An analysis of the half-lives of 12 of the 22 natural aminoacyl-tRNAs used in peptide bond formation shows that the stability of the acyl linkage is determined only by the chemical nature of the amino acid side chain: Proline confers the lowest stability with isoleucine and valine confers the highest to the linkage. Among the variables tested, the protein translation factor EF-Tu is the only one that can protect a weak acyl linkage from hydrolysis.
Keywords: aminoacylation of tRNA, stability of aminoacyl-tRNA, mispaired aminoacyl-tRNA, prolyl-tRNA
Abstract
Aminoacyl-tRNAs are the biologically active substrates for peptide bond formation in protein synthesis. The stability of the acyl linkage in each aminoacyl-tRNA, formed through an ester bond that connects the amino acid carboxyl group with the tRNA terminal 3′-OH group, is thus important. While the ester linkage is the same for all aminoacyl-tRNAs, the stability of each is not well characterized, thus limiting insight into the fundamental process of peptide bond formation. Here, we show, by analysis of the half-lives of 12 of the 22 natural aminoacyl-tRNAs used in peptide bond formation, that the stability of the acyl linkage is effectively determined only by the chemical nature of the amino acid side chain. Even the chirality of the side chain exhibits little influence. Proline confers the lowest stability to the linkage, while isoleucine and valine confer the highest, whereas the nucleotide sequence in the tRNA provides negligible contribution to the stability. We find that, among the variables tested, the protein translation factor EF-Tu is the only one that can protect a weak acyl linkage from hydrolysis. These results suggest that each amino acid plays an active role in determining its own stability in the acyl linkage to tRNA, but that EF-Tu overrides this individuality and protects the acyl linkage stability for protein synthesis on the ribosome.
INTRODUCTION
The synthesis of peptide bonds is central to cellular life. Up to 22 amino acids are used in peptide bond formation, including the 20 canonical ones, as well as selenocysteine (Bock et al. 1991) and pyrrolysine (Hao et al. 2004). Each of these amino acids by itself is not a substrate for peptide bond formation but instead must be transformed into an aminoacyl-tRNA (aa-tRNA). Although aminoacylation can occur on the 2′- or 3′-OH group of the tRNA terminal ribose, depending on the aminoacyl-tRNA synthetase (aaRS) that catalyzes the reaction (Arnez and Moras 1997), rapid trans-esterification between the two groups leads to aminoacyl esterification to the 3′-OH group (Fig. 1A). The aminoacylation reaction proceeds via the formation of an aminoacyl-adenylate intermediate, using ATP as the energy source. In ribosome-dependent peptide bond formation, the aa-tRNA form provides the basis to physically relate the amino acid to the anticodon of the tRNA and to introduce the amino acid to the ribosome at a codon position matching the tRNA anticodon. In the ribosome-independent peptide bond formation on the N-terminal residue of specific acceptor proteins, catalyzed by aminoacyl-tRNA-protein transferases (e.g., L/F transferase, Arg-tRNA-protein transferase), the aa-tRNA form provides the donor amino acid to mark the conjugated protein products for degradation (Leibowitz and Soffer 1971; Balzi et al. 1990). In the synthesis of cyclodipeptides by cyclodipeptide synthases, two aa-tRNA species are sequentially recognized and their aminoacyl moieties linked and cyclized (Gondry et al. 2009). In other cellular biosynthesis pathways, such as the synthesis of dehydrophos (Bougioukou et al. 2013), the aa-tRNA form is the source of the activated amino acid (Ibba and Soll 2004). However, while the stability of the aa-tRNA form is crucial to biology, the ester linkage is susceptible to spontaneous hydrolysis at the physiological pH, primarily due to its proximity to the 2′-OH of the terminal ribose (Bruice et al. 1962; Hentzen et al. 1972). If hydrolysis does occur, the aa-tRNA and the energy of its synthesis would be wasted, requiring the aminoacylation process to be repeated. Thus, the stability of aa-tRNAs toward hydrolysis is an essential factor that determines the speed and efficiency of peptide bond formation.
FIGURE 1.
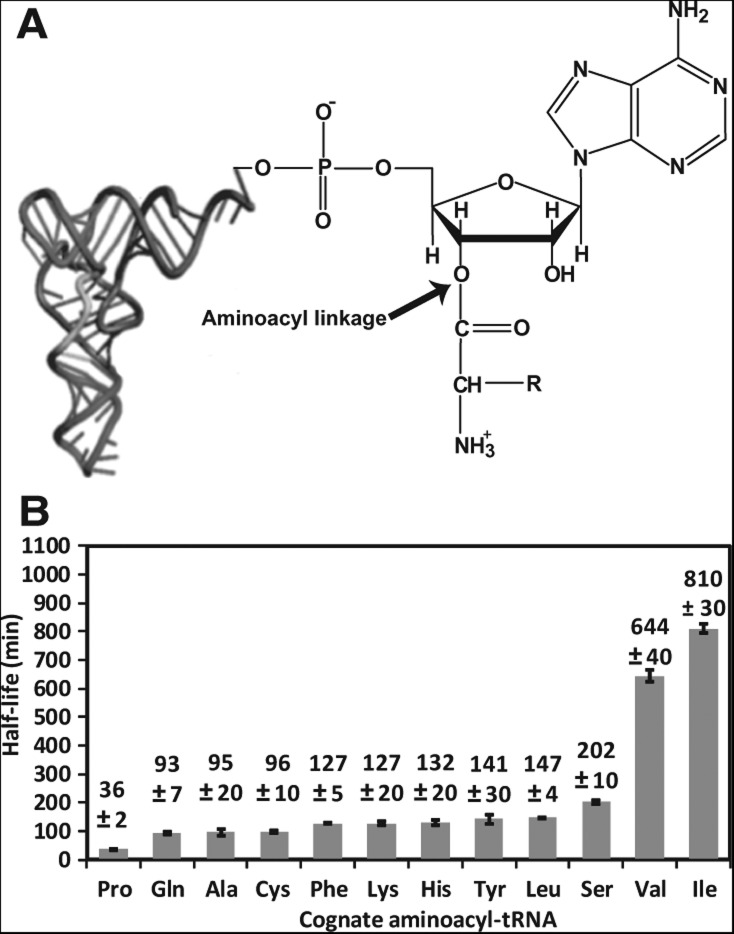
The acyl linkage in aa-tRNA. (A) The chemical structure of the acyl linkage to the 3′-OH of the terminal ribose of A76 in tRNA, with an arrow indicating the location of the acyl linkage. The size of the tRNA L-shape is not drawn to proportion. (B) A summary of T1/2 values of cognate pairs of aa-tRNAs determined in this study, showing the average of each by at least three independent measurements. Error bars are standard deviations. The concentration of aa-tRNA in each reaction was as follows: Pro-tRNAPro (0.18 μM), Gln-tRNAGln (1.51 μM), Ala-tRNAAla (0.85 μM), Cys-tRNACys (0.42 μM), Phe-tRNAPhe (0.29 μM), Lys-tRNALys (1.36 μM), His-tRNAHis (0.67 μM), Tyr-tRNATyr (0.72 μM), Leu-tRNALeu (0.27 μM), Ser-tRNASer (1.98 μM), Val-tRNAVal (0.90 μM), and Ile-tRNAIle (0.51 μM).
Surprisingly, while the acyl linkage is locally identical for all aa-tRNAs, earlier work showed that the half-life of the acyl linkage varied from a few minutes to hours for different cognate pairs of amino acid and tRNA (Hentzen et al. 1972; Schuber and Pinck 1974a,b). This finding was unexpected, suggesting that the determinant for the stability of the acyl linkage was not as simple as the chemical nature of the ester bond. However, some earlier work was performed with unusual components (e.g., 0.5 mM ZnSO4) (Matthaei et al. 1966) or at low pH values (e.g., 4.7) (Chousterman et al. 1966). In one of the most thorough early analyses, by Hentzen et al. (1972), measurements of the acyl linkage stability were performed at pH 7.0 but in buffers with nonphysiological salt concentrations (e.g., 1.5 M potassium phosphate). These unusual conditions make it difficult to evaluate the stability of the acyl linkage in a biological context. Most importantly, even with the determination of the relative stability of the acyl linkage of a series of aa-tRNAs, the earlier work gave no insight into what drives the stability of each linkage, thus leaving open the important question of how and why the stability varies.
To address what determines the stability of each acyl linkage, we considered a number of factors, such as the identity of the amino acid, the sequence of the tRNA near the acceptor end, and the modification state of the tRNA. None of these factors had been rigorously evaluated. Importantly, with the recent development of techniques to generate tRNA molecules with mispaired aa-tRNAs (Murakami et al. 2006), with natural and site-specifically mutated tRNA sequences, and with unmodified transcripts (Sampson and Uhlenbeck 1988), we are in a position to clearly define the determinants of the stability of acyl linkage. We show here, by analysis of 12 of the 22 protein amino acids, that the identity of the amino acid makes the single largest contribution to the stability. Specifically, proline confers the least stability, whereas isoleucine and valine confer the highest, while the tRNA nucleotide sequence near the acceptor end makes little contribution to the stability. The finding that the prolyl linkage to tRNA is the least stable relative to others has implications for translation of proline codons on the ribosome.
RESULTS AND DISCUSSION
Stability of the acyl-linkage in cognate aa-tRNA pairs
While the acyl linkage is the same among cognate pairs of aa-tRNAs, the amino acid side chain differs and the tRNA acceptor end sequence varies. To determine whether the stability of the acyl linkage is the same for all aa-tRNA pairs or varies with the amino acid side chain or with the acceptor end sequence, we prepared 12 pairs of Escherichia coli cognate aa-tRNAs in vitro and determined the half-life of each acyl linkage at pH 7.5 in an aqueous solution mimicking the physiological buffer. These 12 pairs included amino acids with aliphatic, aromatic, basic, amide, and sulfur-containing side chains. To minimize the effect of post-transcriptional modifications and to focus on tRNA primary sequences, we produced each tRNA by template-dependent transcription in vitro. To provide more consistent aminoacylation efficiency, we used the ribozyme dFx to catalyze tRNA aminoacylation with chemically synthesized activated aa-DBE (3,5-dinitrobenzyl ester) derivatives (Falorni et al. 2000; Murakami et al. 2006). The dFx ribozyme is a 46-mer catalytic RNA that attaches an activated amino acid to the 3′-OH of the tRNA terminal ribose (Murakami et al. 2006; Xiao et al. 2008). This ribozyme uses its terminal 5′-GGU-3′ sequence to base-pair with the 5′-ACC-3′ sequence in tRNA from positions 73 to 75, which is particularly appropriate for sequences with A73 as the discriminator base. This simple base-pairing requirement allows dFx to aminoacylate virtually all tRNA species. The level of aminoacylation by dFx was generally twofold lower than those reported previously (Supplemental Fig. S1; Murakami et al. 2006).
To provide a sensitive assay for the stability of the acyl linkage, we labeled each tRNA with 32P at the terminal A76 via the exchange reaction of the CCA-adding enzyme (Shitivelband and Hou 2005), converted the labeled tRNA to aa-32P-tRNA by dFx, and monitored the decay of aa-32P-tRNA into 32P-tRNA at 20°C. The hydrolyzed tRNA product was distinguished from the substrate by digestion with S1 nuclease, generating 32P-AMP and aa-32P-AMP, respectively, which were resolved by TLC. The fraction of aa-32P-AMP radioactivity that remained at each time point relative to the total radioactivity [aa-32P-AMP + 32P-AMP] was then calculated to determine the extent of the acylated state vs. the deacylated state. The advantage of the assay was that the fraction was independent of the input radioactivity. In contrast, the earlier assay of Hentzen et al. used 14C-amino acid to generate 14C-aminoacyl-tRNA (Hentzen et al. 1972), which was then quantified as acid precipitable counts on filter pads. Because the fraction of the acylated state must be measured by a scintillation counter and calculated relative to the total radioactivity, the sensitivity of the earlier assay was highly dependent on the input counts. An example of our 32P-based assay is shown for Gln-tRNAGln (Supplemental Fig. S2), showing the course of deacylation over time. The data were fit to a pseudo-first-order exponential decay equation to determine the half-life T1/2.
We showed that the T1/2 values of the 12 pairs of aa-tRNAs differed significantly over a range of more than 22-fold, from 36 ± 2 min for Pro-tRNAPro to 640 ± 40 min for Val-tRNAVal and to 810 ± 30 min for Ile-tRNAIle (Fig. 1B). Interestingly, while our T1/2 data differed in absolute values from those reported previously (Hentzen et al. 1972), the relative acyl-linkage stability among these cognate pairs was in good agreement (Supplemental Fig. S3). For example, Hentzen et al. reported that the half-life of Pro-tRNAPro was relatively short (T1/2 = 44 ± 6 min), while that of Val-tRNAVal (T1/2 = 690 ± 40 min) and of Ile-tRNAIle (T1/2 = 950 ± 80 min) was long. Also, we showed that the T1/2 values of Phe-tRNAPhe, Lys-tRNALys, and Tyr-tRNATyr were similar (127 ± 5, 127 ± 20, and 141 ± 30 min) (Fig. 1B), consistent with the finding of Hentzen et al. (93 ± 8, 90 ± 6, and 98 ± 8 min, respectively). Thus, despite major differences in buffer compositions and in assay methodologies, the trend of relative stability among aa-tRNA pairs is similar. This result indicates that the buffer composition does not determine the relative stability of aa-tRNAs.
The lack of a buffer effect was further confirmed by analysis of the stability of Tyr-tRNATyr in different buffer conditions (Supplemental Fig. S4). Indeed, we showed that the addition of each individual component (150 mM KCl, 3.5 mM MgCl2, or 0.5 mM spermidine) to the basic buffer (100 mM HEPES-KOH, pH 7.5) had no more than a 1.5-fold effect on the T1/2 value relative to the buffer that contained all of the components. This result further indicates that the ionic strength of the buffer, which would vary with the addition of each component, did not play a major role in determining the stability of the acyl linkage.
Analysis of mispaired aa-tRNA stabilities
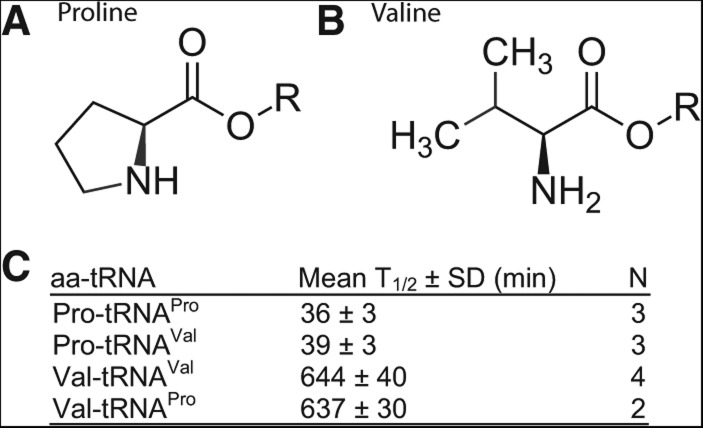
To understand the observed differences among the half-lives of the cognate aa-tRNAs, we considered the chemical structure of the amino acid side chains. In particular, the large differences in the hydrolytic stability between Pro-tRNAPro (least stable) and Val-tRNAVal and Ile-tRNAIle (most stable) may be due to inductive and/or steric effects. Specifically, the proline side chain possesses a cyclic secondary amine moiety (Fig. 2A), which has an appreciably higher pKa than primary congeners (α amine pKa for Pro, Val, Ile = 10.60, 9.72, 9.76, respectively) (Jenks and Regenstein 1976). Lower contribution of the ionized ammonium form under the experimental pH would explain the significantly higher rate of hydrolysis for the proline ester (Wolfenden 1963; Vig et al. 2003). Additionally, both the valine and isoleucine side chains possess secondary substitution at the β carbon (Fig. 2B) and are, therefore, more sterically inhibited toward hydrolytic attack of the ester carbonyl relative to other amino acid side chains. Indeed, among the three aliphatic side chains (Leu, Val, and Ile), while leucine differs from valine by having a γ carbon side-chain, isoleucine differs from valine by having a bulkier branched β carbon side-chain. We showed that the stability of the acyl linkage is Leu-tRNALeu < Val-tRNAVal < Ile-tRNAIle, in an order consistent with the steric effect of the amino acid side chain at the β carbon position. This relative stability was also observed previously (Hentzen et al. 1972), showing T1/2 = 73 ± 7 min for Leu-tRNALeu, 690 ± 40 min for Val-tRNAVal, and 950 ± 80 min for Ile-tRNAIle. Thus, further computational studies of the steric effect at the β carbon position for natural and unnatural amino acids should be beneficial to establish a framework that can be used to predict and test the acyl linkage stability of a wide variety of aa-tRNAs.
FIGURE 2.
Determinants of acyl stability. (A) Structure of a proline side chain with the cyclic secondary amine structure. (B) Structure of a valine side chain with a branched β carbon group. (C) T1/2 values of noncognate pairs of aa-tRNAs compared to the cognate pairs. Error bars are standard deviations. (N) Number of independent measurements. The concentration of aa-tRNA in each reaction was as follows: Val-tRNAVal (0.90 µM), Pro-tRNAPro (0.18 µM), Pro-tRNAVal (0.75 µM), and Val-tRNAPro (0.45 µM).
As an example to test the correlation of the β-side chain steric effect with the stability of the acyl linkage, we focused on proline and valine and used dFx to introduce proline to tRNAVal and valine to tRNAPro (Fig. 2C), such that the most and the least stable linkage was examined in the context of noncognate aa-tRNAs. The results showed that the half-life of Pro-tRNAVal was identical to that of Pro-tRNAPro and that the half-life of Val-tRNAPro was identical to that of Val-tRNAVal. Thus, the attachment of proline to tRNAVal and the attachment of valine to tRNAPro each drove the acyl stability in a different direction. The virtually complete recapitulation of half-lives according to the nature of the amino acid emphasizes that the side chain is the single most important determinant of acyl stability and that the sequence of tRNA plays no major role.
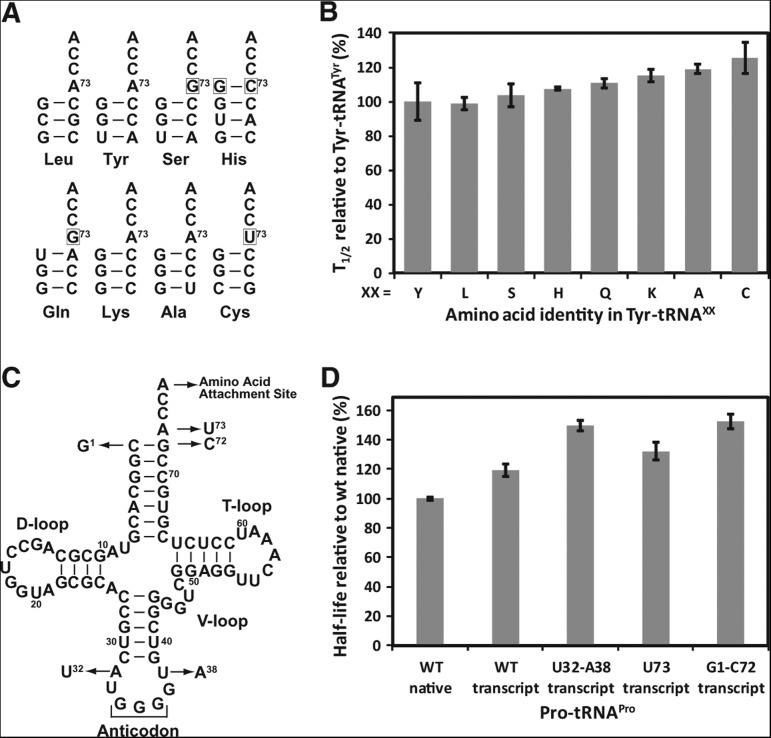
To confirm that the tRNA sequence has no major role, we used Tyr-tRNATyr as a reference and placed Tyr onto eight tRNA species with different sequences near the acceptor end (tRNALeu, tRNATyr, tRNASer, tRNAHis, tRNAGln, tRNALys, tRNAAla, and tRNACys) (Fig. 3A). These tRNA species differ at position 73 and at the first base pair 1-72, both of which have the ability to influence the flexibility of the CCA end (Lee et al. 1993; Hou et al. 1998) to which amino acid is attached. For example, tRNATyr, tRNALys, tRNALeu, and tRNAAla share in common A73 and a G1-C72 pair, whereas tRNASer and tRNAGln share G73 but contain G1-C72 and U1-A72, respectively. The U73 nucleotide, with the ability to confer flexibility to the CCA end, is used by tRNACys, whereas an extra G−1 base is present in tRNAHis to form a base pair with C73. Despite these variations, we showed that the half-lives of different pairs of Tyr-tRNAxx were closely similar within 1.6-fold of each other (Fig. 3B), supporting the notion that the tRNA sequence near the acceptor end plays little role in determining the acyl stability.
FIGURE 3.
No major contribution of tRNA to acyl stability. (A) Analysis of T1/2 in Tyr-tRNAXX, where the amino acid Tyr is charged to tRNAXX by dFx. Each tRNAXX is shown only for the first four base pairs of the acceptor stem. (B) Values of T1/2 of Tyr-tRNAXX. Each tRNA was tested as a transcript. The concentration of aa-tRNAXX in each reaction was as follows: Tyr-tRNALeu (0.20 µM), Tyr-tRNASer (0.70 µM), Tyr-tRNAHis (0.45 µM), Tyr-tRNAGln (0.60 µM), Tyr-tRNALys (0.67 µM), Tyr-tRNAAla (0.60 µM), and Tyr-tRNACys (0.58 µM). (C) The location and nature of the mutations introduced to the sequence of E. coli tRNAPro/GGG as shown by arrows. (D) Values of T1/2 of Pro-tRNAPro/GGG for WT and variants of the tRNA. (WT) Wild-type sequence of the tRNA, (t) transcript of the tRNA, without modifications, (n) native form of the tRNA purified from E. coli, with all natural modifications. Values are the average of at least two independent measurements. Error bars are standard deviations. The concentration of Pro-tRNAPro in each reaction was as follows: WT transcript (0.18 µM), WT native (1.48 µM), U32-A38 transcript (0.37 µM), U73 transcript (0.99 µM), and G1-C72 transcript (0.28 µM).
To further examine the role of tRNA sequences, we analyzed the effect of point substitutions in tRNAPro. Using the GGG isoacceptor of E. coli tRNAPro as an example (Fig. 3C), we showed that the tRNA in the native state isolated from cells and in the transcript state prepared in vitro exhibited no major difference in the acyl stability (Fig. 3D), indicating that the natural base and backbone modifications present in the native state had no effect. In the transcript state, we performed a more global analysis to include substitutions both at the acceptor end and at the distal anticodon end, including the A73U substitution at the discriminator position, the exchange of the first base pair from C1-G72 to G1-C72, and the replacement of A32-U38 in the anticodon loop with U32-A38. None of these sequence replacements had a major effect on the acyl stability (Fig. 3D).
Analysis of amino acid enantiomerism and EF-Tu effects on stabilities
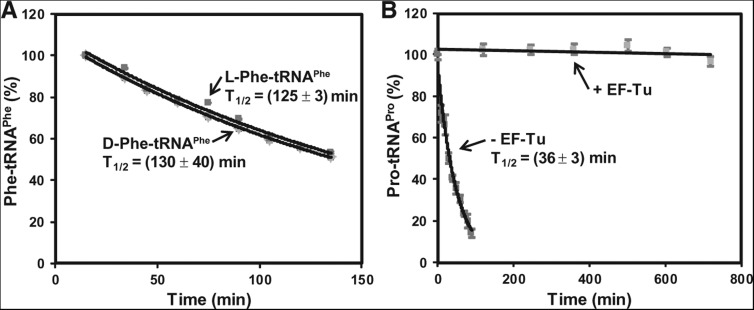
Given that the chemical identity of the amino acid side chain plays the dominant role in the stability of the acyl linkage, we then determined whether the chirality of the side chain has a role. Both D- and L-forms of amino acids can be charged onto the cognate tRNA by natural aaRS enzymes, although only the L-enantiomer is used for peptide bond formation. High concentrations of D-Tyr cause cellular toxicity, in part due to accumulation of D-Tyr-tRNATyr, which limits the pool of tRNATyr available for synthesis of L-Tyr-tRNATyr for the ribosome. This toxicity is ameliorated if cells maintain an active deacylase to remove D-Tyr-tRNATyr, or express tRNATyr to high levels to increase the supply of the L-enantiomer (Soutourina et al. 2004). These observations implied that the D-form of Tyr-tRNATyr is stable enough to exhaust the available pool of tRNATyr and to challenge the synthesis of the L-form. We tested this hypothesis, using the D- and L-forms of Phe-tRNAPhe as an example. Indeed, the two forms showed parallel decay over time with a similar T1/2 value (130 ± 40 min and 125 ± 3 min, respectively) (Fig. 4A), in agreement with an earlier analysis of Tyr-tRNATyr (Calendar and Berg 1967). Even in the study of N-acetyl-Phe on AMP as a model for the 3′-terminal adenosine of tRNA, the D- and L-forms differed in stability by only a small effect (Wickramasinghe and Lacey 1993).
FIGURE 4.
Effects of enantiomerism and EF-Tu on acyl linkage stability. (A) Analysis of the hydrolytic decay of the D- and L-form of Phe-tRNAPhe over time. The calculated T1/2 for each is indicated. The concentration of Phe-tRNAPhe was as follows: L-Phe-tRNAPhe (0.29 µM), D-Phe-tRNAPhe (1.5 µM). (B) Analysis of the hydrolytic decay of Pro-tRNAPro (0.2 µM) over time in the presence and absence of EF-Tu (9 µM). The calculated T1/2 in the absence of EF-Tu is shown. The concentration of Pro-tRNAPro in each reaction was as follows: with EF-Tu (0.3 µM) and without EF-Tu (0.18 µM).
For Pro-tRNAPro, we determined if EF-Tu stabilizes the acyl linkage. EF-Tu is a GTP-dependent bacterial elongation factor (with a homolog EF-1α in eukaryotes), which recognizes all canonical aa-tRNAs and escorts each to the matching codon position on the ribosome and, upon GTP hydrolysis, provides the aa-tRNA as the substrate for protein synthesis. The equivalent protein factor for selenocysteine (Sec)-tRNASec in bacteria is SelB, which possesses similar properties as EF-Tu (Paleskava et al. 2010). In its crystal structures in complex with a cognate aa-tRNA (Nissen et al. 1995, 1999), EF-Tu recognizes the acyl linkage using a conserved β-barrel motif that has the ability to adapt to variations in the amino acid side chain. This recognition mechanism suggests the possibility to protect the acyl linkage of all aa-tRNAs from hydrolysis. Indeed, we showed that the presence of a 30-fold molar excess of EF-Tu-GTP stabilized Pro-tRNAPro up to 800 min (Fig. 4B), indicating protection of the acyl linkage (T1/2 = 36 ± 2 min) by more than 20-fold, to a level similar to that of Ile-tRNAIle (Fig. 1B). Based on high cellular concentrations of EF-Tu (100–200 μM) (Burnett et al. 2013) and the entire population of aa-tRNAs (50–200 μM) (Dong et al. 1996), the protection of EF-Tu-GTP at a high molar ratio to a single Pro-tRNAPro species is physiologically relevant. This protection is consistent with an earlier observation of EF-Tu-GTP protecting the acyl stability of Phe-tRNAPhe by more than 10-fold (from T1/2 of 56 to 800 min) (Beres and Lucas-Lenard 1973).
To determine the significance of the EF-Tu protection of prolyl linkage, we analyzed the stability of the ternary complex EF-Tu-GTP-Pro-tRNAPro. Using an RNase A assay (LaRiviere et al. 2001), in which Pro-tRNAPro released from the complex would be cleaved by the nuclease, we determined the koff of the release as (1.57 ± 0.01) × 10−2 s−1 (Supplemental Fig. S5), ∼10-fold faster compared to the koff of yeast Phe-tRNAPhe [(1.0 ± 0.1) × 10−3 s−1] measured by the same method (data not shown), the latter of which was in agreement with data of others (Schrader et al. 2009). Because the on-rate of Pro-tRNAPro and Phe-tRNAPhe to EF-Tu-GTP is similar between the two (Louie and Jurnak 1985), the off-rate drives the difference in affinity in the ternary complex. Thus, a 10-fold weaker affinity of Pro-tRNAPro in the ternary complex relative to Phe-tRNAPhe would sensitize the former to rapid hydrolysis.
CONCLUSIONS
We show here that, under physiological buffer conditions, the stability of the acyl linkage of cognate pairs of aa-tRNAs differs markedly, with Pro-tRNAPro being most sensitive to hydrolysis and Val-tRNAVal and Ile-tRNAIle being most resistant. While a similar trend was observed in nonphysiological buffer conditions, our study is significant, revealing that it is the amino acid itself that determines the stability of each aa-tRNA. Neither the sequence nor the post-transcriptional modification state of the tRNA contributes to the stability. The finding that only the chemical structure, not even the chirality, of an amino acid is the major determinant of stability emphasizes an active role of the structure in controlling the half-life of the amino acid on the tRNA. Thus, in the aa-tRNA form, a division of labor is evident between the two moieties of the molecule: While the tRNA moiety determines where and when the amino acid is used for protein synthesis on the ribosome, the amino acid moiety, in turn, determines the stability of the tRNA to perform this function. In broader perspectives, this finding provides fundamental insight into the evolution of amino acids and the development of aa-tRNAs for cellular activities. While the great majority of aa-tRNAs are utilized for protein synthesis on the ribosome, a fraction (e.g., Leu-tRNALeu, Arg-tRNAArg, Tyr-tRNATyr, and Phe-tRNAPhe) is also diverted to biosynthesis of metabolites (Ibba and Soll 2004). However, Pro-tRNAPro is so far known only for utilization on the ribosome, perhaps due to the need for protection of its relatively unstable acyl linkage by the large and elaborate protein synthesis machinery. Indeed, Pro-tRNAPro is less favorable relative to other aa-tRNAs in many aspects of peptide bond formation: Upon synthesis it is released from the charging enzyme prolyl-tRNA synthetase directly into solution rather than being channeled to EF-Tu-GTP (Zhang et al. 2006); in solution it has a weaker affinity to bind to EF-Tu-GTP (Supplemental Fig. S5); and on the ribosome it performs peptidyl transfer at a rate slower by five- to 10-fold than others (Pavlov et al. 2009). Thus, the finding that the prolyl linkage, once bound to EF-Tu-GTP, is protected from hydrolysis is significant, indicating that the linkage is stabilized and is enabled to participate in peptide bond formation on ribosome-mRNA complexes. Because EF-Tu is a dedicated factor to the ribosome machinery, this provides an explanation for why Pro-tRNAPro is solely used by the ribosome. It is, therefore, reasonable to expect that, once protein synthesis is initiated on an mRNA sequence, the overall rate of synthesis will depend on the translation of proline codons due to the less reactive nature of Pro-tRNAPro, which, in turn, depends on the stability of the prolyl linkage.
MATERIALS AND METHODS
Substrate tRNAs
Most of the tRNA substrates were prepared by in vitro transcription, using T7 RNA polymerase to transcribe synthetic DNA templates (Hou et al. 1993). In some cases, native tRNA species were prepared by isolation from E. coli cells that overexpressed the tRNA species (Liu et al. 2011).
Preparation of aa-tRNA
Aminoacylation of tRNA was performed using the ribozyme dFx with most aminoacyl-DBE species or using the ribozyme eFx for Phe-DBE (Murakami et al. 2006). The ribozyme charging reaction was performed with radiolabeled tRNA (nominally 0.5 μM), 5 mM aa-DBE, and 18.75 μM ribozyme in 90 mM HEPES-KOH, pH 7.5, 90 mM KCl, and 0.6 M MgCl2 on ice and incubated from 30 min to 12 h, depending on the aa-DBE. The yield of aminoacylation of tRNA was generally ∼10% (Supplemental Fig. S1). The synthesis of the aa-DBEs is described in detail in Supplemental Material. For Pro-tRNAPro, the synthesis of aa-tRNA was also prepared by using the cognate ProRS with similar yields.
Assay for the stability of the acyl linkage
The tRNA substrate in an aa-tRNA was labeled with α-32P-ATP at the A76 nucleotide by the CCA-adding enzyme of Bacillus stearothermophilus (Shitivelband and Hou 2005). The labeled tRNA (0.2–1.5 μM) was then acylated by dFx or eFx to generate the aa-tRNA, which was ethanol-precipitated and used directly for the stability analysis. Analysis of the stability of the acyl linkage was performed at 20°C in a physiological buffer of 100 mM HEPES-KOH, pH 7.5, 150 mM KCl, 3.5 mM MgCl2, 1.0 mM DTT, and 0.5 mM spermidine. Aliquots of 1 μL were taken over time for 1–2 half-lives, quenched in 3 μL of 50 mM NaCl and 200 mM NaOAc, pH 5.0, and stored at −20°C until final processing. The tRNA was digested to mononucleotides by incubation with 4 units of S1 nuclease in the presence of 0.2 mM ZnSO4 for ∼20 min at 37°C. The digested samples were run on 10-cm PEI cellulose plastic TLC sheets with 0.1 M NH4Cl and 5% HOAc buffer until the solvent front reached the end of the sheet. After drying, the TLC sheets were phosphorimaged, and bands of aa-32P-AMP and 32P-AMP were quantified using Image Quant. The ratio of aa-32P-AMP signal to the total signal of [aa-32P-AMP + 32P-AMP] was used to determine the fraction that had been deacylated. The data were fit to the pseudo-first-order exponential decay by least-squares regression to determine the T1/2 from the decay rate constant k.
EF-Tu binding analysis
Stability of the EF-Tu-GTP-Pro-tRNAPro ternary complex was measured with Pro-tRNAPro (0.3 μM) and Thermus thermophilus EF-Tu (9 μM). The protein factor was activated in a buffer (50 mM HEPES-KOH, pH 7.0, 20 mM MgCl2, 5 mM DTT, 0.5 M NH4Cl) containing 20 μM GTP, 1.2 mM PEP (phosphoenolpyruvate), and 50 μg/mL pyruvate kinase for 3 h at 4°C and then incubated with 32P-labeled Pro-tRNAPro (0.3 μM) in an ice bath. A titration of EF-Tu showed that concentrations higher than 5 μM did not further improve the ability of the factor to protect the prolyl linkage, whereas concentrations in the range of 0.5 to 5 μM showed concentration-dependent protection effect, and concentrations at 0.2 μM or below showed limited effect. In the off-rate koff analysis, aliquots were quenched with RNase A (10 μL of 1 mg/mL) over time, and the tRNA was precipitated by acid. Counts that remained acid-precipitable were measured and plotted as a function of time to determine the koff (Schrader et al. 2009). Assays for stability of the acyl linkage and for the off-rate analysis were performed at 20°C and 4°C, respectively.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Virginia Cornish for Ser-DBE; Lee Speight for Cys-DBE, D-Phe-DBE, and L-Phe-DBE; Hiroaki Suga for Cys-DBE; and James Jaynes for discussion and support. This work was supported by grants from NIH (GM081601 and AG042169 to Y.M.H. and GM087605 to M.C.K.) and from NSF (MCB0818118 to James Jaynes).
Glossary
Abbreviations: aa-tRNA, aminoacyl-tRNA; HEPES, 2-[4-(2-hydroxyethyl) piperazin-1-yl] ethanesulfonic acid; TLC, thin layer chromatography; T1/2, half-life in time; DBE, 3,5-dinitrobenzyl ester; PEP, phosphoenol pyruvate; Kd, equilibrium dissociation constant; kon, on rate of binding; koff, off rate from binding
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.044123.113.
REFERENCES
- Arnez JG, Moras D 1997. Structural and functional considerations of the aminoacylation reaction. Trends Biochem Sci 22: 211–216 [DOI] [PubMed] [Google Scholar]
- Balzi E, Choder M, Chen WN, Varshavsky A, Goffeau A 1990. Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J Biol Chem 265: 7464–7471 [PubMed] [Google Scholar]
- Beres L, Lucas-Lenard J 1973. Studies on the binding of bacterial elongation factors EF Tu and EF G to ribosomes. Arch Biochem Biophys 154: 555–562 [DOI] [PubMed] [Google Scholar]
- Bock A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F 1991. Selenocysteine: the 21st amino acid. Mol Microbiol 5: 515–520 [DOI] [PubMed] [Google Scholar]
- Bougioukou DJ, Mukherjee S, van der Donk WA 2013. Revisiting the biosynthesis of dehydrophos reveals a tRNA-dependent pathway. Proc Natl Acad Sci 110: 10952–10957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruice TC, Fife TH, Bruno JJ, Brandon NE 1962. Hydroxyl group catalysis. II. The reactivity of the hydroxyl group of serine. The nucleophilicity of alcohols and the ease of hydrolysis of their acetyl esters as related to their pKa. Biochemistry 1: 7–12 [DOI] [PubMed] [Google Scholar]
- Burnett BJ, Altman RB, Ferrao R, Alejo JL, Kaur N, Kanji J, Blanchard SC 2013. Elongation factor Ts directly facilitates the formation and disassembly of the Escherichia coli elongation factor Tu·GTP·aminoacyl-tRNA ternary complex. J Biol Chem 288: 13917–13928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calendar R, Berg P 1967. D-Tyrosyl RNA: formation, hydrolysis and utilization for protein synthesis. J Mol Biol 26: 39–54 [DOI] [PubMed] [Google Scholar]
- Chousterman S, Herve G, Chapeville F 1966. [Alkaline hydrolysis and the ester linkage of some normal and modified aminoacyl-tRNA]. [Article in French] Bull Soc Chim Biol (Paris) 48: 1295–1303 [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol 260: 649–663 [DOI] [PubMed] [Google Scholar]
- Falorni M, Giacomelli G, Porcheddu A, Dettori G 2000. New oxazole-based conformationally restricted peptidomimetics: design and synthesis of pseudopeptides. Eur J Org Chem 2000: 3217–3222 [Google Scholar]
- Gondry M, Sauguet L, Belin P, Thai R, Amouroux R, Tellier C, Tuphile K, Jacquet M, Braud S, Courcon M, et al. 2009. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat Chem Biol 5: 414–420 [DOI] [PubMed] [Google Scholar]
- Hao B, Zhao G, Kang PT, Soares JA, Ferguson TK, Gallucci J, Krzycki JA, Chan MK 2004. Reactivity and chemical synthesis of L-pyrrolysine—the 22nd genetically encoded amino acid. Chem Biol 11: 1317–1324 [DOI] [PubMed] [Google Scholar]
- Hentzen D, Mandel P, Garel JP 1972. Relation between aminoacyl-tRNA stability and the fixed amino acid. Biochim Biophys Acta 281: 228–232 [DOI] [PubMed] [Google Scholar]
- Hou YM, Westhof E, Giege R 1993. An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc Natl Acad Sci 90: 6776–6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YM, Lipman RS, Zarutskie JA 1998. A tRNA circularization assay: evidence for the variation of the conformation of the CCA end. RNA 4: 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M, Soll D 2004. Aminoacyl-tRNAs: setting the limits of the genetic code. Genes Dev 18: 731–738 [DOI] [PubMed] [Google Scholar]
- Jenks WP, Regenstein J 1976. Ionization constants of acids and bases. In Handbook of biochemistry and molecular biology (ed. Fasman GD), Vol. 1, pp. 305–351 CRC Press, Cleveland, OH [Google Scholar]
- LaRiviere FJ, Wolfson AD, Uhlenbeck OC 2001. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294: 165–168 [DOI] [PubMed] [Google Scholar]
- Lee CP, Mandal N, Dyson MR, RajBhandary UL 1993. The discriminator base influences tRNA structure at the end of the acceptor stem and possibly its interaction with proteins. Proc Natl Acad Sci 90: 7149–7152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz MJ, Soffer RL 1971. Enzymatic modification of proteins. VII. Substrate specificity of leucyl,phenylalanyl-transfer ribonucleic acid-protein transferase. J Biol Chem 246: 5207–5212 [PubMed] [Google Scholar]
- Liu C, Gamper H, Liu H, Cooperman BS, Hou YM 2011. Potential for interdependent development of tRNA determinants for aminoacylation and ribosome decoding. Nat Commun 2: 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie A, Jurnak F 1985. Kinetic studies of Escherichia coli elongation factor Tu-guanosine 5′-triphosphate-aminoacyl-tRNA complexes. Biochemistry 24: 6433–6439 [DOI] [PubMed] [Google Scholar]
- Matthaei JH, Voigt HP, Heller G, Neth R, Schoch G, Kubler H, Amelunxen F, Sander G, Parmeggiani A 1966. Specific interactions of ribosomes in decoding. Cold Spring Harb Symp Quant Biol 31: 25–38 [DOI] [PubMed] [Google Scholar]
- Murakami H, Ohta A, Ashigai H, Suga H 2006. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat Methods 3: 357–359 [DOI] [PubMed] [Google Scholar]
- Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270: 1464–1472 [DOI] [PubMed] [Google Scholar]
- Nissen P, Thirup S, Kjeldgaard M, Nyborg J 1999. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure 7: 143–156 [DOI] [PubMed] [Google Scholar]
- Paleskava A, Konevega AL, Rodnina MV 2010. Thermodynamic and kinetic framework of selenocysteyl-tRNASec recognition by elongation factor SelB. J Biol Chem 285: 3014–3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC 2009. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci 106: 50–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JR, Uhlenbeck OC 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci 85: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader JM, Chapman SJ, Uhlenbeck OC 2009. Understanding the sequence specificity of tRNA binding to elongation factor Tu using tRNA mutagenesis. J Mol Biol 386: 1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuber F, Pinck M 1974a. On the chemical reactivity of aminacyl-tRNA ester bond. 3. Influence of ionic strength, spermidine and methanol on the rate of hydrolysis. Biochimie 56: 397–403 [DOI] [PubMed] [Google Scholar]
- Schuber F, Pinck M 1974b. On the chemical reactivity of aminoacyl-tRNA ester bond. I. Influence of pH and nature of the acyl group on the rate of hydrolysis. Biochimie 56: 383–390 [DOI] [PubMed] [Google Scholar]
- Shitivelband S, Hou YM 2005. Breaking the stereo barrier of amino acid attachment to tRNA by a single nucleotide. J Mol Biol 348: 513–521 [DOI] [PubMed] [Google Scholar]
- Soutourina O, Soutourina J, Blanquet S, Plateau P 2004. Formation of D-tyrosyl-tRNATyr accounts for the toxicity of D-tyrosine toward Escherichia coli. J Biol Chem 279: 42560–42565 [DOI] [PubMed] [Google Scholar]
- Vig BS, Lorenzi PJ, Mittal S, Landowski CP, Shin HC, Mosberg HI, Hilfinger JM, Amidon GL 2003. Amino acid ester prodrugs of floxuridine: synthesis and effects of structure, stereochemistry, and site of esterification on the rate of hydrolysis. Pharm Res 20: 1381–1388 [DOI] [PubMed] [Google Scholar]
- Wickramasinghe NS, Lacey JC Jr 1993. N-Acetyl-D- and L-esters of 5′-AMP hydrolyze at different rates. Chirality 5: 150–153 [DOI] [PubMed] [Google Scholar]
- Wolfenden R 1963. The mechanism of hydrolysis of amino acyl RNA. Biochemistry 2: 1090–1092 [DOI] [PubMed] [Google Scholar]
- Xiao H, Murakami H, Suga H, Ferre-D'Amare AR 2008. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature 454: 358–361 [DOI] [PubMed] [Google Scholar]
- Zhang CM, Perona JJ, Ryu K, Francklyn C, Hou YM 2006. Distinct kinetic mechanisms of the two classes of aminoacyl-tRNA synthetases. J Mol Biol 361: 300–311 [DOI] [PubMed] [Google Scholar]