, and methylomes. Considering the enormous heterogeneity of cell populations in nervous systems and the complexity of developmental, neurodegenerative, and aging processes, the current advances in semiconductor sequencing offer novel opportunities for many fields of experimental biology and clinical studies: from whole genome sequencing in personalized medicine down to targeted probing of dynamic genomic and epigenomic organizations of individual cells (see additional details in ref. 1).
Acknowledgements
We would like to thank our collaborators Drs. Thomas Foster, C. Jason Frazier, Scott Harden and Mrs Elena Bobkiva (University of Florida) for the single neuron isolations and help with dissections. We also want to thank Alexander Fodor for help in single-cell library construction. We thank Mr. James Netherton for reading and commenting on the manuscript. We thank Dr. Manfred Lee for his technical advice and guidance with the Ion PGM sequencing process. We also thank Miss Brandi McLaughlin for help with her guidance and updates for semiconductor sequencing technologies and novel chips as well as for providing photos and schematic diagrams for Figs. 1a-c and 5. This work is supported by NIH grants 1R01GM097502, R21RR025699, 5R21DA030118, R01MH097062, McKnight Brain Research Foundation, Florida Biodiversity Institute as well as NSF-0744649, NSF CNS-0821622, and UF Opportunity Fund awards to LLM.
Footnotes
From our experience with more than 200 different invertebrate species, if RNA cannot be isolated immediately from tissue or even single cells, we prefer placing the tissue in 100 % ethanol rather than Ambion’s RNAlater® for shipping or storage. We have obtained mixed results with RNAlater® based on the quality of the RNA isolated. Tissue and/or single cells can be stored in lysis buffer and frozen at −20 °C as long as the cells or tissues are totally lysed before freezing. The kits with a lysis buffer that we find work best for this procedure are Ambion/Life Technologies, RNAqueous® Kits. We use the Ambion RNAqueous-Micro™ Kit for individual Aplysia cells and hippocampal neurons and the regular RNAqueous® Kit for larger parts of the rat brain and developmental tissues. We did not note any substantial RNA degradation for up to a month using the described storage.
A number of RNA isolation methods and protocols are widely available and can be classified into four general techniques: organic extraction methods, spin basket or column formats, magnetic particle methods, and direct lysis methods. For the organic extraction process, the sample is homogenized in a phenol-containing solution and the sample is then centrifuged. This method cannot be efficiently used for the small amounts of starting material in single cells and is not considered here. The spin bucket or column protocol is best for small samples. Two kits have proven to work best for our samples: Ambion/Life Technologies RNAqueous™ and the RNAqueous-Micro™ kits. Qiagen’s RNeasy Plus Mini Kit with gDNA Eliminator Spin Columns (Cat # 74134) as well as the QIAshredder (Cat # 79654) tissue shredders are also convenient. The QIAshredder can be used for simple and rapid homogenization of cell and tissue lysates. There is also the mini version of RNeasy Micro Kit (Cat # 74004) for small amounts of tissue, like single cells. We also efficiently used Qiagen kits, especially the columns for removal of the gDNA, and have gotten very good results.
Nevertheless, the kits described above cannot be used effectively with pigmented tissue like retina or chromatophores. For these purposes, we employed NUCLEOSPIN RNA II from Macherey-Nagel (Fisher Cat # NC9581114). Here, we have used the RNAqeuous™ lysis buffer to lyse pigmented tissue then followed the column spin protocol in this kit to get excellent RNA quality and yield.
Many invertebrates including gastropod molluscs such as Aplysia have rRNA that does not have the same electrophoretic mobility as vertebrate rRNA, in which the intact total RNA has a larger (28s) rRNA band that is twice as intense as the smaller (18s) band when denatured. Aplysia’s 28s rRNA has a true break in the middle of the rRNA molecule, which is called the “hidden break,” and under non-denaturing conditions, the rRNA molecule is held in one piece by the hydrogen bonding between its secondary structure elements [18]. However when heated, the two halves of the 28s rRNA separate and have similar electrophoretic mobility to the 18s rRNA, thus producing only one peak on the Bioanalyzer. The electrophoretic mobility pattern is species specific, creating many unique patterns (see Fig. 6 for several examples).
We have successfully employed RiboMinus™ on as little as 20 ng of total RNA starting material.
Ribosomal-depleted starting material will ensure reduced ribosomal “contamination” in the final sequencing library. This is an optional step and is useful to reduce nonspecific capture of rRNAs, which is very common for any existing protocol. Clearly, this step is essential if random primers are to be used.
We like the convenience of one kit having all the cDNA synthesis components, as with Clontech’s Marathon cDNA amplification kit. However, cheaper and just as effective alternatives are available. Any of the MMLV reverse transcriptases can be used for first-strand synthesis, see Note 6. Recently NEB came out with NEBNext® mRNA second strand synthesis module (NEB Cat # E6111) that is compatible with this protocol. These alternatives reduce the library construction costs by a quarter of the price of the Marathon cDNA amplification kit.
Currently, we are unable to sequence RNA directly; therefore RNA must be converted to cDNA, then to double stranded DNA (dsDNA) for sequencing library constructions. The starting point of all sequencing comes in a form of RNA amplification for which there are two major strategies. First is the linear amplification of in vitro transcription (IVT) which was widely employed by the microarray industry. The second approach is PCR-based amplification. Both strategies have their advantages and disadvantages. For ultra-small amounts of material such as a single cell, PCR methodology is far more applicable. PCR amplification methods still require generation of cDNA, and several options for reverse transcriptases (RT) are commercially available [avian myeloblastosis virus (AMV) and Moloney murine leukemia virus (MMLV)]. With the more traditional AMV RT, first-strand cDNA is synthesized then the second strand is synthesized with a cocktail of enzymes. Another popular method of cDNA generation is the template-switch method with an MMLV RT such as SMARTScribe™ (Cat # 634925, Clontech). This template-switch method uses the unique properties of the MMLV enzyme to synthesize additional nucleotides at the 3′ end of the cDNA, then uses a template-switch oligo to reverse direction and make the complementary second strand. Fragmentation of sequencing material is critical for all technologies. One can start RNA fragmentation using one of several techniques (heat, NaOH, RNaseIII), followed by ligation of RNA adaptors, then cDNA can be generated and PCR amplified (as in the Illumina, SOLiD, and the new Ion Torrent RNA-seq libraries). This approach does generate directionality to the library, but requires μg quantities of starting material and is not applicable to low amounts of starting material. For single cells, the template-switch method is attractive and what we use for the second protocol presented here. We use a commercially produced kit for reproducibility, consistency, and quality control, Marathon cDNA Amplification Kit (Cat # 634913, Clontech) which contains an AMV reverse transcriptase and a DNA polymerase I/RNAseH/DNA ligase enzyme cocktail. Once the second strand cDNA is produced, we polish the ends with T4 DNA polymerase, making it suitable for sequential blunt-end library-specific adaptor ligations followed by PCR amplification.
We used a shorter than usual oligo dT (13Ts compared to 30Ts) primer in our reverse transcription because we were promoting a more promiscuous priming of the oligo dT in hopes of priming upstream of the terminal oligo dT tail, thus obtaining less of the 3′UTR. This method was efficiently used for our EST collections as described [14]. Also the oligo dT portion of the primer was designed to target invertebrate RNA.
These controls allow us to monitor the quality of the library. The directionality was confirmed with the spike-ins (only 0.7 % was sequenced in the antisense direction). Also, their spiked-in copy number showed a linear relation to their specific number of reads [1].
The Adaptors A and P1 are made with one primer having a large overhang. This overhang helps prevent adaptors ligating to each other and forming primer/dimers. To produce the adaptors, individual primers are annealed together by mixing equal molar concentrations of each primer, then heating the mixtures at 72 °C for 2 min, 42 °C for 2 min, then 37 °C for 30 min.
Sequential addition of adaptors with purification between steps ensures strandedness of the fragments and thus directionality to our libraries. This also promotes reduced representation of the library. Purification between steps does cause loss of material, but insures directionality as well as reduced representation of the overall library (see Comment 2). Thus, the same amount of sequencing coverage captures more distinct transcripts from a cell. Second, by preserving strandedness, this approach unbiasedly represents both sense and antisense RNAs expressed in a given cell. Some potential drawbacks of using reduced representation methods (versus methods with random fragmentation of all transcripts) might be a reduced ability to observe certain events such as alternative splicing from the 5′end of very long transcripts. However, in contrast to the current reduced representation method, conventional random fragmentation protocols do require a significantly deeper sequence coverage and inability to separate sense and antisense transcriptional events.
The time and combination of restriction enzymes can be varied for the technology used. Both AluI and RsaI are 4 base bluntend cutters and in theory should cut every 256 bases if the DNA was random. By combining the enzymes, you can obtain smaller fragmentation of the library or vice versa.
The number of cycles of PCR amplification is dependent on the amount of starting material. We have started with as little as 40 pg of total RNA. Most times we use between 15 and 22 cycles of PCR amplification.
We like LA Taq™ (Cat # RR002M, Clontech) and Advantage® 2 Polymerase Mix (Cat # 639201, Clontech) just as well as Platinum Taq. All are interchangeable and give similar results with slight variations in price.
The design of this library is to target the 5′ end of a transcript. A complementary 3′ targeted library can be constructed in which the timing of the ligation of adaptors and digestion is adjusted. Here, after second strand synthesis the dsDNA is digested with the same enzyme pair as above, then purified with AMPure XP Reagent. Then the 5′ adaptor A is ligated. The PCR amplification uses the A primer and a P1 primer extension of the cDNA synthesis primer. This library maintains directionality and quantification. When working with very low amounts of starting material (picograms), this method is preferred because it requires the least amount of manipulation and steps. 15–21 PCR cycles are typically required in stage 21 of library construction. However, in a case of small cells (40–50 μm, such as Aplysia sensory neurons and interneurons), we successfully used up to 23 cycles.
A cheaper method of library construction is to make your own template-switch oligo and use your preferred MMLV reverse transcriptase such as SuperScript® (Invitrogen/ Life technologies) or SMARTScribe Reverse Transcriptase (Clontech). This will cut the cost of a library by 75 %. However, the SMARTer® PCR cDNA Synthesis Kit (Clontech) is convenient and great for less-experienced people to work with.
The SMARTer II A Oligonucleotide can be synthesized by substituting three ribo Guanosine (rG) for the proprietary Xs (Table 1). We did side-by-side comparisons of the commercial and our oligo, and they both produce the same amount of amplified cDNA under the same conditions.
The amplification product for the cDNA will appear as a smear on an agarose gel. It is best to check several time points so as to not over amplify. Overcycled cDNA will have large molecular weight fragments.
New England BioLabs Inc. recently introduced their line of library construction kits for Ion Torrent, NEBNext® Fast DNA Library Prep Set for Ion Torrent™ (Cat # E6270S, NEB). We did side-by-side comparisons and got comparable results from the Ion Torrent/Life Technologies and NEB kits.
E-Gel® technology is a quick and easy 15 min size separation and recovery system for library construction. However, it is critical to collect more than one sample for the library process. Getting the exact size and concentration of amplified library can be more complicated. We do four different sample extractions for each library around the size of interest on the gel, also making sure to replace the liquid in the wells (each sample should be run on the Bioanalyzer for RNA quality). For 200 bp templating and sequencing kits, we start collecting samples at ~280 bp, then 300 and 320 bp, and finally reverse to 300 bp again. This ensures that we get the best combination of size and concentration for described libraries.
Using the Agilent Bioanalyzer™ High Sensitivity DNA Kit can be complicated and time consuming and remains the current and substantial bottleneck (ironically, one almost needs to know the concentration of a sample before it can be run for the assay). We usually obtain the concentration of a sample with Qubit if we have enough material, but for single cells it is not an option. It is also important not to load over 500 pg/μL in each well, since it might clog the Bioanalyzer pins. Recently, the Agilent TapeStation 2200 was introduced and may be complementary to the Bioanalyzer. The TapeStation 2200 allows a laboratory to work with a few samples for faster assays. We have demonstrated that both machines produce comparable results.
The Ion OneTouch™ Reagent Mix has been problematic in the past. Small, undissolved particles can cause incomplete injection of the sample into the OneTouch™ machine. We let the reagent tube come to room temperature for 1 h and vortex for at least 1 min several times during that hour.
In principle, the emulsion PCR is one of the most critical steps in the process. At the beginning of the amplification, each droplet should (ideally) contain a single progenitor molecule at the outset of the emulsion PCR process (Fig. 4). These are amplified within the droplets, such that the result is that each bead contains many clonal copies of the progenitor molecule. A noticeable disadvantage of this approach is that it is necessary to carefully titrate the library prior to emulsion PCR in order to obtain no more than one library molecule per droplet. In practice, we often chose to make suggested calculations (as in Subheading 3.5.1) for initial screenings and, unfortunately, must tolerate up to 20–50 % of polyclonal reads that can be automatically removed by the Ion Torrent server. At the time of the writing, Life Technology announced that there is a possibility to eliminate the emulsion PCR step in the future, which will make the entire procedure more robust and straightforward. Finally, for the proposed single-cell RNA-seq, we noted that homopolymers at the scale of 4–5 bases do not represent a problem in the initial mapping and annotation of transcripts. However, they might cause a problem for genotyping of repetitive and homopolymer regions in clinical applications. The expected greater coverage of the Ion Proton should (Fig. 2) also improve the accuracy of sequencing.
It is really difficult to visualize the Ion Spheres. So when centrifuging the Ion Spheres, it is always suggested to leave a certain volume in the tube and not disturb the pelleted spheres. For practical purposes, we fill the same size centrifuge tube with the appropriate volume then cap and tape them to shelves in front of the bench for quick eye level comparisons. This simple procedure speeds up the process for all the steps requiring this type of comparison.
Calibration of the Ion OneTouch™ ES magnetic enrichment machine is critical to avoid carryover of the brown MyOne™ Beads in the pelleted template-positive particles (ISPs). We found that if this happens it is best to redo the process from the preparation of template-positive particles (ISPs) on the Ion OneTouch™ instrument.
The Ion Sphere™ Quality Control Kit is used to perform templated bead quality control using the Qubit fluorometer. The process labels a sample by hybridizing the Ion Sphere™ Particles (ISPs) 200 with two different fluorophores: Alexa Fluor® 488 and Alexa Fluor® 647. The probe labeled Alexa Fluor® 488 anneals to primer B sites, or all of the ISPs present, and the probe labeled Alexa Fluor® 647 anneals to primer A sites, or only the ISPs with extended templates. Then the ratio of the Alexa Fluor® 488 fluorescence (all ISPs present) to the Alexa Fluor® 647 fluorescence (templated ISPs) yields the % templated ISPs. The protocol is a short hybridization of the labeled probes and templated ISPs followed by several washes to remove the excess probes. Unenriched templated ISPs are collected after the emPCR with the Ion OneTouch™ instrument, and enriched templated ISPs are collected after enrichment on the Ion OneTouch™ ES magnetic enrichment machine. An excel format of the Qubit® 2.0 Easy Calculator containing the Calibration Factor (obtained from the ISP lot used) is used to calculate Percent Templated ISPs. Optimal values are between 10 and 30 % for an unenriched templated ISPs and >50 % for an enriched templated ISPs. It can be determined if a sample has been properly templated and/or enriched. We use this on questionable samples.
Removing liquid from the chip gives better loading; however, removing all liquid from the 314 chip is very challenging. Even if a small amount of liquid remains, our yields are above company specifications.
The Ion Torrent system recommends performing two sequencing runs per instrument initialization. We always sequence two samples consecutively for cost benefits.
Cost as of July 2012 for sequencing one sample is the following: 314 chip—$99.00, 316 chip—$150.00, and 318 chip—$499.00. The OneTouch™ and sequencing reagents are the same for all chips, $125.00 for the OneTouch™ and $125.00 for sequencing per sample.
References
- 1.Moroz LL, Kohn AB. Single-neuron transcriptome and methylome sequencing for epigenomic analysis of aging. In: Tollefsbol Trygve O., editor. Biological Aging: Methods and Protocols, Methods in Molecular Biology: Methods and Protocols. Vol. 1048. # Springer Science +Business Media; New York: 2013. DOI 10.1007/978-1-62703-556-9_21. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, Park PJ, Walsh CA. Single-neuron sequencing analysis of l1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang F, Lao K, Surani MA. Development and applications of single-cell transcriptome analysis. Nat Methods. 2011;8:S6–S11. doi: 10.1038/nmeth.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moroz LL, Kohn AB. Do different neurons age differently? Direct genome-wide analysis of aging in single identified cholinergic neurons. Front Aging Neurosci. 2010;2:1–18. doi: 10.3389/neuro.24.006.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merriman B, Rothberg JM. Progress in ion torrent semiconductor chip based sequencing. Electrophoresis. 2012;33:3397–3417. doi: 10.1002/elps.201200424. [DOI] [PubMed] [Google Scholar]
- 7.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 8.Walters ET, Bodnarova M, Billy AJ, Dulin MF, Diaz-Rios M, Miller MW, Moroz LL. Somatotopic organization and functional properties of mechanosensory neurons expressing sensorin-A mRNA in Aplysia californica. J Comp Neurol. 2004;471:219–240. doi: 10.1002/cne.20042. [DOI] [PubMed] [Google Scholar]
- 9.Kandel ER. Cellular basis of behavior. W.H. Freeman and Company; San Francisco: 1976. [Google Scholar]
- 10.Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 11.Moroz LL. Aplysia. Curr Biol. 2011;21:R60–R61. doi: 10.1016/j.cub.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ha TJ, Kohn AB, Bobkova YV, Moroz LL. Molecular characterization of NMDA-like receptors in Aplysia and Lymnaea: relevance to memory mechanisms. Biol Bull. 2006;210:255–270. doi: 10.2307/4134562. [DOI] [PubMed] [Google Scholar]
- 13.Lovell P, Moroz LL. The largest growth cones in the animal kingdom and dynamics of neuronal growth in cell culture of Aplysia. Integr Comp Biol. 2006;46:847–870. doi: 10.1093/icb/icl042. [DOI] [PubMed] [Google Scholar]
- 14.Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Ha T, Heyland A, Knudsen B, Sahni A, Yu F, Liu L, Jezzini S, Lovell P, Iannucculli W, Chen M, Nguyen T, Sheng H, Shaw R, Kalachikov S, Panchin YV, Farmerie W, Russo JJ, Ju J, Kandel ER. Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell. 2006;127:1453–1467. doi: 10.1016/j.cell.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyland A, Vue Z, Voolstra CR, Medina M, Moroz LL. Developmental transcriptome of Aplysia californica. J Exp Zool B Mol Dev Evol. 2011;316B:113–134. doi: 10.1002/jez.b.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moroz LL. Localization of putative nitrergic neurons in peripheral chemosensory areas and the central nervous system of Aplysia californica. J Comp Neurol. 2006;495:10–20. doi: 10.1002/cne.20842. [DOI] [PubMed] [Google Scholar]
- 17.Heyland A, Moroz LL. Signaling mechanisms underlying metamorphic transitions in animals. Integr Comp Biol. 2006;46:743–759. doi: 10.1093/icb/icl023. [DOI] [PubMed] [Google Scholar]
- 18.Karlstedt KA, Paatero GI, Makela JH, Wikgren BJ. A hidden break in the 28.0S rRNA from Diphyllobothrium dendriticum. J Helminthol. 1992;66:193–197. doi: 10.1017/s0022149x00014553. [DOI] [PubMed] [Google Scholar]
- 19.Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 23.Girardo DO, Citarella MR, Kohn AB, Moroz LL. Automatic transcriptome analysis and quest for signaling molecules in basal metazoans. Society for integrative and comparative biology abstracts; Charleston: Jan 3-7, 2012. [Google Scholar]
- 24.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]