This method paper presents a novel strategy based on the use of transcription activator-like effector nucleases (TALENs) together with homologous recombination (HR) to disrupt miRNA loci in cell lines. As such, it introduces a new tool that will be useful in many studies of miRNA function.
Keywords: double knockout, gene-editing, homology-directed repair (HDR), microRNA, transcription activator-like effector nucleases (TALENs)
Abstract
Significant progress in the functional understanding of microRNAs (miRNAs) has been made in mice, but a need remains to develop efficient tools for bi-allelic knockouts of miRNA in the human genome. Transcription activator-like effector nucleases (TALENs) provide an exciting platform for targeted gene ablation in cultured human cells, but bi-allelic modifications induced by TALENs alone occur at low frequency, making screening for double knockouts a tedious task. Here, we present an approach that is highly efficient in bi-allelic miRNA ablation in the human genome by combining TALENs targeting to the miRNA seed region with a homologous recombination donor vector and a positive selection strategy. A pilot test of this approach demonstrates bi-allelic miR-21 gene disruption at high frequency (∼87%) in cultured HEK293 cells. Analysis of three independent clones showed a total loss of miR-21 expression. Phenotypical analysis revealed increased miR-21 target gene expression, reduced cell proliferation, and alterations of global miRNA expression profiles. Taken together, our study reveals a feasible and efficient approach for bi-allelic miRNA ablation in cultured human cells and demonstrates its usefulness in elucidating miRNA function in human cells.
INTRODUCTION
MicroRNAs (miRNAs) are short noncoding RNAs that negatively regulate gene expression in mammalian cells by targeting the 3′ untranslated region of mRNAs. In recent years, it has become increasingly apparent that miRNAs are of crucial functional importance for normal development and physiology, as well as a factor in various diseases in mammalians (Svoboda and Flemr 2010; Pauli et al. 2011; Amiel et al. 2012; Gommans and Berezikov 2012; Iorio and Croce 2012b). For instance, miR-21 was found to be overexpressed in virtually all types of human cancers and thus has emerged as an important therapeutic target in cancer treatment (Huang et al. 2012; Iorio and Croce 2012a; Li et al. 2012a,b). Recently, miR-21 has been shown to play crucial regulatory roles in cell growth, proliferation, and apoptosis, as well as in autoimmune and cardiovascular diseases (Buscaglia and Li 2011; Liu et al. 2013; Xu et al. 2013).
Despite their fundamental importance, the exact functions of miRNAs in the context of human development and disease processes remain largely unknown. This is, in part, due to a lack of effective methods for completely abolishing the expression of miRNAs in human cells and disease-relevant models (Park et al. 2010, 2012). Although targeted gene knockdown by short-interfering RNAs (siRNAs) provides a rapid and inexpensive tool to functionally study most protein-coding genes, it cannot be used to reduce mature miRNAs in a sensible way at the cellular level. Alternatives to siRNAs include small molecule inhibitors, antisense oligonucleotides, anti-miR vectors (miRZips), and miRNA sponges (Krutzfeldt et al. 2005; Zhu et al. 2011; Hu et al. 2013b). The major limitations of these methods are (1) the transient nature of their effects, and (2) a high risk of off-target effects and resulting toxicity (Jackson et al. 2003; van Dongen et al. 2008; Khan et al. 2009), limitations highlighted by reports on discrepancies between the effect of miRNA inhibitors and genetic knockouts (Patrick et al. 2010; Park et al. 2012).
Transcription activator-like effector nucleases (TALENs) are powerful gene editing tools for uncovering gene functions (Bogdanove and Voytas 2011; Clark et al. 2011; Miller et al. 2011; Joung and Sander 2013; Sun and Zhao 2013). Though relatively new, TALENs have been successfully employed in a broad variety of systems (Christian et al. 2010; Hockemeyer et al. 2011; Carlson et al. 2012; Tong et al. 2012; Sung et al. 2013; Zhang et al. 2013). TALENs are custom endonucleases that work as dimers to create double-strand breaks in their target DNA sequences (Clark et al. 2011; Miller et al. 2011; Joung and Sander 2013; Sun and Zhao 2013). Each TALEN typically recognizes a binding site of ∼20 bp, providing a high specificity of gene targeting. Coupled with the ability of context-independent binding (Cermak et al. 2011; Moore et al. 2012; Ansai et al. 2013; Kim et al. 2013a), TALENs are uniquely suited to precisely edit very small genes such as miRNAs (Hu et al. 2013a; Kim et al. 2013b).
A number of approaches have been developed for rapid assembly of custom TALENs (Cermak et al. 2011; Briggs et al. 2012; Sanjana et al. 2012; Sakuma et al. 2013; Uhde-Stone et al. 2013). With these advances, TALEN pairs can be generated easily and economically in a matter of days. Many protein-coding genes or genetic loci with long genome sequences have been documented for the purpose of gene editing, but the short and noncoding nature of miRNA presents different challenges. Hu et al. have reported disruption of miRNA genes in human cells by TALENs. While it is possible to disrupt genes in the human genome with TALENs alone at a frequency of typically 2%–40%, averaging around 16% for mono-allelic disruptions (Kim et al. 2013a; Sakuma et al. 2013), isolating cells carrying rare bi-allelic disruptions requires time-consuming single cell-derivation and subsequent screening. Our approach differs from other strategies (Hu et al. 2013a; Kim et al. 2013b) in that we combine TALENs targeting to the miRNA seed region with a homologous recombination (HR) donor vector carrying a selectable marker. Our strategy enables convenient positive selection, and the combination of NHEJ with stem–loop deletions results in efficient bi-allelic miRNA gene ablation, which is especially valuable for loci that may be difficult to target. Additionally, by using HR donors, endogenous loci can be potentially modified with custom sequences (such as IRES-fluorescent proteins) to allow functional assessment of endogenous gene expression and regulation (Hockemeyer et al. 2011).
As proof of concept, we targeted the human miR-21 seed region in cultured human HEK293 cells, and successfully selected bi-allelic miRNA knockouts with high efficiency (87%). Quantitative RT-PCR analysis of three independent clones confirmed complete loss of mature miR-21 expression. Phenotypical analysis confirmed increased protein levels of the miR-21 target gene PDCD4, reduced cell proliferation, and changes in global miRNA expression profiles. Re-expression of miR-21 in a miR21-knockout line restored miR-21 function, demonstrated by a decrease in target gene expression, further supporting the validity of our approach. Taken together, the high efficiency and ease of a positive selection protocol for bi-allelic miRNA deletions in the human genome provides a powerful tool for elucidating miRNA function in humans to realize, in the long-term, their full therapeutic potential.
RESULTS
System design and knockout strategies
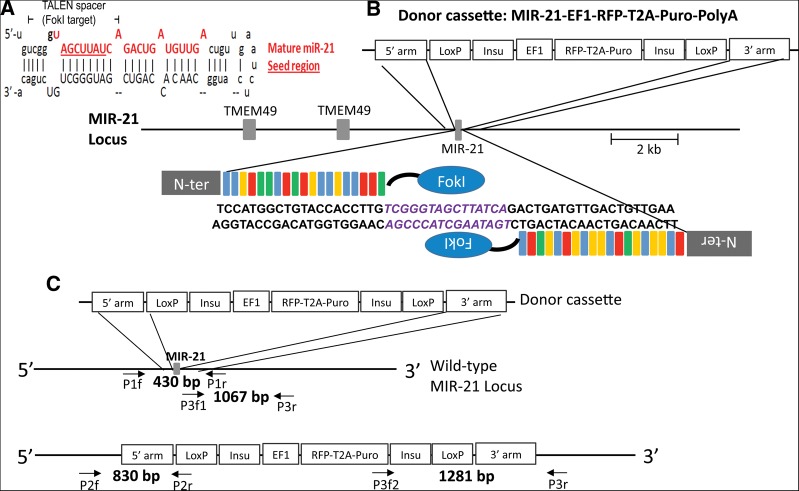
TALENs have almost no restriction in regard to their target sequences. To take advantage of this feature, we designed a TALEN pair flanking the ∼6-bp seed region of the human miR-21, directing FokI cleavage to the seed region (Fig. 1A). TALEN-induced double-strand breaks can achieve gene knockouts in two main ways: by homologous recombination of exogenous donor DNA, or by nonhomologous end joining (NHEJ) repair. In the case of HR events, the donor replaces the entire miR-21 precursor with an RFP and a puromycin-resistance expression cassette (Fig. 1B). To completely knock out a gene of interest in diploid cells, the other allele must be disrupted by a simultaneous HR event or, more likely, by NHEJ-induced mutations. Due to the two selectable marker genes embedded in the HR donor vector (RFP and puromycine resistance), we can easily isolate the clonal populations of cells in which an HR event occurred. Because NHEJ is the predominant repair mechanism induced by double-strand breaks, selecting for HR events will most likely identify clones that harbor bi-allelic modifications, with the second allele carrying a NHEJ-mediated mutation.
FIGURE 1.
Schematic overview of the targeting strategy against miR-21 in the human genome using TALENs in combination with an HR donor vector. (A) miR-21 stem–loop structure, the mature miR-21 sequence is shown in red; the seed region is underlined. TALENs were designed to position the miRNA seed region in the central portion of the spacer, directing cleavage to this functionally essential miRNA region. (B) A HR donor plasmid was created corresponding to the cleavage location of the TALEN pair and carried 509-bp (5′ arm) and 600-bp (3′ arm) regions of homology to the miR-21 sequence, which, in the native genome, are separated by 202 bp that include the miR21 stem–loop structure. Two LoxP sites are flanking the insulated expression cassette, which is composed of an EF1α promoter-driven RFP and puromycin-resistant gene (Puro), separated by a T2A linker (self-cleaving peptide sequence). (C) Locations of primers used for genotyping of HEK293 cells targeted with TALENs and HR donor. Primer pairs P2f, P2r and P3f, P3r were designed to amplify the junctions between genome and inserted HR donor cassette (830 bp and 1281 bp, respectively). Triple primers including two forward primers (P3f1 and P3f2) and one common reverse primer (P3r) were designed to co-amplify the HR knockout allele (P3f2 and P3r; 1281 bp) and the wild-type or NHEJ-modified allele (P3f1 and P3r; 1067 bp). Primer pair P1f and P1r was designed to amplify a 430-bp portion of the miR-21 region for subsequent sequence analysis to detect possible NHEJ events.
Junction PCR and sequencing confirms seamless donor integration by HR at miR-21 locus
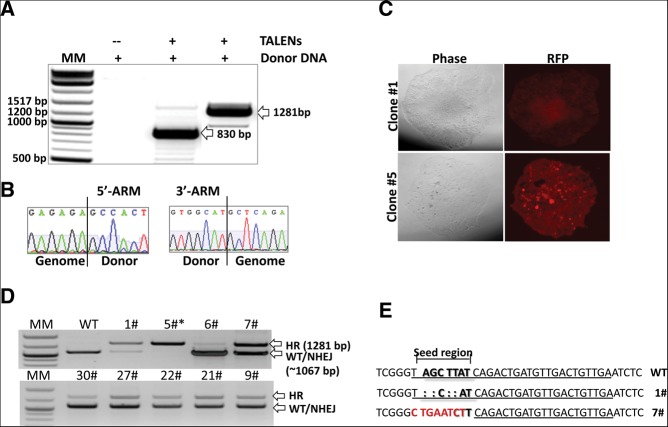
To evaluate correct TALEN-mediated HR donor integration in the human miR21 genomic locus (chromosome 17q23.2; 55273409–55273480, adjacent to the coding gene TMEM49), we conducted transient transfection studies using TALENs in combination with an HR donor vector in HEK293 cells. Junction PCR with a combination of genome and vector-specific primers (Fig. 1C) was performed as specified below. The successful amplification of the correct PCR products was obtained only in cells cotransfected with both TALENs and HR donor vectors (Fig. 2A) but not in cells transfected with the HR donor alone, demonstrating the importance of TALENs in mediating the HR event. Sequence analysis of the junction-PCR products demonstrated the seamless fusion of donor DNA with miR-21 genomic sequences (Fig. 2B), confirming the gene targeting ability of the designed TALENs and the HR donor vector.
FIGURE 2.
Genotyping analysis of HEK293 cells targeted with miR-21-directed TALENs and HR donor. (A) Combination of genome-specific and donor-cassette-specific primers amplified expected PCR products of 830 bp (5′ arm) and 1281 bp (3′ arm) in cells cotransfected with TALENs and the HR donor vector. Transfection with only the HR donor vector did not result in any detectable PCR product. (B) Subsequent sequencing of the amplified PCR products confirmed seamless integration of both HR donor vector arms. (C) Selection reveals RFP-positive puromycin-resistant single cell-derived colonies. (D) A triple primer PCR strategy was used to determine whether mono- or bi-allelic HR events had occurred in single cell-derived clones. A donor-specific and a wild type-specific forward primer were combined with a wild type-specific reverse primer. Alleles with donor cassette integration were recognized by a 1281-bp amplicon, while WT or NHEJ events result in a 1067-bp amplicon. (E) Examples of seed region modification by NHEJ, compared to the wild type.
Enrichment and isolation of miR-21 knockout candidates from single cell-derived clones
To enrich and isolate miR-21 precursor-deleted cells, HEK293 cells cotransfected with the miR-21-specific TALEN pair and HR donor vector were put under positive selection. Potential miR-21 HR-candidate cells were selected by puromycin resistance and RFP expression (Fig. 2C). Out of 96 single cell-derived clones, 30 lines survived in the presence of puromycin and remained RFP-positive and puromycin-resistant for >2 mo. In the group transfected solely with the HR donor vector, only six colonies were initially formed, and none of these clones survived after prolonged puromycin treatment.
Genotyping after selection reveals highly efficient generation of bi-allelic knockouts
We next determined the genotypes of the selected, single cell-derived clones. As shown in Figure 2D, correctly targeted colonies were revealed by the presence of longer PCR products, while short PCR products indicate either WT or small changes induced by NHEJ. Out of 23 successful PCR amplifications, we found that ∼96% (22/23) were mono-allelic for HR-induced miR-21 deletion and ∼4% (1/23) were bi-allelic for HR-induced miR-21 deletion, demonstrating that double miRNA gene ablation occurs in HEK293 cells via HR when coupled to TALEN-directed cleavage.
To determine whether a simultaneous modification of the other allele by NHEJ had occurred, we sequenced the PCR products spanning the targeted seed region of miR-21. Among 11 mono-allelic HR clones analyzed, a significant portion of the non-HR alleles (91%; 10/11) were NHEJ-modified (Fig. 2E). Important, and consistent with previous reports (Cermak et al. 2011), almost all modifications were centered on the targeted region, resulting in at least 4-bp deletions or complete changes of the seed sequences of miR-21. Taken together, these results show an 87% efficiency of our approach in generating bi-allelic modifications of miR-21 in the human genome.
Bi-allelic disruption abolishes miR-21 expression but does not affect expression of the neighboring gene TMEM49
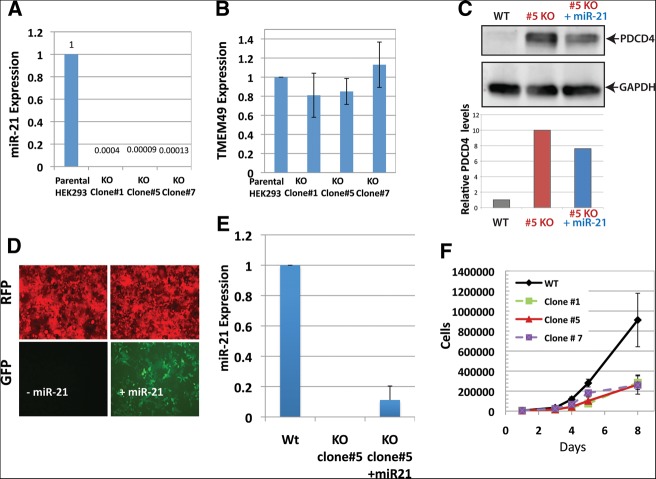
We first investigated the loss of miR-21expression in three independent bi-allelic clones by quantitative RT-PCR. Clone #5 bears deletions of the miR-21 precursor on both alleles (bi-allelic HR events). Clones #1 and #7 have one deletion of the miR-21 precursor (via HR) on one allele and another small deletion or mutations (via NHEJ) of the seed sequence on the other allele (Fig. 2E). As shown in Figure 3A, there was a total depletion of miR-21expression in all three cell lines, compared to those of the parental control. We next examined whether custom TALEN-mediated miR-21 targeting may lead to any changes in the neighboring gene TMEM49 (also referred to as VMP1). We performed quantitative RT-PCR to examine the mRNA levels of TMEM49 in three miR-21 knockout cell lines and the parental control. We found that there was no significant change in the mRNA levels of TMEM 49, compared to the control (Fig. 3B), confirming that neither TALEN-mediated gene replacement nor small deletions caused significant changes in the expression of this neighboring gene.
FIGURE 3.
Expression of miR-21 and associated changes in TALEN-mediated knockout lines. (A) Complete loss of miR-21expression in three independent lines. The constitutive expression of miR-21 in parental HEK293 cells was high with a CT value of 18∼21. Quantitative PCR confirmed a >1000-fold decrease in miR-21 levels in all three lines tested compared to those of the parental control. (B) No significant change in the mRNA expression level of TMEM49, the closest neighbor of MIR21 on chromosome 17, as assessed by real-time RT-PCR. (C) Expression of miR-21 is associated with corresponding changes in the expression of its targeted gene PDCD4; GAPDH served as loading control. Western blot was conducted with equal amount of proteins extracted from parental HEK293, miR-21 double knockout line, and double knockout line re-expressing miR-21. (D,E) Confirmation of miR-21 re-expression in the miR-21-knockout line. (D) The upper panel shows cells positive for RFP due to presence of the HR donor cassette (bi-allelic). Re-expression of miR-21 was confirmed by co-expression of a GFP marker (lower panel) and real-time RT-PCR (E). (F) Reduced proliferations in all three miR-21 knockout lines tested. Growth curves were plotted for the parental HEK293 cells as well as the three independent knockout lines.
Loss of miR-21 causes an increase in target gene expression that can be rescued by re-expression of miR-21
To test whether miR-21 ablation caused corresponding changes in the expression of tumor suppressor gene PDCD4 (Programmed cell death 4), a known target of miR-21 (Liu et al. 2012), we performed Western blot analyses. As expected, while the basal PDCD4 protein level is barely detectable, it was drastically elevated following miR-21 knockout (Fig. 3C, lanes 1,2). To further validate whether the elevated expression of PDCD4 was due to the loss of miR-21, we transduced miR-21 knockout cells with lentiviral particles containing a cassette to express miR-21 and GFP. As shown in Figure 3E, more than ∼80% of miR-21 knockout cells (RFP-positive) were also GFP-positive, suggesting expression of miR-21 in most transduced cells (Fig. 3D). This was confirmed by quantitative RT-PCR showing an ∼3000-fold increase in miR-21 RNA expression in the transduced cells vs. the control (Fig. 3E). Following restoration of miR-21 expression in miR-21 knockout cells, PDCD4 protein was partially reduced (Fig. 3C).
miR-21 ablation results in inhibition of cell proliferation and alterations in global miRNA expression
We next analyzed cell proliferation rates following miR-21 ablation in HEK 293. As shown in Figure 3F, the three independent miR-21-ablated lines tested all exhibited reduced cell proliferation compared to the parental control. The inhibition in cell proliferation occurred as early as Day 3 after plating and became more prominent with time, lasting as long as 8 d. We further tested proliferation of cell line KO#5 with reintroduced miR-21. However, proliferation of KO#5 expressing miR-21 was not restored to the wild-type level, possibly due to the relatively lower expression levels of miR-21 compared to wild type (Fig. 3E), or to complex, nonreversible changes in the KO cell line. Nevertheless, we can't rule out that the miR21 phenotype is due to possible off-target effects.
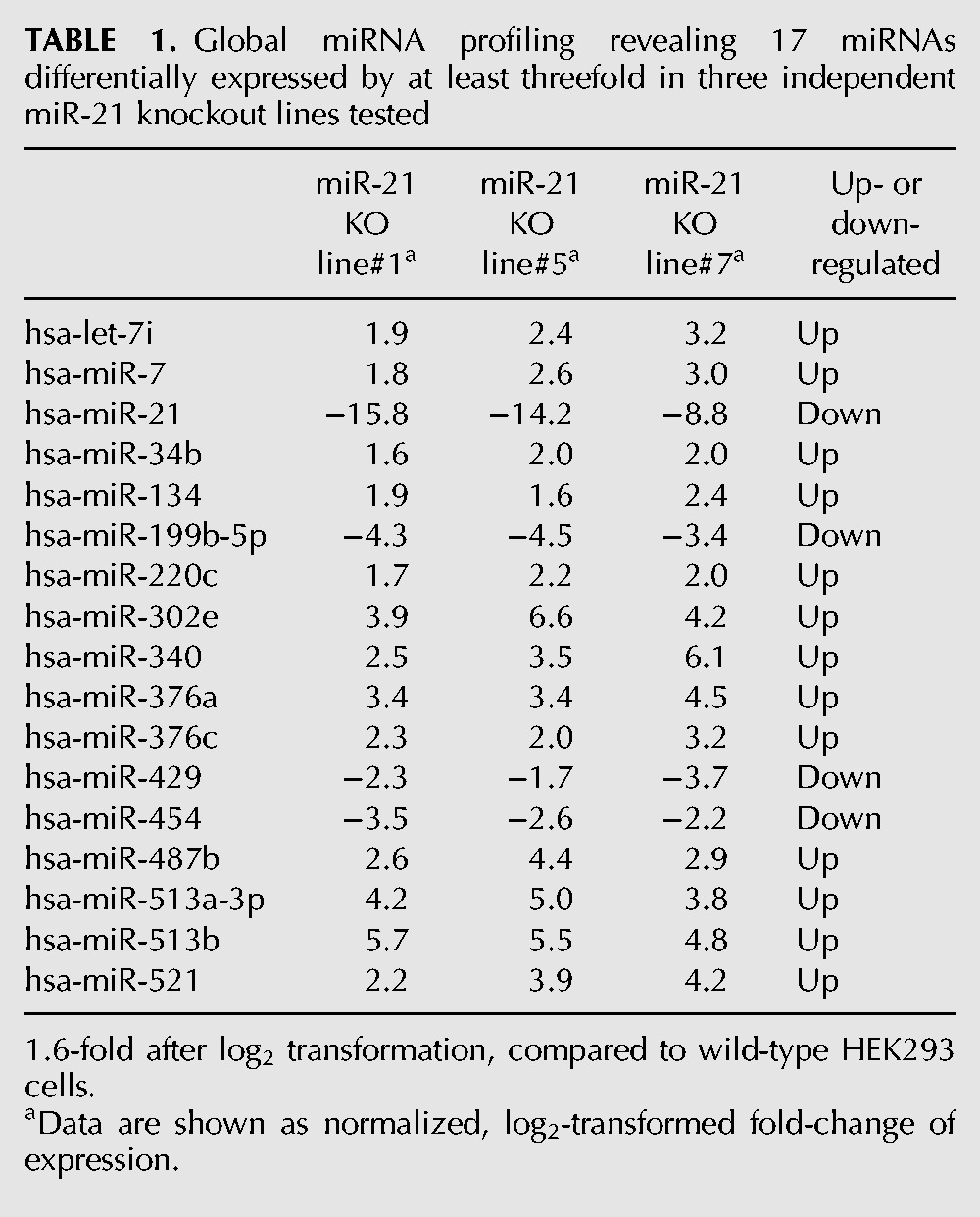
To examine whether the reduced proliferation was associated with changes in other miRNAs, we profiled global miRNA expression in three independent bi-allelic miR-21 knockout lines (clone #1, #5, #7), compared to the parental control. Seventeen out of 760 miRNAs tested (∼2%) showed more than threefold up- or down-regulation in all three mutant lines, demonstrating strong concordance between the independent lines (Table 1). A BLASTn search against the NCBI nucleotide database confirmed that these miRNA do not display sequence homology to the TALEN target sequences, indicating that the observed changes are secondary in nature and not off-target effects.
TABLE 1.
Global miRNA profiling revealing 17 miRNAs differentially expressed by at least threefold in three independent miR-21 knockout lines tested
DISCUSSION
Gene targeting of miRNAs in mammalian cells by homologous recombination is inefficient (Bollag et al. 1989), which has limited the use of human disease models in elucidating miRNA functions and exploring their therapeutic potential. To overcome these limitations, we developed an approach for efficient bi-allelic miRNA ablation combining TALENs with a selectable HR donor vector. We demonstrate that this approach robustly abolished miR-21 expression both via seed region disruption and precise stem–loop structure removal. In addition, the HR-added marker genes allowed for efficient selection (87%) of cells carrying bi-allelic modifications. These findings established a feasible approach for bi-allelic miRNA ablation in cultured human HEK293 cells, which should advance the study of miRNA function in cell culture model systems.
Programmable nucleases, such as zinc finger nucleases (ZFNs), meganucleases, and, more recently, TALENs and CRISPR/Cas9-guided DNA endonucleases have emerged as powerful tools for targeted genome editing (Rahman et al. 2011; Hafez and Hausner 2012; Perez-Pinera et al. 2012; Cho et al. 2013; Cong et al. 2013; Gaj et al. 2013; Mali et al. 2013). Among these, TALENs offer several advantages for miRNA perturbation, including the ability to bind any sequence in the genome, which enables specific targeting of small sequences, such as the miRNA seed sequence (∼6 bp). Several recent studies indicate high specificity and low cytotoxicity of TALENs, compared to ZFNs and the CRISPR/Cas9 system (Fu et al. 2013; Kim et al. 2013b; Sun and Zhao 2013). In addition, their simple, modular design is easy to implement, due to their availability as an open-source technology.
However, there are some key limitations in the use of programmable nucleases to achieve gene ablation in diploid mammalian cells. While TALENs have the potential to induce mutations in the human genome at a frequency averaging around 16% (Kim et al. 2013a; Sakuma et al. 2013) by creating targeted double-strand breaks and subsequent NHEJ, the majority of mutations are mono-allelic and require time-consuming single cell-derivation and subsequent screening (Hu et al. 2013a). For example, an average mono-allelic efficiency of 16% would result in ∼2.6% bi-allelic modifications. A low-efficiency TALEN of 2% mono-allelic disruption would result in rare bi-allelic modifications of ∼0.04%. To overcome these limitations, we employed an approach of combining TALENs targeting the miRNA seed region with an HR donor vector for deleting the entire miRNA stem–loop structure. The HR donor vector was engineered to include an insulated cassette for RFP and puromycin markers to allow positive selection by drug treatment and/or fluorescent-activated cell sorting. Our combined approach of miRNA gene targeting via TALEN and HR donor vectors was highly efficient and produced some intriguing results:
First, we achieved robust bi-allelic gene modifications with an efficiency of 87%. By simple puromycin selection, we were able to establish more than 20 bi-allelic modified clones from one transfection experiment with a starting cell number of 2 × 105. Although most of the bi-allelic clones harbor one complete hairpin deletion and one seed region disruption, we speculated that the expression of miRNAs was effectively abolished due to the precise disruption of the miRNA seed region. In fact, both bi-allelic modified clones tested showed a complete loss of miR-21 expression, similar to those of the bi-allelic hairpin deletion. Another advantage of our HR donor is the fact that the HR selection cassette can be removed via its loxP sites, enabling a second round of screening for even higher frequency of bi-allelic hairpin deletions, if desired.
Second, targeted miRNA inactivation is a powerful method capable of providing conclusive information about gene function in a cell-type or disease-specific manner. This will help to establish the crucial link between genotype and phenotype, thus yielding functional- and clinical-relevant knowledge. As expected, miR-21 knockout lines displayed an increased protein level of the miR-21 target gene PDCD4. Re-expression of miR-21 in miR-21-knockout cells restored its functional role, demonstrated by a decrease in protein expression of the target gene PDCD4, further confirming the validity of miR-21 knockout lines for functional analysis. Moreover, we observed a pronounced attenuation of cell proliferation in all three independent miR-21-ablated cell lines tested. Strikingly, miRNA profiling by qRT-PCR revealed large-scale changes of miRNA expression in the three independent knockout lines tested, highlighting the promise of this approach for future dissection of miRNA regulatory circuits. With the ease and efficiency of bi-allelic mutant generation, combined with the advantage of a stable phenotype, we envision that this approach will broaden our knowledge in deciphering the role of miRNAs in human physiology and disease.
MATERIALS AND METHODS
Cell culture and transfection
The human embryonic kidney cell line HEK293 cells were maintained in high glucose Dulbecco's Minimal Essential Medium (DMEM) supplemented with 10% FBS, 2 mM GlutaMax (Life Technologies), 100 units/mL penicillin and 100 units/mL streptomycin. All transfections were performed using Purefection transfection reagent according to the manufacturer's manual (Cat# LV750A-1; System Biosciences Inc.). Transfected cells were incubated in a 37°C incubator with 5% CO2.
TALEN design and TALEN expression plasmids
To target the seed region of the human miR-21 locus (Fig. 1A), a TALEN pair was designed using an online tool, TAL Effector Nucleotide Targeter 2.0 (Doyle et al. 2012). To streamline the design of miRNA targeted TALENs, we followed these criteria: (1) TALEN binding sites were set to 20 bp, including the first T, to ensure high specificity of gene targeting; (2) spacer lengths of 15–25 bp were chosen to maximize cleavage efficiency; (3) the miRNA seed sequence was centrally situated within the spacer to direct cleavage to the seed region. Following these criteria, designed TALENs were assembled into a CMV-driven expression cassette using the EZ-TAL Assembly Kit (Cat# GE100A-1; System Biosciences Inc.), and the final constructs were confirmed by DNA sequencing.
miR-21 gene targeting and screening
A knockout HR donor vector that bears homologous arms of 509 bp (left arm) and 600 bp (right arm) was designed and constructed (Fig. 1B). In the genome, the regions represented on the vector arms are spaced 202 bp apart, flanking the miR-21 stem–loop region. The TALEN cut site is located 52 bp downstream from the left vector arm, within the miR-21 seed region. In the HR donor vector, the two homologous arms flank an insulated cassette to express dual selectable markers, RFP and puromycin resistance proteins.
After transfection, RFP-positive and puromycin-resistant single cells were isolated and expanded. Briefly, a total of ∼4 × 105 cells were transfected with 1 µg of donor plasmid and 1 µg of each TALEN-encoding plasmid. Cells were trypsinized 2 d after transfection and subsequently plated on 10-cm culture dishes for 24 h. Puromycin was then added to the culture medium (final concentration 4 µg/mL), to allow single cell-derived colony formation. After 2 wk of puromycin selection, RFP-positive clones were picked and expanded for further analysis.
Genotyping for HR and NHEJ events
To confirm miR-21 gene disruption, the targeted gene loci were PCR-amplified from genomic DNA prepared from transfected cells or candidate clones. To detect HR events, two pairs of primers were used to perform junction PCR. PCR products with sizes of 830 bp and 1281 bp were expected in the case of successful HR, using the following primers—P2f: 5′-TGATGCATTGCACACCCTCTGG-3′; P2r: 5′-GAAAGATCTGATGCATTCGCGAGG-3′; P3f2: 5′-AGCCCCGGAATTGACTGGATTC-3′; P3r: 5′-TGTATTGCACTGCCTGACAGTCAC-3′ (Fig. 1C).
To determine whether mono- or bi-allelic HR events occurred in single cell-derived clones, a triple-primer PCR strategy was devised. As shown in Figure 1C, two forward primers (P3f1: 5′-TAGCTTATCAGACTGATGTTGACTGTTGA-3′ and P3f2: 5′-TAGCTTATCAGACTGATGTTGACTGTTGA-3′) and a common reverse primer (P3r: 5′-TGTATTGCACTGCCTGACAGTCAC-3′), were used to amplify a predicted 1067-bp fragment of the wild-type allele and a 1281-bp fragment in the case of HR, respectively.
Even cells that carry only one HR event may carry bi-allelic modifications of the miRNA if the miRNA seed region of the other allele is modified by NHEJ. To test for small NHEJ-induced sequence changes on the other allele, genomic PCR was performed to amplify the targeted sites using the primers P1f and P1r. The amplified PCR products were subjected to agarose gel electrophoresis and DNA purification using the QIAquick Gel Extraction Kit (QIAGEN), and the purified DNA was sequenced.
Western blot analysis
Whole-cell extracts were prepared using M-PER lysis buffer (Pierce). The concentration of proteins was measured by Bradford assay (Amresco), and equal protein amounts were used for SDS-PAGE. Briefly, proteins from whole-cell extracts were separated and transferred onto nitrocellulose membranes. The membranes were blocked with 1× TBS-T with 5% nonfat dry milk (Bio-Rad), followed by incubation overnight at 4°C with 1:1000-diluted primary anti-rabbit PDCD4 antibody (Cell Signaling). For protein loading controls, rabbit anti-human GAPDH antibody (Abcam) was used at 1:2500 dilution in Superblock T20 buffer (Pierce). The blot was then probed with 1:10,000-diluted goat anti-rabbit IgG secondary-HRP antibody (Pierce). The signal was detected by Super Signal West Femto ECL (Pierce).
miR-21 rescue
To re-express miR-21 in a knockout line, we used a lentiviral gene delivery system. Briefly, an expression vector containing the miR-21 precursor construct (PMIRH21PA-1; SBI) was packaged into pseudo-lentiviral particles using LentiSuite (LV300A-1; SBI) (Mendenhall et al. 2012). The lentiviral particles (MOI = 5) were transduced into miR-21-knockout cells in the presence of the virus-transduction reagent Transdux (LV850A-1, SBI). The re-expression of miR-21 was confirmed by both co-expression of GFP and quantitative RT-PCR measuring mature miR-21 expression levels as follows.
Quantitative RT-PCR and global miRNA profiling
Total cellular RNA was prepared from cells using Trizol Reagent (Life Technologies) according to the manufacturer's instructions. Total RNA was converted to cDNA using RNA-Quant (Cat# RA430A-1; SBI), and qRT-PCR was performed. A miR-21-specific forward primer (CSRA 640A-1, SBI) was used in combination with a universal reverse primer (cat# RA420AU-3, SBI). For the miR-21-neighboring gene TMEM49 (also referred to as VMP1), gene-specific forward and reverse primers (TMEM49-F: 3′-CGGCATAGGTCCATCTCTGCAG-5′ and TMEM49-R: 5′-TCAAACATCCAGGACAACCAG-3′) were used to evaluate mRNA expression levels. To perform global expression profiling of cellular miRNAs, miRNA-specific primers were obtained from the hsa-miRNome miRNA Profiler kit (Cat# RA660A-1; SBI) in combination with a universal reverse primer (SBI) according to the manufacturer's instructions. We used the comparative threshold cycle (Ct) method to quantify the expression levels. The Cts were normalized to three housekeeping RNAs (human U6 snRNA, RNU43 snoRNA, and Hm/Ms/Rt U1 snRNA).
Phenotypic growth analysis
Parental HEK-293 cells and miR-21-knockout cell lines were seeded at 10,000 cells/well in a 24-well plate in culture medium as described. Growth was monitored by counting cells with a hemocytometer from each cell line on Day 3, 4, 5, and 8. Culture medium was changed at Day 3 and 6. The experiments were performed in triplicate.
Data collection and presentation
For live cell monitoring, cultured cells were monitored at various times under a fluorescent microscope. RFP or GFP live cell images were taken using the same exposure conditions and magnification within the group of comparison. For qPCR assays, all data are presented as mean ± SD (n = 3), unless stated otherwise.
ACKNOWLEDGMENTS
We thank members of the System Biosciences, Inc. for their technical assistance, discussion, and support.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.042010.113.
REFERENCES
- Amiel J, de Pontual L, Henrion-Caude A 2012. miRNA, development and disease. Adv Genet 80: 1–36 [DOI] [PubMed] [Google Scholar]
- Ansai S, Sakuma T, Yamamoto T, Ariga H, Uemura N, Takahashi R, Kinoshita M 2013. Efficient targeted mutagenesis in medaka using custom-designed transcription activator-like effector nucleases. Genetics 193: 739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF 2011. TAL effectors: customizable proteins for DNA targeting. Science 333: 1843–1846 [DOI] [PubMed] [Google Scholar]
- Bollag RJ, Waldman AS, Liskay RM 1989. Homologous recombination in mammalian cells. Annu Rev Genet 23: 199–225 [DOI] [PubMed] [Google Scholar]
- Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, Church GM 2012. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res 40: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaglia LE, Li Y 2011. Apoptosis and the target genes of microRNA-21. Chin J Cancer 30: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC 2012. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci 109: 17382–17387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS 2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230–232 [DOI] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KJ, Voytas DF, Ekker SC 2011. A tale of two nucleases: gene targeting for the masses? Zebrafish 8: 147–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, Bogdanove AJ 2012. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res 40: W117–W122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF III 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommans WM, Berezikov E 2012. Controlling miRNA regulation in disease. Methods Mol Biol 822: 1–18 [DOI] [PubMed] [Google Scholar]
- Hafez M, Hausner G 2012. Homing endonucleases: DNA scissors on a mission. Genome 55: 553–569 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. 2011. Genetic engineering of human pluripotent cells using tale nucleases. Nat Biotechnol 29: 731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Wallace J, Dahlem TJ, Grunwald DJ, O'Connell RM 2013a. Targeting human microRNA genes using engineered tal-effector nucleases (TALENS). PLoS One 8: e63074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Guo J, Zheng L, Li C, Zheng TM, Tanyi JL, Liang S, Benedetto C, Mitidieri M, Katsaros D, et al. 2013b. The heterochronic microRNA let-7 inhibits cell motility by regulating the genes in the actin cytoskeleton pathway in breast cancer. Mol Cancer Res 11: 240–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li XQ, Chen X, Che SM, Chen W, Zhang XZ 2012. Inhibition of microRNA-21 increases radiosensitivity of esophageal cancer cells through phosphatase and tensin homolog deleted on chromosome 10 activation. Dis Esophagus 26: 823–831 [DOI] [PubMed] [Google Scholar]
- Iorio MV, Croce CM 2012a. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4: 143–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Croce CM 2012b. MicroRNA involvement in human cancer. Carcinogenesis 33: 1126–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21: 635–637 [DOI] [PubMed] [Google Scholar]
- Joung JK, Sander JD 2013. TALENS: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS 2009. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 27: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E, et al. 2013a. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol 31: 251–258 [DOI] [PubMed] [Google Scholar]
- Kim YK, Wee G, Park J, Kim J, Baek D, Kim JS, Kim VN 2013b. TALEN-based knockout library for human microRNAs. Nat Struct Mol Biol 20: 1458–1464 [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438: 685–689 [DOI] [PubMed] [Google Scholar]
- Li S, Liang Z, Xu L, Zou F 2012a. MicroRNA-21: a ubiquitously expressed pro-survival factor in cancer and other diseases. Mol Cell Biochem 360: 147–158 [DOI] [PubMed] [Google Scholar]
- Li T, Li RS, Li YH, Zhong S, Chen YY, Zhang CM, Hu MM, Shen ZJ 2012b. miR-21 as an independent biochemical recurrence predictor and potential therapeutic target for prostate cancer. J Urol 187: 1466–1472 [DOI] [PubMed] [Google Scholar]
- Liu CZ, Liu W, Zheng Y, Su JM, Li JJ, Yu L, He XD, Chen SS 2012. PTEN and PDCD4 are bona fide targets of microRNA-21 in human cholangiocarcinoma. Chin Med Sci J 27: 65–72 [PubMed] [Google Scholar]
- Liu F, Zheng S, Liu T, Liu Q, Liang M, Li X, Sheyhidin I, Lu X, Liu W 2013. MicroRNA-21 promotes the proliferation and inhibits apoptosis in Eca109 via activating ERK1/2/MAPK pathway. Mol Cell Biochem 381: 115–125 [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall A, Lesnik J, Mukherjee C, Antes T, Sengupta R 2012. Packaging HIV- or FIV-based lentivector expression constructs and transduction of VSV-G pseudotyped viral particles. J Vis Exp 62: e3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. 2011. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, Khayter C, Ramirez CL, Joung JK, Langenau DM 2012. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENS). PLoS One 7: e37877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Choi YS, McManus MT 2010. Analysis of microRNA knockouts in mice. Hum Mol Genet 19: R169–R175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Jeker LT, Carver-Moore K, Oh A, Liu HJ, Cameron R, Richards H, Li Z, Adler D, Yoshinaga Y, et al. 2012. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep 1: 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN 2010. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest 120: 3912–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF 2011. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12: 136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Ousterout DG, Gersbach CA 2012. Advances in targeted genome editing. Curr Opin Chem Biol 16: 268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman SH, Maeder ML, Joung JK, Cathomen T 2011. Zinc-finger nucleases for somatic gene therapy: the next frontier. Hum Gene Ther 22: 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Hosoi S, Woltjen K, Suzuki K, Kashiwagi K, Wada H, Ochiai H, Miyamoto T, Kawai N, Sasakura Y, et al. 2013. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells 18: 315–326 [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F 2012. A transcription activator-like effector toolbox for genome engineering. Nat Protoc 7: 171–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Zhao H 2013. Transcription activator-like effector nucleases (TALENS): a highly efficient and versatile tool for genome editing. Biotechnol Bioeng 110: 1811–1821 [DOI] [PubMed] [Google Scholar]
- Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW 2013. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol 31: 23–24 [DOI] [PubMed] [Google Scholar]
- Svoboda P, Flemr M 2010. The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. EMBO Rep 11: 590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C, Huang G, Ashton C, Wu H, Yan H, Ying QL 2012. Rapid and cost-effective gene targeting in rat embryonic stem cells by TALENS. J Genet Genomics 39: 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhde-Stone C, Gor N, Chin T, Huang J, Lu B 2013. A do-it-yourself protocol for simple transcription activator-like effector assembly. Biol Proced Online 15: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen S, Abreu-Goodger C, Enright AJ 2008. Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods 5: 1023–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu WD, Pan HF, Li JH, Ye DQ 2013. MicroRNA-21 with therapeutic potential in autoimmune diseases. Expert Opin Ther Targets 17: 659–665 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang F, Li X, Baller JA, Qi Y, Starker CG, Bogdanove AJ, Voytas DF 2013. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol 161: 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Sun W, Okano K, Chen Y, Zhang N, Maeda T, Palczewski K 2011. Sponge transgenic mouse model reveals important roles for the microRNA-183 (mir-183)/96/182 cluster in postmitotic photoreceptors of the retina. J Biol Chem 286: 31749–31760 [DOI] [PMC free article] [PubMed] [Google Scholar]