Abstract
Our body is colonized by more than a hundred trillion commensals, represented by viruses, bacteria and fungi. This complex interaction has shown that the microbiome system contributes to the host’s adaptation to its environment, providing genes and functionality that give flexibility of diet and modulate the immune system in order not to reject these symbionts. In the intestine, specifically, the microbiota helps developing organ structures, participates of the metabolism of nutrients and induces immunity. Certain components of the microbiota have been shown to trigger inflammatory responses, whereas others, anti-inflammatory responses. The diversity and the composition of the microbiota, thus, play a key role in the maintenance of intestinal homeostasis and explain partially the link between intestinal microbiota changes and gut-related disorders in humans. Tight junction proteins are key molecules for determination of the paracellular permeability. In the context of intestinal inflammatory diseases, the intestinal barrier is compromised, and decreased expression and differential distribution of tight junction proteins is observed. It is still unclear what is the nature of the luminal or mucosal factors that affect the tight junction proteins function, but the modulation of the immune cells found in the intestinal lamina propria is hypothesized as having a role in this modulation. In this review, we provide an overview of the current understanding of the interaction of the gut microbiota with the immune system in the development and maintenance of the intestinal barrier.
Keywords: Microbiota, Immune system, Lamina propria, Intestinal barrier
Core tip: Each of our bodies is colonized by more than a hundred trillion commensals, which include viruses, bacteria and fungi. The association between microbiota and their hosts is complex and has important repercussions for both. The diversity and the composition of the microbiota thus play a key role in the maintenance of intestinal homeostasis and the induction of immunity. These features partially explain the link between alterations in intestinal microbiota and gut-related disorders in humans. In this review, we provide an overview of the current understanding of the interaction between gut microbiota and the immune system in the development and maintenance of the intestinal barrier.
INTRODUCTION
Each of our bodies is colonized by many commensals, such as viruses, bacteria and fungi, which are called microbiota. If we consider only the bacterial fraction, we will be examining more than a hundred trillion cells, spread all over our skin and mucosal surfaces. This quantity makes explicit the clear mutual benefit for both the microbiota and the host[1]. Due to the complex and specific demands of symbiotic and commensal organisms to survive, it is quite difficult to culture them in the lab and, therefore, to understand their contribution to the host’s biological processes. However, with current genomic sequencing techniques, a significantly greater understanding of the microbiome has been achieved.
It has become clear that adaptation of the host is influenced by the microbiome, adding new genes and functions that allow flexibility in the diet, which explains why so much effort is spent by the immune system to balance this genetic modulation. Therefore, it is reasonable to state that the increased capacity of accommodating new symbionts correlates with the increasing of the complexity of diet[2].
Although the microbiota may encompass both Eukarya and Archaea members, their relative abundance in their niche is low compared to bacteria. The highest number and most diverse microbial population is found in the colon, where there are 1010-1012 organisms per gram of luminal content[3]. Most of the bacteria found in the colon belong to the phyla Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria, and Verrucomicrobia[4]. The relationship between microbiota and host is complex, having important repercussions for both. It is now understood that microbiota contribute to physiological processes of the host, whereas the host provides the necessary nutritional environment for its survival[1].
Interestingly, in the host’s gastrointestinal tract, microbiota may have different effects. The microbiome has an important role in facilitating the development of gut-associated lymphoid tissues and participating in the metabolism of nutrients. On the other hand, under certain circumstances, the microbiota can also trigger diseases in genetically susceptible individuals[5]. Recent studies have suggested that commensal microbiota influence the host’s intestinal immune response[1,6,7]. For example, certain components of the gut microbiota are capable of inducing immunoglobulin A (IgA)-mediated responses and developing Th1/Th17 effector T cells and regulatory T (Treg) cells[8-12]. Moreover, Bacteroides fragilis mediates the development of Foxp3+ Treg cells through the activation of Toll-like receptor (TLR)2[13-15]. In the large intestine, Clostridium species induce Foxp3+ Treg cells independently of TLRs through the induction of transforming growth factor-β (TGF-β)[16]. Thus, various types of bacteria influence intestinal T cell development.
Moreover, gut microbiota have an important role in the development of Foxp3+ Treg-mediated CD4+ T cell homeostasis[17] and in the acquisition of antigen repertoire of the Foxp3+ Treg cells[18]. Although the mechanism is not clear, other cells from the immune system have important roles in the maintenance of the intestinal homeostasis[19]. Tr1 cells, for instance, do not express Foxp3 transcription factor and are induced by cytokines such as interleukin (IL)-10 and IL-27[20,21], which can be produced by CD103+ dendritic cells (DCs) when exposed to Bifidobacterium breve (B. breve)[22]. However, the mechanism by which CD103+ CX3CR1- DCs sense B. breve is not clear because CX3CR1 is required for dendrite extension[22].
In this review, we provide an overview of the current understanding of the role of the gut microbiome in the development and maintenance of the intestinal barrier.
DISTINGUISHING ENEMIES AND FRIENDS: A VISCERAL CHALLENGE
Interestingly, the intestinal immune system is able to distinguish commensals from pathogenic microorganisms. Hosts can sense commensals differently than pathogens even though they have the same immunostimulatory molecules as pathogenic bacteria and are capable of triggering inflammation if they penetrate the intestinal epithelial barrier. Many studies have shown that this sensing of commensals is important for the development and functionality of the immune system because germ-free mice have reduced cellularity and impaired functionality of the immune system in the lamina propria of the small intestine[23].
Under normal conditions, the immune system is instructed by commensal microbiota to not respond to luminal antigens. Furthermore, commensal microbiota secrete metabolites by nutrient processing, prevent infections by pathogenic microbes, provide signals to induce healthy immune development, and stimulate innate and adaptive immune responses to maintain homeostasis. However, when dysbiosis occurs, non-invasive bacteria are transported to key immune inductive sites, the mesenteric lymph nodes (MLN)[24-30]. This abnormal situation leads to aberrant immune responses against microorganisms that otherwise would not be considered a threat.
The most important difference that distinguishes pathogens from commensals is the outcome of their interaction with the host. In the intestine, an infectious process usually starts with adhesion to the brush border of intestinal cells[31,32]. After the adhesion phase, pathogenic bacteria produce virulence factors that are secreted in the external environment or injected into the cytosol of host cells. Non-invasive bacterial pathogens are able to inject virulence factors that contribute to the remodeling of the cytoskeleton of the host, leading to the formation of pedestal structures, which facilitate enhanced adhesion. Other pathogens include invasive and facultative intracellular bacteria, which secrete virulence factors that enable these pathogens to cross the epithelial barrier[33] by remodeling the actin cytoskeleton. Thus, these bacteria are able to penetrate into host cells and form a specialized niche that increases their survival[34]. Importantly, invasive pathogens need to resist innate immune defenses, survive phagocytosis and, in some cases, manipulate adaptive immunity to cross the epithelial barrier and establish infection.
Certain components of the microbiota have been shown to lead to inflammatory responses, whereas others lead to anti-inflammatory mechanisms. The diversity and the composition of the microbiota thus play key roles in the maintenance of intestinal homeostasis and partially explain the link between intestinal microbiota changes and gut-related disorders in humans[3,12,13,16,35-37].
Indeed, an association has been established between changes in the relative abundance of certain bacterial groups and the unexpected responses of the human immune system leading to diseases. The opposite situation is also observed, in which introducing a bacterial type restores homeostasis[38]. For example, Faecalibacterium prausnitzii, a member of the normal human microbiota, has been associated with the extension of the period of remission in patients with Crohn’s disease[39].
Gram-positive bacteria have microbe-associated molecular patterns (MAMPs), such as cell wall polysaccharides, peptidoglycans, lipoprotein anchors, lipoteichoic acids (LTA) and wall bound teichoic acids (WTA), that are capable of influencing pattern recognition receptor (PRR) recognition of known MAMPs, leading, for instance, to a shield effect[40,41]. These MAMPs interact with PRRs, such as theTLRs, C-type lectin receptors (CLRs) and nucleotide oligomerization domain (NOD)-like receptors (NLRs), driving the induction of innate immune responses, with immune activation, antigen presentation, and expression of antimicrobial factors[42,43].
Commensal bacterial components are usually recognized by TLRs, which is important for protection against gut injury and associated mortality. Impairment in the interaction between commensal bacteria and TLRs have been reported to promote chronic inflammation and tissue damage, e.g., inflammatory bowel disease[44]. There are two possible mechanisms by which TLR activation mediates this interaction: (1) steady-state induction of protective factors via constitutive detection of lumen-derived microbial products by TLR2 expressed on colonic epithelium or (2) upon epithelial damage, commensal-derived TLR ligands induce the production of protective factors. Recent studies have shown a role for CpG DNA, which is an agonist of TLR9, in mediating the beneficial effects of probiotics in the gastrointestinal tract[28].
Interestingly, a study has shown that non-pathogenic bacteria may modify immune responses by activating peroxisome proliferator-activated receptor gamma (PPARγ), a protein that promotes the export of the nuclear factor kappa B (NF-κB) subunit RelA from the nucleus to the cytosol, downregulating the transcriptional activity of NF-κB[45]. For instance, Bacteroides thetaiotaomicron induces PPARγ expression, leading to an anti-inflammatory profile in the intestinal compartment. This effect was not observed with a related strain, B. vulgatus[45]. It has also been suggested that commensal bacteria induce the expression of PPARγ through activation of the TLR4 pathway[46]. Additionally, the administration of an exogenous source of PPARγ by local gene therapy results in decreased inflammation in an experimental colitis model[47].
Another interesting mechanism by which commensal bacteria inhibit the NF-κB pathway occurs through stabilization of IκBα, a key inhibitor of the NF-κB pathway. Studies have shown that certain strains of bacteria, such as nonpathogenic Salmonella and Lactobacillus casei, inhibit IκBα degradation by the ubiquitin/proteasome system[48,49].
Although MAMPs appear to be identical between different species, there are variations in their chemical structure in regards to polymer composition, length and substitutions[4]. Some studies in several lactobacilli have targeted the D-alanylation of LTA as having a role in the immunogenicity of these MAMPs. Loss of D-alanylation of LTA in Lactobacillus plantarum, for instance, leads to a decrease in the capacity of the molecule to initiate TLR2-dependent proinflammatory responses[50]. In a mouse model of colitis, this mutation leads to a more protected phenotype compared with the WT[50]. In addition, other strain- or species-specific variations in the chemical modification (acetylation or pyruvylation) of the conserved peptidoglycan polymer backbones may lead to altered immunomodulatory capacities in the intestine[51].
The pilin-encoding spaABC operon found in probiotic Lactobacillus rhamnosus (LGG) leads to the production of SpaC protein, which can bind to mucus, explaining why it is more persistent in the human intestine than a closely related strain L. rhamnosus that lacks pili. Other protein effector molecules produced by LGG have been identified that prevent apoptosis induced by proinflammatory cytokines[52-54].
Another study demonstrated that protein glycosylation of the S-layer protein produced by Lactobacillus acidophilus (L. acidophilus) North Carolina Food Microbiology is essential for its interaction with the CLR DC-SIGN (DC-specific ICAM3-grabbing non-integrin) as it influences cytokine response in DCs and T cell priming[55].
The absence of the microbiota in germ-free mice causes developmental defects in the immune system. These mice have fewer plasma cells and intraepithelial lymphocytes, lower IgA levels, and smaller Peyer’s patches and MLNs than conventional animals and exhibit increased susceptibility to pathogenic bacteria[56].
The intestinal epithelial barrier is composed of tightly attached epithelial cells, antimicrobial products, and a mucus layer. Commensal microbiota maintain the integrity of epithelial cells, stimulate them to secrete mucus and anti-microbial peptides, and thereby contribute to maintaining a basal level of steady-state host defense. Goblet cells secrete mucin-2, which forms a net-like mucus layer that physically separates most of the microbiota from the epithelium. In the colon, the lower layer is dense, relatively free of bacteria, and has concentrated levels of alpha-defensins; the upper layer contains some commensal bacteria. In the small intestine, the mucus is only one layer thick, and the epithelium is protected from microbiota by antibacterial proteins such as primarily regenerating islet-derived 3-gamma (RegIIIγ)[57].
In the innate immunity scenario, antimicrobial peptides, such as alpha-defensins, lysozyme C, phospholipases, C-type lectin, and RegIIIγ are produced by Paneth cells or by enterocytes[1,58]. In the adaptive immunity scenario, system effectors are secreted into the intestinal lumen, restricting bacterial penetration into the host’s mucosal tissue. An example of this is IgA[59]. With these peptides, the host shapes the gut microbiome and controls the interaction between the host and microbiota (Figure 1).
Figure 1.
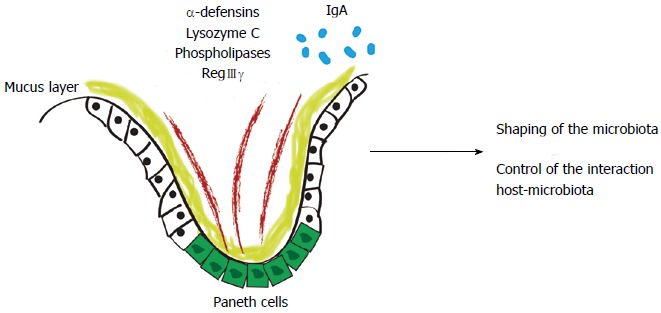
First line of defense of the intestinal barrier shapes the gut microbiota. Antimicrobial peptides are produced by Paneth cells, such as alpha-defensins, lysozyme C, phospholipases and C-type lectin, primarily regenerating islet-derived 3-gamma (RegIIIγ) or by enterocytes (RegIIIγ). In the adaptive immunity scenario, system effectors are secreted into the intestinal lumen, restricting bacterial penetration into the host mucus and mucosal tissue.
INTESTINAL DENDRITIC CELLS AND MACROPHAGES: A COMPLEX DISTINCTION
Mononuclear phagocytes such as macrophages and DCs are the main cells involved in the maintenance of tissue integrity as well as in the initiation and control of innate and adaptive immune responses. Thus, they are crucial[60] to preserving homeostasis and preventing infections through the maintenance of tolerance to dietary antigens and control of commensal microorganisms and pathogens in the intestinal mucosa[61]. These phagocytes are distributed in lymphoid organs such as Peyer’s patches and MLNs and are also very abundant in the gut lamina propria[62], but their phenotypic characterization is not completely understood.
DC populations definition was initially proposed based on the expression of the markers CX3CR1 (fractalkine receptor) and CD103 (αE integrin)[63], but the complexity of markers has increased over time. Rivollier et al[64] has shown that CD11c+ DCs can be divided into three populations: CD103+CX3CR1-CD11b- DCs, CD103+CX3CR1-CD11b+ DCs, and CD103-CX3CR1intCD11b+ DCs. Particularly, CD103+CX3CR1- DCs. These three populations, which also express CD11c and major histocompatibility complex II (MHC II), have been well characterized[61,65] and have generated great interest. Currently, there appears to be a consensus that CD11c+CD103+ MHCII+ cells are the “bona fide” DCs of the lamina propria[66] because of their contribution to intestinal health, as described below.
DCs constantly survey the microenvironment and coordinate a balance of maintaining immune tolerance to harmless antigens while mounting immune responses against enteric pathogens. Depending on from which bacterial strain components were derived, DCs can be stimulated, leading to either IL-12 secretion and a Th1 response, or IL-10 secretion and a Th2 response, as will be detailed below. However, a controversy remains regarding whether the CX3CR1-expressing cell line is DCs or macrophages. Many groups still refer to these as DCs, whereas others categorize them as mononuclear phagocytes and others as macrophages[67]. Increasing evidence has shown that there are numerous subsets of DCs and macrophages in the lamina propria[64,67].
In addition, it has been demonstrated that CD103+CX3CR1- DCs develop independently of macrophage colony-stimulating factor (M-CSF) but expand in response to fms-like tyrosine kinase 3 ligand and GM-CSF[68]. These DCs appear to be the primary, if not the only, population of DCs that migrate to the MLNs through a CCR7-dependent mechanism, and they are important for the induction of oral tolerance and suppression of the development of colitis through the induction of Treg cells[62,63,68-70]. These DCs have also been described to have the ability to generate and activate CD8+ T cells[71] with TGF-β production[70]. In addition, these cells produce the vitamin A metabolite retinoic acid (RA) in the gut[72]. RA production by DCs is enhanced by inflammatory stimuli and plays a role in immune homeostasis and maintenance of intestinal tolerance in the steady-state[73].
Kinnebrew et al[74] showed that the CD103+CD11b+ DCs from the lamina propria promote tolerance against food antigens and can rapidly produce IL-23 in response to flagellin in the lamina propria. In addition, Rivollier et al[64] demonstrated that in ulcerative colitis, Ly6Chi monocytes infiltrate into the colon and differentiate into pro-inflammatory DCs that express CD103-CX3CR1intCD11b+ and secrete high levels of IL-12, IL-23, iNOS, and tumor necrosis factor-α (TNF-α). This work showed that Ly6Chi monocytes have the ability to differentiate into regulatory mononuclear phagocytes or inflammatory DCs in the colon. Zigmond et al[75], using an acute innate model of colitis, showed that infiltrating Ly6Chi monocytes acquire two functionally distinct fates in the inflamed colonic lamina propria. Rather than giving rise to resident CX3CR1hi macrophages as in the healthy colon, the monocyte infiltrate initially differentiates into CX3CR1intLy6Chi effector cells that sense bacterial products via TLRs and NOD2. The monocyte infiltrate gives rise to a phenotypically and functionally distinct CX3CR1intLy6Clo population that displays migratory DC hallmarks such as uptake and processing of orally acquired antigens and priming of naive CD4+ T cells. This process occurs with C-C chemokine receptor type 7 (CCR7) expression, which enables these cells to emigrate from the colonic lamina propria towards the draining lymph nodes. Recently, Cerovic et al[76] demonstrated the presence of two distinct subpopulations of CD103- DCs in the intestine. Similar to what is observed in CD103+ DCs, intestinal-derived CD103- DCs appear to be responsive to Flt3 and able to activate naive T lymphocytes, giving them a migratory phenotype. This presents a new mechanism for the rapid activation of T effector responses in the intestine.
In summary, CD103+ DCs act as sentinels. They sense inflammatory signals, capture luminal antigens, and migrate to MLNs to interact with T cells. DCs are key players in the intestinal mucosa, promoting tolerance, and immunity. Their plasticity and motility allows them to play multiple roles as they move from the lamina propria to the epithelium and, subsequently, towards the MLNs.
In contrast, CX3CR1+ cells that do not express CD103 were initially described as DCs in the distal ileum. These cells were shown by several studies to play a key role in capturing and transporting intestinal antigens to MLNs[63,77,78]. Furthermore, CX3CR1+ cells have an ontogeny that is distinct from CD103+ DCs and appear to be derived in an M-CSF-dependent manner[68]. Pro- and anti-inflammatory properties have been linked to CX3CR1+ cells from the lamina propria. Importantly, the CX3CR1+ cells from the lamina propria represent a heterogeneous group of cells, which express high and low levels of CX3CR1[63].
CX3CR1hi cells from the lamina propria were defined as macrophages because they did not have the ability to migrate to the MLNs[61]. Therefore, CX3CR1+ macrophages were thereafter known as residents of the lamina propria. CX3CR1hi macrophages have been shown to contribute to intestinal homeostasis through commensal bacteria recognition and the production of anti-inflammatory cytokines[79]. The absence of CX3CR1 led to failure to establish oral tolerance; in other words, they cannot efficiently suppress local and systemic antigen-specific immune responses upon exposure to food antigens. These cells also appear to play an important role in the induction of oral tolerance by expanding Foxp3+ Treg cells[80]. Both CX3CR1+ macrophages and Foxp3+ Treg cells are mostly abundant in the colon, whereas Foxp3+ Treg cells are scarce in the duodenum. The interactions between these cells remain to be elucidated[81].
Medina-Contreras et al[82] demonstrated an important role for maintaining CX3CR1+ macrophage populations in the lamina propria preventing commensal bacteria translocation to MLNs, these cells limit Th17 responses in colitis. CX3CR1 knockout mice (KO) had reduced frequencies of lamina propria macrophages and exhibited markedly increased translocation of commensal bacteria to MLNs. In addition, the severity of dextran sodium sulfate (DSS)-induced colitis was drastically increased in the KO compared with the control mice. These cells appear to be important for protection against intestinal inflammation and gut barrier integrity. Interestingly, Diehl et al[83] showed that the CX3CR1hi mononuclear phagocytes of the intestine, which had previously been shown to be non-migratory, were able to migrate into MLNs in the absence of MyD88 or under conditions of antibiotic-induced dysbiosis in a CCR7-dependent manner, carrying non-invasive bacteria captured from the intestinal lumen and inducing both T lymphocyte responses and IgA production to avoid inflammatory bowel disease. The microbiota seem to instruct the immune system to inhibit migration of bacteria to MLNs via CX3CR1hi cells. This mechanism leads to tolerance to commensal bacteria. Recently, using the expression of CD64, Tamoutounour et al[84] also managed to distinguish macrophages from DCs in the lamina propria and in the MLNs. The authors identified the gamma chain IgG receptor high affinity FcyRI (CD64) as a marker to label intestinal macrophages. The authors showed that macrophages and DCs could clearly be discriminated by CD64 expression, even when the macrophages express CD11cint (CD64+) or when the DCs express CX3CR1int (CD64-). The expression of CD64 in macrophages is induced by interferon (IFN)-γ and suppressed by IL-4. However, on the other hand, IL-10 also upregulates CD64 and might sustain CD64 expression on macrophages. In the last stage of development in the lamina propria, macrophages express CD64+CD11b+CX3CR1hi. More importantly, it has been demonstrated that CD64 can be used as a reliable marker of macrophages in both the small and large intestine under steady-state conditions and inflammatory responses[85].
CX3CR1+ macrophages and CD103+ DCs in the intestinal lamina propria have developed mechanisms to prevent exacerbated responses to commensal bacteria, but they can also respond to infection by pathogens. The effects of gut microbiota in the cells of the lamina propria, which are crucial in recognizing bacterial tolerance induction and orientation of T cell responses, appear to be essential for the maintenance of intestinal immune homeostasis. The plasticity of dendritic cells, for example, is extremely important for their ability to respond to microbial stimuli and the ability to capture luminal bacteria and migrate to MLN. In the lamina propria, macrophages are educated to acquire non-inflammatory characteristics. Interestingly, however, the expression of CX3CR1+ in macrophages that were isolated from colon differs considerably from those isolated from the duodenum, jejunum and ileum, suggesting that the instructions that macrophages receive from these regions are variable. This makes it clear that distinct commensal populations in different regions of the intestine give signals to these cells, influencing their profiles[86].
The role of gut microbiota in macrophage and DC development is not clear. It is known that these cells participate in the regulation of intestinal immune responses against various microorganisms and diseases by producing several pro- and anti-inflammatory cytokines in an attempt to maintain intestinal homeostasis. This is an important topic for further investigation. A summary of these findings is illustrated in Figure 2A and B.
Figure 2.
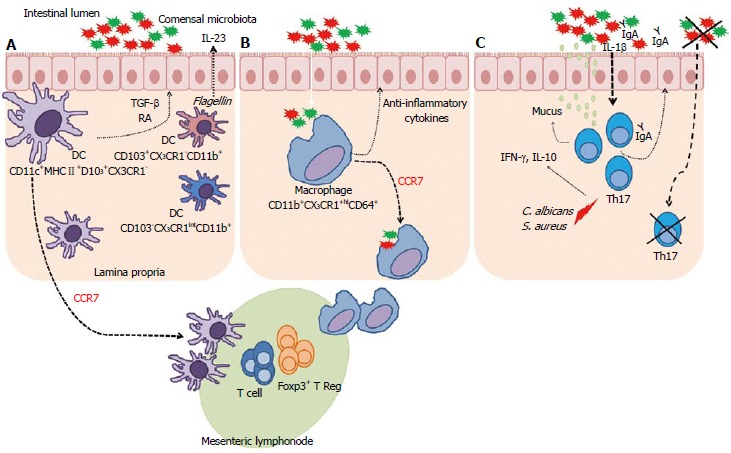
Complex interaction between microbiota, dendritic cells and macrophages. A: CD11c+MHCII+D103+CX3CR1 cells migrate to the mesenteric lymph nodes (MLN) through a C-C chemokine receptor type 7 (CCR7) dependent mechanism. These dendritic cells (DCs) can generate and activate CD8+ T cells and Treg Foxp3+ cells. These DCs also have the ability to produce transforming growth factor-β (TGF-β) and retinoic acid (RA). CD103+CX3CR1-CD11b+ DCs can produce interleukin (IL)-23 in response to flagellin. Other DCs in lamina propria are CD103-CX3CR1intCD11b+; B: CD11b+CX3CR1hiCD64+ macrophages in the lamina propria contribute to the intestinal homeostasis through the production of anti-inflammatory cytokines and are able to recognize commensal microbiota beyond the epithelial barrier. These macrophages had previously been described as non-migratory were able to migrate into the MLNs, in a CCR7-dependent manner, carrying non-invasive bacteria captured in the intestinal lumen induces T lymphocyte responses; C: Under steady state, Th17 cells are usually found in the lamina propria of the small intestine, where its development depends on the presence of dietary antigens and commensal microbiota. These cells have important effects on the intestinal epithelium, improving the barrier function by stimulating mucin production, function of tight junction proteins, and increasing the transport of IgA to lumen. Candida albicans and Staphylococcus aureus induce Th17 cells, which produce interferon (IFN)-γ and IL-10. In the absence of microbiota, Th17 cells are not found. IL-1β, induced by commensal bacteria, is critical for differentiation of Th17 cells in the intestine.
MICROBIOTA AND ITS ROLE ON TH17 ACTIVATION
Th17 cells are a prominent population among the T cells present in the intestinal lamina propria that cooperate in maintaining intestinal homeostasis[87]. These Th17 cells play a key role in mucosal host defenses as well as in the development of autoimmune diseases[88]. Under steady-state conditions, Th17 cells are usually found in the lamina propria of the small intestine, where Th17 cells development depends on the presence of dietary antigens and commensal flora[12]. These cells are a subset of CD4+ T cells and primarily secrete IL-17, which has important effects on the intestinal epithelium, through improving the barrier function and stimulating mucin production, as well as on the function of tight junctions and transport of IgA to the lumen[89,90].
While accumulating evidence shows that Th17 cells play a role in the pathogenesis of a variety of inflammatory conditions, there is considerable controversy concerning whether they also contribute to the maintenance of intestinal immune homeostasis. Both protective and pathogenic roles of IL-17 have been reported in patients with inflammatory bowel disease (IBD) and in experimental colitis in mice[91,92]. Patients with IBD often have increased levels of IL-17, and IL-17 specific inhibition protected them from this disease[93].
It is important to note that during inflammatory conditions, such as experimental autoimmune encephalomyelitis (EAE), the induction of Th17 cells requires the following cytokines: IL-1β, IL-6, IL-23 and TGF-β1[88]. In addition to being present during the inflammatory response, a population of T cells that expresses retinoic acid receptor, RORγT (which is a specific transcription factor of Th17 cells), was also found under steady-state conditions (sTh17) in the lamina propria of the small intestine[94], where they accumulate in the presence of luminal commensal microbiota.
An important role for these cells in the digestive tract has been shown in RORγT KO mice, which lack both innate and Th17 cells. These mice displayed a large expansion of lymphoid follicles in the intestine, had an increased number of Th1 and IgG+ B cells and were extremely susceptible to DSS-induced colitis[95]. Moreover, Th17 cells are not found in the gastrointestinal tract of germ-free mice, suggesting that this cell population is generated in response to the gut microbiota[96]. Segmented filamentous bacteria (SFB) are potent inducers of Th17 in the intestine, despite being found in low frequency in the intestine[12]. Other components of the microbiota can also stimulate Th17 cells in the intestine, including the “Altered Schaedler Flora” (ASF), which comprises L. acidophilus (strain ASF 360), Lactobacillus salivarius (strain ASF 361), and Bacteroides distasonis (strain ASF 519) and several other species[14]. This stimulation depends on the host immune response and the exposure time. The induction of Th17 cells in the intestinal lamina propria by SFB protects against Citrobacter infection by stimulating the production of RegIIIγ defensins[12]. Nevertheless, SFB also increases the susceptibility to EAE, arthritis[97], colitis[98] and diabetes[99]. The exact mechanism by which SFB are able to induce Th17 differentiation in the intestine is not understood. Flagellins are potentially involved[100]. Colonization with SFB leads to increased IgA production and secretion; moreover, the colonization of germ-free mice with SFB also increases the expression of Th17 cells in the intestine[12,101].
A recent study has shown that Candida albicans and Staphylococcus aureus induce the expression of Th17 cells and that these cells are able to produce IFN-γ and IL-10[102]. Furthermore, Shaw et al[103] showed that IL-1β induced by commensal bacteria is critical for the differentiation of Th17 cells in the intestine under steady-state conditions. It is clear that the differentiation of Th17 cells is extremely complex and triggered by various ligands, such as microbial cells and innate cytokines. Th17 cells are double-edged swords: they can act as both protectors and aggressors, depending on the context. They are generated in response to microbiota, and they are able to induce the secretion of pro- and anti-inflammatory cytokines with important effects on the intestine epithelium. Th17 cells are also important for maintaining homeostasis between the host and microbiota. A summary of these findings is illustrated in Figure 2C.
GUT PERMEABILITY: AN UNCLEAR CONNECTION BETWEEN ALTERED GUT MICROBIOTA AND THE IMMUNE SYSTEM
The gastrointestinal tract is considered the largest surface of the human body that is in contact with the environment. The mucosal barrier plays an important role in the selection of luminal factors that are allowed to enter the body and those that are forbidden to enter because of the danger they may pose.
The mucosal barrier is composed of a mucus layer, epithelial cells, and intercellular tight-junction proteins between these cells[104]. Tight junction proteins are key molecules for determining paracellular permeability; they form complex protein systems, which are organized by the transmembrane proteins occludin and claudins interacting with zonula occludens proteins that bind to the actin cytoskeleton. When actin contracts, it leads to increased permeability to electrolytes and small molecules[105].
In the context of inflammatory bowel syndrome (IBS), some studies have shown that the intestinal barrier is compromised, and decreased expression and differential distribution of tight-junction proteins are observed[106-110]. The nature of the luminal or mucosal factors that affect the function of tight junction proteins is still unclear.
There is some evidence suggesting a role for the mast cell enzyme tryptase in the degradation of the tight-junction proteins and increased permeability because the infiltration and activation of these cells are increased in IBS patients in association with higher output of tryptase from their mucosal biopsies[111]. Therefore, it is possible that these proteins are both expressed less because of transcriptional/translational regulation and destroyed because of increased tryptase output. Understanding the predominant mechanism involved may present a possibility for interference as a potential therapy by improving the intestinal barrier in IBS. However, it is still unclear whether the altered gut microbiota found in IBS or the modulation of intestinal immune cells may trigger detrimental effects on the gut barrier. Recent findings suggest that there is a complex interaction between alterations in microbiota and immune cell recruitment, which lead to physiological responses such as an altered gut barrier.
Some probiotic molecules appear to modulate changes in host cell signaling. This scenario can be illustrated by the p40 and p75 proteins produced by LGG: they comodulate phosphoinositide 3-kinase (PI3K)/Akt signaling[112]. When TNF-α, IL-1β and IFN-γ are secreted, p40 protein and unidentified epidermal growth factor receptor ligands stimulate the production of Bcl2, stabilizing tight-junction proteins and promoting epithelial barrier function and cell survival[113].
TLR and NLR signaling triggered by MAMPs are likely to have roles in the production of physical and chemical defenses in the small intestine, limiting the numbers of mucosa-associated bacteria and preventing bacterial penetration of host tissues. Some bacterial strains can also stimulate regulatory immune mechanisms through the activation of DCs and CD4+Foxp3+ T cells[114]. This phenomenon has been shown by a study in which Bifidobacterium breve led to the induction of IL-10-producing regulatory Tr1 cells in the colon via TLR2/MYD88-dependent production of IL-10 and IL-27 in CD103+ DCs[22]. WTA and LTA have also been shown to shift IL-10/IL-12 ratios in macrophages towards IL-10 via the TLR2/Extracellular signal-regulated kinase (ERK) signaling pathway[115].
Functional changes of epithelial cells can also be triggered by bacterial components. LGG proteins p40 and p75 increase the resilience of intestinal epithelial cells to cytokine-induced proapoptotic signals and induce a strengthening of the epithelial barrier function involving the EGFR/PI3K/Akt/PKC pathway[113]. Another study has shown that the expression of tight-junction in humans is modulated through TLR2 signaling[116]. Moreover, B-cell lymphoma-9, ERK3, c-Jun N-Terminal Protein Kinase and poly(Adenosine diphosphate-ribose) polymerase (PARP)14 have also been implicated in the signaling events induced by LGG consumption, leading to the induction of IFN/STAT4 pathway activation and production of T helper 1-type cytokines[117].
In the context of obesity and metabolic syndrome, it is unclear how immune modulation occurs in the intestine, despite numerous lines of evidence showing that intestinal barrier disruption is associated with alterations in the gut microbiota[118-121]. Conversely, other models, such as colitis and inflammatory bowel disease, have shed light on mechanisms that potentially orchestrate the modulation of the immune system by the microbiota, which may be very useful for understanding gut barrier alterations in models of obesity.
Other studies have shown that the endocannabinoid system is also involved in the regulation of the gut barrier and inflammation. Metabolic endotoxemia and systemic inflammation are suppressed by 2-arachidonoylglycerol; these phenomena are potentiated by 2-palmitoylglycerol. In addition, 2-oleoylglycerol leads to the release of gut peptides from intestinal L-cells, such as the glucagon-like peptide 2, which is associated with the regulation of gut barrier function[120].
Although some investigations have led to the hypothesis that Gram-negative bacteria may be involved in triggering metabolic endotoxemia and, therefore, in worsening the condition of the intestinal barrier[119-123], it is plausible that mechanisms other than lipopolysaccharide (LPS) are responsible for this. This is illustrated by the study that showed that Akkermansia muciniphila, a Gram-negative bacterium, decreased metabolic endotoxemia, which was induced by a high-fat diet, through increasing levels of endocannabinoids that control inflammation, the gut barrier and the gut peptide secretion[121]. A summary of these findings is illustrated in Figure 3.
Figure 3.
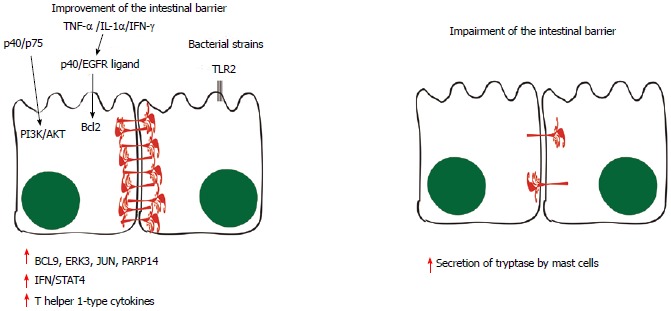
Pathways involved in the improvement and in the impairment of the intestinal barrier. Some probiotic molecules seem to modulate changes in host cell signaling, such as the p40 and p75 proteins, which comodulate phosphoinositide 3-kinase (PI3K)/Akt signaling. When tumor necrosis factor-α (TNF-α), interleukin (IL)-1α and interferon (IFN)-γ are secreted, the p40 protein and unidentified epidermal growth factor receptor (EGFR) ligands stimulate the production of Bcl2, stabilizing tight junctions and promoting epithelial barrier function and cell survival. Toll-like receptor (TLR) and NOD-like receptor (NLR) signaling triggered by microbe-associated molecular patterns (MAMPs) are likely to have roles in the production of physical and chemical defenses in the small intestine, limiting numbers of mucosa-associated bacteria and preventing bacterial penetration of host tissues. Moreover, BCL-9, ERK3, JUN and poly(ADP-ribose) polymerase (PARP)14 have also been implicated in the signaling events induced by probiotics, leading to induction of IFN/STAT4 pathway activation and to the production of T helper 1-type cytokines. On the other hand, evidences suggest that the mast cell tryptase is involved in the degradation of the tight-junction proteins and increased permeability, since the infiltration and activation of these cells are increased in inflammatory bowel syndrom patients in association with higher output of tryptase from their mucosal biopsies. BCL9: B-cell lymphoma-9; JNK: c-Jun N-Terminal Protein Kinase; ERK: Extracellular signal-regulated kinase.
PROBIOTICS: EPITHELIUM, IMMUNE RESPONSES AND THERAPEUTICS
Probiotics have been described as a “beneficial live microbial supplement which improves the intestinal microbial balance”[124]. The mechanisms of action of probiotics have been thoroughly discussed. It has been demonstrated that they are capable of modulating the permeability of epithelial barriers, changing the inflammatory potential of epithelial cells, or directly modulating the activity of immune cells[125-127].
The immune system of the intestinal mucosa plays a key role in defending against pathogens. The potential role of probiotics in the function of immune cells, such as DCs, suggests that certain species of probiotics could be used to modify T lymphocyte responses[128]. Certain probiotics that have the property of inhibiting IL-12 secretion can be extremely important in Th1-mediated diseases due to their ability to restore the homeostasis of the intestinal immune system[129,130]. Probiotics have also been described as being capable of inducing Foxp3+ Treg cells or developing TGF-β-bearing Treg cells[131,132]. Furthermore, stimulation of the immune system with probiotics can contribute to the production of IL-10, an essential cytokine for intestinal homeostasis maintenance[22,115,132]. Moreover, probiotics have been described as antagonists of pathogenic bacteria because they trigger effects such as reduction of luminal pH, inhibition of bacterial adherence, and production of antimicrobial molecules[4].
The use of probiotics can promote improvement in several diseases; for example, they cause diminished symptoms of IBD[124]. Most of the currently used probiotics belong to the genera Lactobacillus and Bifidobacterium. In EAE mouse models, L. paracasei and L. plantarum induced CD4 + CD25 + Foxp3 + T cells in the mesenteric lymph nodes, leading to increased TGF-β expression and reduced inflammation in the central nervous system[132]. Other studies have confirmed this immunomodulatory effect of Lactobacillus, showing that it leads to augmentation of IL-10 levels[129,132,133] and to a reduction of pro-inflammatory cytokines such as IL-6 and TNF-α[134]. Other probiotics are able to inhibit NF-κB, such as L. plantarum, suggesting that it induces tolerance to food antigens[135].
Some studies also highlight Lactobacillus as an activator of conventional DCs and Bifidobacterium as an activator of CD103+ DCs[70]. Bermudez-Brito et al[136] showed that Lactobacillus paracasei Collection Nationale de cultures de microorganismes I-4034 treatment led to a suppressed pro-inflammatory cytokine and chemokine profile in human intestinal DCs challenged with Salmonella in a TLR2-dependent manner. In addition, probiotics may induce functional changes in epithelial cells. It is not clear which soluble factors are released in the conditioned medium by LGG, but they are suggested as regulators of the expression of heat shock proteins 25 and 72 in intestinal epithelial cells in vitro[137], conferring protection against oxidative stress-mediated apoptosis. Another probiotic, L. plantarum WCFS1, modulates the expression of tight junction proteins in humans via TLR2 signaling pathways[116]. Probiotics may also lead to increased production and secretion of IgA through modulating cytokine expression in the intestine[138].
INTERACTIONS BETWEEN MICROBIOTA, THE IMMUNE SYSTEM AND ORGANS
Despite our growing understanding about the consequences of the host-microbiota interaction for the immune function in the intestine, the extent to which the intestinal flora contribute to immunity at distal sites remains an enigma.
The skin provides the first line of defense by the host immune system against invading pathogens. There are several commensal communities residing on the skin[139]; inflammatory skin diseases, such as psoriasis and dermatitis, have been associated with imbalanced skin microbiota[140,141].
Naik et al[142] showed that the commensal microbiota of the skin is necessary for an appropriate immune response. Protective immunity to a pathogen on the skin was considered critically dependent on the microbiota of the skin, and not of the intestine. These cutaneous commensal microorganisms exert their effects by increasing IL-1 signaling and amplifying responses according to the site of inflammation. Therefore, through their ability to promote IL-1 signaling and, thus, the function of effector T cells, commensals of the skin are likely drivers and amplifiers of pathologies of the skin[142].
Commensal bacteria, such as Streptococcus epidermidis, produce ligands that are capable of activating the TLR pathway. To investigate whether commensal bacteria influence the skin inflammatory response, Lai et al [143] treated primary human keratinocytes with a TLR ligand, poly(I:C), which was able to activate TLR3 signaling, causing an increase in the expression of TNF-α and IL-6. The authors also observed that staphylococcal lymphotoxin is a selective suppressor of TLR3-mediated inflammation in the skin.
The investigation of lung microbiota is relatively new and may lead to new discoveries about respiratory diseases. The lungs of healthy humans were previously believed to be sterile. However, studies have shown that the lungs of healthy patients are colonized by some communities of bacteria[144,145]. The results of published studies differ, but Proteobacteria, Firmicutes and Bacteroidetes are commonly identified at the phylum level. At the genus level, Pseudomonas, Streptococcus, Prevotella, Veillonella and Fusobacteria predominate with minor contributions from potential pathogens, including Haemophilus and Neisseria[146].
Low levels of bacterial products can be detected in systemically infected patients and, to a lesser degree, in healthy people, suggesting that the products of intestinal microbiota can activate TLR and NLR in the liver. Numerous studies indicate that macrophages are also sensitive to physiologically relevant levels of microbial products reaching the liver as these cells respond to low concentrations of LPS through the activation of NF-κB and production of pro-inflammatory cytokines[147].
Alteration of the permeability of the intestine is the primary means by which intestinal microbiota alterations activate innate immunity in the liver. Therefore, liver injury mediated by endotoxin can be reversed by removing Kupffer cells or by neutralizing TNF-α with anti-TNF-α antibody[148]. Recent evidence also demonstrated the involvement of microorganisms in less severe forms of liver disease. More specifically, intestinal microbiota can have a central role in liver fibrosis as evidenced by results with mice showing that chemically induced fibrosis is associated with increased bacterial translocation[149].
CONCLUSION
Understanding the interaction between commensal microorganisms and the host contributes to comprehending the functionality of a new organ, the microbiota, which is responsible for the maturation and modulation of many systems, such as the immune and metabolic systems. Although many of these microorganisms perform functions that are essential for maintaining the homeostasis of the immune system, they pose a threat if the intestinal barrier is impaired and may lead to numerous pathologies, such as inflammatory bowel disease and metabolic syndrome. Further investigations are necessary to increase the understanding of how the microbiota influence the development of the immune system and cell differentiation as well as how these changes are able to stimulate responses in distant organs.
Footnotes
P- Reviewers: Bailey MT, Bakke AM, Ukena SN S- Editor: Zhai HH L- Editor: A E- Editor: Wu HL
References
- 1.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 2.Harvill ET. Cultivating our “frienemies”: viewing immunity as microbiome management. MBio. 2013;4 doi: 10.1128/mBio.00027-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 4.van Baarlen P, Wells JM, Kleerebezem M. Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol. 2013;34:208–215. doi: 10.1016/j.it.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung H, Kasper DL. Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Curr Opin Immunol. 2010;22:455–460. doi: 10.1016/j.coi.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MA, Geuking MB, Beutler B, Tedder TF, Hardt WD, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 14.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 18.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnes MJ, Powrie F. Regulatory T cells reinforce intestinal homeostasis. Immunity. 2009;31:401–411. doi: 10.1016/j.immuni.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 20.O’Garra A, Vieira PL, Vieira P, Goldfeld AE. IL-10-producing and naturally occurring CD4+ Tregs: limiting collateral damage. J Clin Invest. 2004;114:1372–1378. doi: 10.1172/JCI23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pot C, Apetoh L, Awasthi A, Kuchroo VK. Molecular pathways in the induction of interleukin-27-driven regulatory type 1 cells. J Interferon Cytokine Res. 2010;30:381–388. doi: 10.1089/jir.2010.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeon SG, Kayama H, Ueda Y, Takahashi T, Asahara T, Tsuji H, Tsuji NM, Kiyono H, Ma JS, Kusu T, et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012;8:e1002714. doi: 10.1371/journal.ppat.1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 24.Macpherson AJ, Uhr T. Compartmentalization of the mucosal immune responses to commensal intestinal bacteria. Ann N Y Acad Sci. 2004;1029:36–43. doi: 10.1196/annals.1309.005. [DOI] [PubMed] [Google Scholar]
- 25.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 26.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 27.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 28.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Abt MC, Artis D. The dynamic influence of commensal bacteria on the immune response to pathogens. Curr Opin Microbiol. 2013;16:4–9. doi: 10.1016/j.mib.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyle EC, Finlay BB. Bacterial pathogenesis: exploiting cellular adherence. Curr Opin Cell Biol. 2003;15:633–639. doi: 10.1016/s0955-0674(03)00099-1. [DOI] [PubMed] [Google Scholar]
- 32.Torres AG, Vazquez-Juarez RC, Tutt CB, Garcia-Gallegos JG. Pathoadaptive mutation that mediates adherence of shiga toxin-producing Escherichia coli O111. Infect Immun. 2005;73:4766–4776. doi: 10.1128/IAI.73.8.4766-4776.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cossart P, Sansonetti PJ. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 34.Gruenheid S, Finlay BB. Crowd control: quorum sensing in pathogenic E. coli. Trends Microbiol. 2000;8:442–443. doi: 10.1016/s0966-842x(00)01818-7. [DOI] [PubMed] [Google Scholar]
- 35.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 38.Reid G, Younes JA, Van der Mei HC, Gloor GB, Knight R, Busscher HJ. Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat Rev Microbiol. 2011;9:27–38. doi: 10.1038/nrmicro2473. [DOI] [PubMed] [Google Scholar]
- 39.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bron PA, Tomita S, van Swam II, Remus DM, Meijerink M, Wels M, Okada S, Wells JM, Kleerebezem M. Lactobacillus plantarum possesses the capability for wall teichoic acid backbone alditol switching. Microb Cell Fact. 2012;11:123. doi: 10.1186/1475-2859-11-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remus DM, van Kranenburg R, van Swam II, Taverne N, Bongers RS, Wels M, Wells JM, Bron PA, Kleerebezem M. Impact of 4 Lactobacillus plantarum capsular polysaccharide clusters on surface glycan composition and host cell signaling. Microb Cell Fact. 2012;11:149. doi: 10.1186/1475-2859-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol. 2010;8:171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 43.Wells JM, Rossi O, Meijerink M, van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4607–4614. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbara G, Stanghellini V, Brandi G, Cremon C, Di Nardo G, De Giorgio R, Corinaldesi R. Interactions between commensal bacteria and gut sensorimotor function in health and disease. Am J Gastroenterol. 2005;100:2560–2568. doi: 10.1111/j.1572-0241.2005.00230.x. [DOI] [PubMed] [Google Scholar]
- 45.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG, Pettersson S, Conway S. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 46.Dubuquoy L, Jansson EA, Deeb S, Rakotobe S, Karoui M, Colombel JF, Auwerx J, Pettersson S, Desreumaux P. Impaired expression of peroxisome proliferator-activated receptor gamma in ulcerative colitis. Gastroenterology. 2003;124:1265–1276. doi: 10.1016/s0016-5085(03)00271-3. [DOI] [PubMed] [Google Scholar]
- 47.Katayama K, Wada K, Nakajima A, Mizuguchi H, Hayakawa T, Nakagawa S, Kadowaki T, Nagai R, Kamisaki Y, Blumberg RS, et al. A novel PPAR gamma gene therapy to control inflammation associated with inflammatory bowel disease in a murine model. Gastroenterology. 2003;124:1315–1324. doi: 10.1016/s0016-5085(03)00262-2. [DOI] [PubMed] [Google Scholar]
- 48.Neish AS, Gewirtz AT, Zeng H, Young AN, Hobert ME, Karmali V, Rao AS, Madara JL. Prokaryotic regulation of epithelial responses by inhibition of IkappaB-alpha ubiquitination. Science. 2000;289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 49.Tien MT, Girardin SE, Regnault B, Le Bourhis L, Dillies MA, Coppée JY, Bourdet-Sicard R, Sansonetti PJ, Pédron T. Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J Immunol. 2006;176:1228–1237. doi: 10.4049/jimmunol.176.2.1228. [DOI] [PubMed] [Google Scholar]
- 50.Grangette C, Nutten S, Palumbo E, Morath S, Hermann C, Dewulf J, Pot B, Hartung T, Hols P, Mercenier A. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA. 2005;102:10321–10326. doi: 10.1073/pnas.0504084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleerebezem M, Hols P, Bernard E, Rolain T, Zhou M, Siezen RJ, Bron PA. The extracellular biology of the lactobacilli. FEMS Microbiol Rev. 2010;34:199–230. doi: 10.1111/j.1574-6976.2010.00208.x. [DOI] [PubMed] [Google Scholar]
- 52.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc Natl Acad Sci USA. 2009;106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lebeer S, Claes I, Tytgat HL, Verhoeven TL, Marien E, von Ossowski I, Reunanen J, Palva A, Vos WM, Keersmaecker SC, et al. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol. 2012;78:185–193. doi: 10.1128/AEM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan F, Polk DB. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J Biol Chem. 2002;277:50959–50965. doi: 10.1074/jbc.M207050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, Molle D, Lortal S, Altermann E, Klaenhammer TR, et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci USA. 2008;105:19474–19479. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 57.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pott J, Hornef M. Innate immune signalling at the intestinal epithelium in homeostasis and disease. EMBO Rep. 2012;13:684–698. doi: 10.1038/embor.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Macpherson AJ, Geuking MB, Slack E, Hapfelmeier S, McCoy KD. The habitat, double life, citizenship, and forgetfulness of IgA. Immunol Rev. 2012;245:132–146. doi: 10.1111/j.1600-065X.2011.01072.x. [DOI] [PubMed] [Google Scholar]
- 60.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 61.Varol C, Vallon-Eberhard A, Elinav E, Aychek T, Shapira Y, Luche H, Fehling HJ, Hardt WD, Shakhar G, Jung S. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009;31:502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 62.Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nat Rev Immunol. 2008;8:435–446. doi: 10.1038/nri2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, Pabst O. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009;206:3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med. 2012;209:139–155. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niess JH, Adler G. Enteric flora expands gut lamina propria CX3CR1+ dendritic cells supporting inflammatory immune responses under normal and inflammatory conditions. J Immunol. 2010;184:2026–2037. doi: 10.4049/jimmunol.0901936. [DOI] [PubMed] [Google Scholar]
- 66.Rescigno M. Intestinal dendritic cells. Adv Immunol. 2010;107:109–138. doi: 10.1016/B978-0-12-381300-8.00004-6. [DOI] [PubMed] [Google Scholar]
- 67.Pabst O, Bernhardt G. The puzzle of intestinal lamina propria dendritic cells and macrophages. Eur J Immunol. 2010;40:2107–2111. doi: 10.1002/eji.201040557. [DOI] [PubMed] [Google Scholar]
- 68.Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, et al. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Worbs T, Bode U, Yan S, Hoffmann MW, Hintzen G, Bernhardt G, Förster R, Pabst O. Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J Exp Med. 2006;203:519–527. doi: 10.1084/jem.20052016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coombes JL, Siddiqui KR, Arancibia-Cárcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johansson-Lindbom B, Svensson M, Pabst O, Palmqvist C, Marquez G, Förster R, Agace WW. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J Exp Med. 2005;202:1063–1073. doi: 10.1084/jem.20051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guilliams M, Crozat K, Henri S, Tamoutounour S, Grenot P, Devilard E, de Bovis B, Alexopoulou L, Dalod M, Malissen B. Skin-draining lymph nodes contain dermis-derived CD103(-) dendritic cells that constitutively produce retinoic acid and induce Foxp3(+) regulatory T cells. Blood. 2010;115:1958–1968. doi: 10.1182/blood-2009-09-245274. [DOI] [PubMed] [Google Scholar]
- 73.Pino-Lagos K, Guo Y, Brown C, Alexander MP, Elgueta R, Bennett KA, De Vries V, Nowak E, Blomhoff R, Sockanathan S, et al. A retinoic acid-dependent checkpoint in the development of CD4+ T cell-mediated immunity. J Exp Med. 2011;208:1767–1775. doi: 10.1084/jem.20102358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, Flavell RA, Littman DR, Pamer EG. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, Mack M, Shpigel N, Boneca IG, Murphy KM, et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 2012;37:1076–1090. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 76.Cerovic V, Houston SA, Scott CL, Aumeunier A, Yrlid U, Mowat AM, Milling SW. Intestinal CD103(-) dendritic cells migrate in lymph and prime effector T cells. Mucosal Immunol. 2013;6:104–113. doi: 10.1038/mi.2012.53. [DOI] [PubMed] [Google Scholar]
- 77.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 78.Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 79.Bain CC, Mowat AM. Intestinal macrophages - specialised adaptation to a unique environment. Eur J Immunol. 2011;41:2494–2498. doi: 10.1002/eji.201141714. [DOI] [PubMed] [Google Scholar]
- 80.Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, Müller W, Sparwasser T, Förster R, Pabst O. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 81.Denning TL, Norris BA, Medina-Contreras O, Manicassamy S, Geem D, Madan R, Karp CL, Pulendran B. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J Immunol. 2011;187:733–747. doi: 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Medina-Contreras O, Geem D, Laur O, Williams IR, Lira SA, Nusrat A, Parkos CA, Denning TL. CX3CR1 regulates intestinal macrophage homeostasis, bacterial translocation, and colitogenic Th17 responses in mice. J Clin Invest. 2011;121:4787–4795. doi: 10.1172/JCI59150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tamoutounour S, Henri S, Lelouard H, de Bovis B, de Haar C, van der Woude CJ, Woltman AM, Reyal Y, Bonnet D, Sichien D, et al. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol. 2012;42:3150–3166. doi: 10.1002/eji.201242847. [DOI] [PubMed] [Google Scholar]
- 85.De Calisto J, Villablanca EJ, Mora JR. FcγRI (CD64): an identity card for intestinal macrophages. Eur J Immunol. 2012;42:3136–3140. doi: 10.1002/eji.201243061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mann ER, Landy JD, Bernardo D, Peake ST, Hart AL, Al-Hassi HO, Knight SC. Intestinal dendritic cells: their role in intestinal inflammation, manipulation by the gut microbiota and differences between mice and men. Immunol Lett. 2013;150:30–40. doi: 10.1016/j.imlet.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 87.Ohnmacht C, Marques R, Presley L, Sawa S, Lochner M, Eberl G. Intestinal microbiota, evolution of the immune system and the bad reputation of pro-inflammatory immunity. Cell Microbiol. 2011;13:653–659. doi: 10.1111/j.1462-5822.2011.01577.x. [DOI] [PubMed] [Google Scholar]
- 88.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 89.Feng T, Elson CO. Adaptive immunity in the host-microbiota dialog. Mucosal Immunol. 2011;4:15–21. doi: 10.1038/mi.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 91.O’Connor W, Kamanaka M, Booth CJ, Town T, Nakae S, Iwakura Y, Kolls JK, Flavell RA. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol. 2009;10:603–609. doi: 10.1038/ni.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, Bamba T, Fujiyama Y. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weaver CT, Elson CO, Fouser LA, Kolls JK. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol. 2013;8:477–512. doi: 10.1146/annurev-pathol-011110-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lochner M, Bérard M, Sawa S, Hauer S, Gaboriau-Routhiau V, Fernandez TD, Snel J, Bousso P, Cerf-Bensussan N, Eberl G. Restricted microbiota and absence of cognate TCR antigen leads to an unbalanced generation of Th17 cells. J Immunol. 2011;186:1531–1537. doi: 10.4049/jimmunol.1001723. [DOI] [PubMed] [Google Scholar]
- 96.Sawa S, Cherrier M, Lochner M, Satoh-Takayama N, Fehling HJ, Langa F, Di Santo JP, Eberl G. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010;330:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 97.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, Sharma VK, Kim SW, Takahashi M, Saitou N, et al. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell Host Microbe. 2011;10:273–284. doi: 10.1016/j.chom.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 101.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, Monticelli S, Lanzavecchia A, Sallusto F. Pathogen-induced human TH17 cells produce IFN-γ or IL-10 and are regulated by IL-1β. Nature. 2012;484:514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- 103.Shaw MH, Kamada N, Kim YG, Núñez G. Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 105.Nusrat A, Turner JR, Madara JL. Molecular physiology and pathophysiology of tight junctions. IV. Regulation of tight junctions by extracellular stimuli: nutrients, cytokines, and immune cells. Am J Physiol Gastrointest Liver Physiol. 2000;279:G851–G857. doi: 10.1152/ajpgi.2000.279.5.G851. [DOI] [PubMed] [Google Scholar]
- 106.Camilleri M, Gorman H. Intestinal permeability and irritable bowel syndrome. Neurogastroenterol Motil. 2007;19:545–552. doi: 10.1111/j.1365-2982.2007.00925.x. [DOI] [PubMed] [Google Scholar]
- 107.Rao AS, Camilleri M, Eckert DJ, Busciglio I, Burton DD, Ryks M, Wong BS, Lamsam J, Singh R, Zinsmeister AR. Urine sugars for in vivo gut permeability: validation and comparisons in irritable bowel syndrome-diarrhea and controls. Am J Physiol Gastrointest Liver Physiol. 2011;301:G919–G928. doi: 10.1152/ajpgi.00168.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Piche T, Barbara G, Aubert P, Bruley des Varannes S, Dainese R, Nano JL, Cremon C, Stanghellini V, De Giorgio R, Galmiche JP, et al. Impaired intestinal barrier integrity in the colon of patients with irritable bowel syndrome: involvement of soluble mediators. Gut. 2009;58:196–201. doi: 10.1136/gut.2007.140806. [DOI] [PubMed] [Google Scholar]
- 109.Dunlop SP, Hebden J, Campbell E, Naesdal J, Olbe L, Perkins AC, Spiller RC. Abnormal intestinal permeability in subgroups of diarrhea-predominant irritable bowel syndromes. Am J Gastroenterol. 2006;101:1288–1294. doi: 10.1111/j.1572-0241.2006.00672.x. [DOI] [PubMed] [Google Scholar]
- 110.Bertiaux-Vandaële N, Youmba SB, Belmonte L, Lecleire S, Antonietti M, Gourcerol G, Leroi AM, Déchelotte P, Ménard JF, Ducrotté P, et al. The expression and the cellular distribution of the tight junction proteins are altered in irritable bowel syndrome patients with differences according to the disease subtype. Am J Gastroenterol. 2011;106:2165–2173. doi: 10.1038/ajg.2011.257. [DOI] [PubMed] [Google Scholar]
- 111.Lee JW, Park JH, Park DI, Park JH, Kim HJ, Cho YK, Sohn CI, Jeon WK, Kim BI. Subjects with diarrhea-predominant IBS have increased rectal permeability responsive to tryptase. Dig Dis Sci. 2010;55:2922–2928. doi: 10.1007/s10620-009-1094-8. [DOI] [PubMed] [Google Scholar]
- 112.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132:562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yan F, Cao H, Cover TL, Washington MK, Shi Y, Liu L, Chaturvedi R, Peek RM, Wilson KT, Polk DB. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J Clin Invest. 2011;121:2242–2253. doi: 10.1172/JCI44031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kwon HK, Lee CG, So JS, Chae CS, Hwang JS, Sahoo A, Nam JH, Rhee JH, Hwang KC, Im SH. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci USA. 2010;107:2159–2164. doi: 10.1073/pnas.0904055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kaji R, Kiyoshima-Shibata J, Nagaoka M, Nanno M, Shida K. Bacterial teichoic acids reverse predominant IL-12 production induced by certain lactobacillus strains into predominant IL-10 production via TLR2-dependent ERK activation in macrophages. J Immunol. 2010;184:3505–3513. doi: 10.4049/jimmunol.0901569. [DOI] [PubMed] [Google Scholar]
- 116.Karczewski J, Troost FJ, Konings I, Dekker J, Kleerebezem M, Brummer RJ, Wells JM. Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am J Physiol Gastrointest Liver Physiol. 2010;298:G851–G859. doi: 10.1152/ajpgi.00327.2009. [DOI] [PubMed] [Google Scholar]
- 117.van Baarlen P, Troost F, van der Meer C, Hooiveld G, Boekschoten M, Brummer RJ, Kleerebezem M. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4562–4569. doi: 10.1073/pnas.1000079107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Caricilli AM, Picardi PK, de Abreu LL, Ueno M, Prada PO, Ropelle ER, Hirabara SM, Castoldi Â, Vieira P, Camara NO, et al. Gut microbiota is a key modulator of insulin resistance in TLR 2 knockout mice. PLoS Biol. 2011;9:e1001212. doi: 10.1371/journal.pbio.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120.Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, Geurts L, Naslain D, Neyrinck A, Lambert DM, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Carvalho BM, Guadagnini D, Tsukumo DM, Schenka AA, Latuf-Filho P, Vassallo J, Dias JC, Kubota LT, Carvalheira JB, Saad MJ. Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia. 2012;55:2823–2834. doi: 10.1007/s00125-012-2648-4. [DOI] [PubMed] [Google Scholar]
- 123.Son G, Kremer M, Hines IN. Contribution of gut bacteria to liver pathobiology. Gastroenterol Res Pract. 2010;2010 doi: 10.1155/2010/453563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dongarrà ML, Rizzello V, Muccio L, Fries W, Cascio A, Bonaccorsi I, Ferlazzo G. Mucosal immunology and probiotics. Curr Allergy Asthma Rep. 2013;13:19–26. doi: 10.1007/s11882-012-0313-0. [DOI] [PubMed] [Google Scholar]
- 125.Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr. 2011;141:769–776. doi: 10.3945/jn.110.135657. [DOI] [PubMed] [Google Scholar]
- 126.Klaenhammer TR, Kleerebezem M, Kopp MV, Rescigno M. The impact of probiotics and prebiotics on the immune system. Nat Rev Immunol. 2012;12:728–734. doi: 10.1038/nri3312. [DOI] [PubMed] [Google Scholar]
- 127.Jirillo E, Jirillo F, Magrone T. Healthy effects exerted by prebiotics, probiotics, and symbiotics with special reference to their impact on the immune system. Int J Vitam Nutr Res. 2012;82:200–208. doi: 10.1024/0300-9831/a000112. [DOI] [PubMed] [Google Scholar]
- 128.Matsuzaki T, Nagata Y, Kado S, Uchida K, Kato I, Hashimoto S, Yokokura T. Prevention of onset in an insulin-dependent diabetes mellitus model, NOD mice, by oral feeding of Lactobacillus casei. APMIS. 1997;105:643–649. doi: 10.1111/j.1699-0463.1997.tb05066.x. [DOI] [PubMed] [Google Scholar]
- 129.Di Giacinto C, Marinaro M, Sanchez M, Strober W, Boirivant M. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-beta-bearing regulatory cells. J Immunol. 2005;174:3237–3246. doi: 10.4049/jimmunol.174.6.3237. [DOI] [PubMed] [Google Scholar]
- 130.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lyons A, O’Mahony D, O’Brien F, MacSharry J, Sheil B, Ceddia M, Russell WM, Forsythe P, Bienenstock J, Kiely B, et al. Bacterial strain-specific induction of Foxp3+ T regulatory cells is protective in murine allergy models. Clin Exp Allergy. 2010;40:811–819. doi: 10.1111/j.1365-2222.2009.03437.x. [DOI] [PubMed] [Google Scholar]
- 132.Lavasani S, Dzhambazov B, Nouri M, Fåk F, Buske S, Molin G, Thorlacius H, Alenfall J, Jeppsson B, Weström B. A novel probiotic mixture exerts a therapeutic effect on experimental autoimmune encephalomyelitis mediated by IL-10 producing regulatory T cells. PLoS One. 2010;5:e9009. doi: 10.1371/journal.pone.0009009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gad M, Ravn P, Søborg DA, Lund-Jensen K, Ouwehand AC, Jensen SS. Regulation of the IL-10/IL-12 axis in human dendritic cells with probiotic bacteria. FEMS Immunol Med Microbiol. 2011;63:93–107. doi: 10.1111/j.1574-695X.2011.00835.x. [DOI] [PubMed] [Google Scholar]
- 134.Resta-Lenert S, Barrett KE. Probiotics and commensals reverse TNF-alpha- and IFN-gamma-induced dysfunction in human intestinal epithelial cells. Gastroenterology. 2006;130:731–746. doi: 10.1053/j.gastro.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 135.van Baarlen P, Troost FJ, van Hemert S, van der Meer C, de Vos WM, de Groot PJ, Hooiveld GJ, Brummer RJ, Kleerebezem M. Differential NF-kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc Natl Acad Sci USA. 2009;106:2371–2376. doi: 10.1073/pnas.0809919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bermudez-Brito M, Muñoz-Quezada S, Gomez-Llorente C, Matencio E, Bernal MJ, Romero F, Gil A. Human intestinal dendritic cells decrease cytokine release against Salmonella infection in the presence of Lactobacillus paracasei upon TLR activation. PLoS One. 2012;7:e43197. doi: 10.1371/journal.pone.0043197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tao Y, Drabik KA, Waypa TS, Musch MW, Alverdy JC, Schneewind O, Chang EB, Petrof EO. Soluble factors from Lactobacillus GG activate MAPKs and induce cytoprotective heat shock proteins in intestinal epithelial cells. Am J Physiol Cell Physiol. 2006;290:C1018–C1030. doi: 10.1152/ajpcell.00131.2005. [DOI] [PubMed] [Google Scholar]
- 138.Mathias A, Duc M, Favre L, Benyacoub J, Blum S, Corthésy B. Potentiation of polarized intestinal Caco-2 cell responsiveness to probiotics complexed with secretory IgA. J Biol Chem. 2010;285:33906–33913. doi: 10.1074/jbc.M110.135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gallo RL, Nakatsuji T. Microbial symbiosis with the innate immune defense system of the skin. J Invest Dermatol. 2011;131:1974–1980. doi: 10.1038/jid.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, Murray PR, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, Wu ZR, Hooper LV, Schmidt RR, von Aulock S, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dickson RP, Erb-Downward JR, Huffnagle GB. The role of the bacterial microbiome in lung disease. Expert Rev Respir Med. 2013;7:245–257. doi: 10.1586/ers.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Su GL, Klein RD, Aminlari A, Zhang HY, Steinstraesser L, Alarcon WH, Remick DG, Wang SC. Kupffer cell activation by lipopolysaccharide in rats: role for lipopolysaccharide binding protein and toll-like receptor 4. Hepatology. 2000;31:932–936. doi: 10.1053/he.2000.5634. [DOI] [PubMed] [Google Scholar]
- 148.French SW. Mechanisms of alcoholic liver injury. Can J Gastroenterol. 2000;14:327–332. doi: 10.1155/2000/801735. [DOI] [PubMed] [Google Scholar]
- 149.Gómez-Hurtado I, Santacruz A, Peiró G, Zapater P, Gutiérrez A, Pérez-Mateo M, Sanz Y, Francés R. Gut microbiota dysbiosis is associated with inflammation and bacterial translocation in mice with CCl4-induced fibrosis. PLoS One. 2011;6:e23037. doi: 10.1371/journal.pone.0023037. [DOI] [PMC free article] [PubMed] [Google Scholar]


