Abstract
Helicobacter pylori (H. pylori) is widely adaptable for colonization in human stomachs in more than half of the world’s population. The microorganism is characterized by an unusual capability of arranging itself in both genotypic and phenotypic ways. Stressing conditions, including antimicrobial agents in sub-inhibitory concentrations, facilitate entering the viable but nonculturable state in which bacterial cells acquire the coccoid form. This morphotype represents an important strategy for bacterial survival in unsuitable conditions and also allows escape from the immune system. H. pylori is capable of forming biofilm outside and inside the host. For the bacterial population, the sessile growth mode represents an ideal environment for gene rearrangement, as it allows the acquiring of important tools aimed to improve bacterial “fitness” and species preservation. Biofilm formation in H. pylori in the human host also leads to recalcitrance to antibiotic treatment, thus hampering eradication. These lifestyle changes of H. pylori allow for a “safe haven” for its survival and persistence according to different ecological niches, and strongly emphasize the need for careful H. pylori surveillance to improve management of the infection.
Keywords: Helicobacter pylori, Viable But Non Culturable state, Route of transmission, Biofilm in vitro, Biofilm in vivo
Core tip: Helicobacter pylori (H. pylori) is a Gram negative bacterium that colonizes the human stomach early in the life of the host and tends to persist. The present review is focused on the general phenomenon of the fickleness in H. pylori and analyses the significance and role of this “chameleon-like” approach to life in the persistence of this fastidious bacterium outside and inside the host.
INTRODUCTION
Helicobacter pylori (H. pylori) is a Gram negative bacterium that colonizes the human stomach early in the life of the host and tends to persist. It is estimated that the bacterium is present in the gastric mucosa of half of the world population, but disease only occurs in about 15% of colonized individuals[1].
Today, H. pylori is recognized as the most common cause of gastritis, peptic, and duodenal ulcers, and is responsible for an increasing incidence of gastric cancer[2-4]. The natural habitat of the microorganism is the mucus layer of the stomach, but it may also need to survive in other environments to become a life-long infection threat[5]. In fact, a large number of studies support evidence of the microorganism in dental plaque (detected by culture and PCR techniques), in houseflies, in human and animal feces[6-10], and in natural environmental waters[11-17]. Therefore, water supplies contaminated by sewage containing fluids or feces from infected people have been considered as a potential route of H. pylori transmission[13,14,18,19].
H. pylori is able to overcome environmental stressed conditions, such as sub-inhibitory concentrations of drugs or non-permissive atmosphere, by entering the viable but nonculturable (VBNC) state, in which the microorganism modifies its morphology from a spiral to coccoid (spherical) form with a loss of cultivability[20,21]. This important strategy of survival is emphasized when bacterial cells organize themselves into microbial communities, establishing a sort of “free multicellularity” forming biofilm[22-26].
Moreover, as a species, H. pylori possesses one of the most fluid genomes within the prokaryotic kingdom[27,28], with many investigators asserting that H. pylori polymorphisms reflect human phylogeography and historical migrations[29-31], as it is virtually impossible to find two identical DNA patterns in microorganisms isolated from different hosts[28,32,33]. Furthermore, a host individual can harbor more than one isolate, which can derive either from a micro-evolutionary change among strains coming from a unique host microorganism or from a multi-strain infection. This typology of colonization may offer a condition for a more efficacious bacterium-host association during long-term harboring colonization[33,34].
The present review is focused on the general phenomenon of the fickleness in H. pylori and analyzes the significance and role of this “chameleon-like” approach to life in the persistence of this fastidious bacterium outside and inside the host.
VBNC STATE: A GENERAL VIEW
Bacteria, when subjected to inauspicious environmental conditions such as an insufficient supply of nutrients, non-permissive temperature, oxygen, or pH conditions, irradiation, or toxic chemicals, can survive by entering the VBNC state[35-37]. In this “survival state” that is well-documented in both Gram negative and Gram positive bacteria (including those of medical interest and widely-recognized in aquatic environments), the bacteria are not detectable by conventional culture techniques, and can undergo changes in morphology[38], cell wall composition[39], gene expression[36], and protein synthesis[40].
This protective condition, first described by the Rita Colwel group[35], represents a viable survival strategy in unsuitable situations that has contributed to the formation of environmental reservoirs of non sporulating bacteria[39].
It has been demonstrated that bacteria in the VBNC state are able to maintain their metabolic activity and pathogenicity, as well as, in some cases, the ability to revert to active re-growth conditions[41-44].
The broad distribution of VBNC cells among bacterial species underlines their significance in the ability to cope with stresses, and also draws attention to their potential risk for human health[37].
VBNC STATE IN H. PYLORI
H. pylori, which can be defined “a master of adaptation”, is able to overcome stressed conditions, such as sub-inhibitory drug concentrations or non-permissive atmospheres which occur outside and inside the host, by entering the VBNC state[45]. This protective state occurs when the microorganism modifies its elongated, spiral morphology to transform into the coccoid morphotype through a U-shaped intermediate form (Figure 1), which results in it becoming unculturable[20]. Thus, H. pylori essentially displays three different cellular types: the spiral cells which grow under optimum conditions for replication and are both virulent and capable of inducing inflammation in experimental models; the viable coccoid forms that are unable to grow on solid media and are characteristically more persistent in the host and environment; and the degenerative spiral and coccal dead forms[21].
Figure 1.
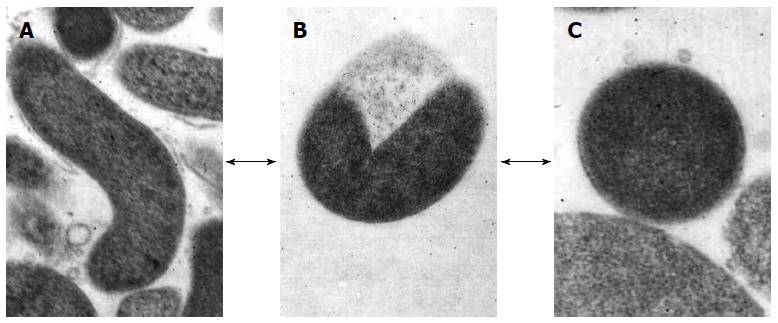
Morphological appearance of Helicobacter pylori. A: Rod-shaped; B: U-shaped; C: Coccoid form. Arrows show the hypothetical alternative pathway between the different forms. Transmission electron microscope: original magnification × 20000.
The conversion into the VBNC state represents an active process in which the microorganism switches on their adaptive machinery as a protection mechanism. In support of this, a study by Costa et al[46] demonstrated that the change in shape in H. pylori was related to its more resistant condition, due to a significant modification in the cell wall which resembled those of endospores. In fact, the peptidoglycan of H. pylori coccoid cells was similar to those of sporulating Bacillus sphaericus.
In another work, Chaput et al[47] demonstrated the ability of H. pylori coccoid cells to escape detection by the immune system because of a significant modification of the cell wall peptidoglycan which had no IL-8 stimulatory activity in gastric epithelial cells. Thus, H. pylori in the VBNC state may be able to escape or modulate the host response and thereby persist in the human stomach.
The capability of viable H. pylori coccoid cells to be persistent outside the host has been demonstrated in many works. In a study by Shahamat et al[48], it was demonstrated that VBNC H. pylori cells could be present for up to 1 year in fresh water. In another work, the authors[49] verified the entrance of H. pylori into a VBNC state as the cells aged in a natural freshwater environment by using viability assays, also confirming that coccoid viable cells continued to transcribe several genes, including those responsible for its virulence.
Regarding this last aspect, Wang et al[50] obtained a coccoid H. pylori population by exposure to sub-inhibitory concentration of antibiotics, with the target fragment of the cagA gene of these cells being amplified and cloned into a plasmid, and then transformed in Escherichia coli. By sequence analysis, the authors demonstrated that coccoid H. pylori contained a completed cagA gene that displayed a homology with the reported original sequence of vegetative forms of H. pylori (99.7%), thus supporting speculation about the pathogenicity of these cells.
The contamination of drinking water by human feces has been suggested as one of the possible routes of H. pylori transmission, and it has been demonstrated that the microorganism is present in the VBNC state in this unsuitable environment[51], meaning that their role in fecal-oral transmission via contaminated water sources cannot be disregarded.
In our study[19], we demonstrated, by Nested-PCR, the presence of H. pylori DNA in seawater both free and bound to zooplanktonic organisms, such as copepod and cladocerans. Considering that the intensive activity of enzymes produced by prokaryotic and eukaryotic cells in seawater favors the instability and degradation of nucleic acids[52], we assumed that the detected nucleic acids were part of viable resistant coccal cells able to survive in marine environments. Indeed, H. pylori was isolated by culture from marine zooplankton, supporting speculation about the potential role of zooplankton in H. pylori survival and transmission[15]. This isolated microorganism, named H. pylori MDC1, harbored a genotype coding for the most important virulence markers and, in particular, contained cagPAI, which could both exert a role in adapting to the marine environment and also be acquired by different species. In this regard, a cagA like gene of H. pylori was found to be present in environmental isolates of Aeromonas spp. from different water samples in India[53].
All of these considerations strongly underline that the morphological fickleness of H. pylori is in response to external stimuli entering the VBNC state, and representing, during the lifespan of the microorganism, a powerful response to improve bacterial “fitness” and species preservation.
MICROBIAL BIOFILM: A GENERAL VIEW
Bacterial biofilms may be considered an ancestral selective event used by prokaryotes to adapt to the environment. In this way, microorganisms are organized in communities that settle and proliferate on biotic and abiotic surfaces embedded in a highly hydrated self-produced matrix constituted of extracellular polymeric substances[54-57].
Bacteria aggregated in biofilm represent a complex dynamic system that could be considered the best program of survival in unsuitable conditions[57]. Many bacterial species match their lifecycle to the human host and environment, and thus change their regulatory processes to adapt to this new niche[54].
It is widely recognized that an ever increasing number of infections arise from biofilm-producing microorganisms that are extremely difficult to eradicate. Infections caused by sessile bacteria are characterized both by a strong tolerance to antimicrobial/biocidal agents and by an extraordinary resistance to phagocytosis, which allows them to evade the hosts’ defenses[58,59]. These processes are thought to be the major contributors to the etiology and the persistence of infectious diseases.
Biofilm growing bacteria represent a major cause of exacerbating chronic infections with persistent inflammation and damage of tissue[60].
Many signals and gene products are involved in biofilm development under a cyclic and dynamic process depending on different bacteria and surfaces[61,62]. Into these microbial communities, bacteria may convey their presence to one another by producing, detecting, and responding to small diffusible signal molecules referred to as autoinducers, which carry out the Quorum-Sensing[63].
Moreover, bacteria organized in a biofilm can find a protected environment to facilitate horizontal gene transfer, thus providing a bacterial population with newly-modified genomes[64].
The biofilm represents an ideal environment for gene rearrangement and also for the horizontal bacteriophage and plasmid transfer that contributes significantly to strain variability and adaptability[65].
BIOFILM FORMATION IN H. PYLORI
It is well known that H. pylori is capable of forming biofilm both outside and inside the human host, which likely provides greater protection under stressful conditions.
The first evidence of biofilm formation in H. pylori was provided by Stark et al[66] in 1999, which characterized the water-insoluble biofilm accumulated at the air/liquid interface of a continuous culture of H. pylori NCTC 11637 by gas chromatography and mass spectrometry. Fucose, glucose, galactose, glycero-manno-heptose, N-acetylglucosamine, and N-acetylmuramic acid were identified, suggesting their role in enhancing resistance to host defense factors, antibiotics, and micro-environmental pH homeostasis, which facilitates the growth and survival of H. pylori in vivo.
When studied on abiotic surfaces, H. pylori forms a structured biofilm with an extracellular polymeric substance (EPS) matrix in which are mixed exogenous DNA fragments (eDNA)[67]. This extracellular DNA (eDNA), detected in the 2 d-old EPS biofilm matrix of H. pylori strains, showed some remarkable differences when compared by RAPD-PCR analysis to the intracellular DNA (iDNA). The different profiles of eDNA and iDNA indicated that lysed cells were not the primary source of eDNA release, which suggested a role in the active dynamic flow of information, such as recombination processes (via transformation), and contributing to the wide genomic variability of this microorganism that has been defined as a “quasi-species”. Moreover, promotion of genetic transfer was studied by our group[27] between two clinical H. pylori strains when grown in the biofilm mode. Two co-cultured H. pylori clinical strains were analyzed for their cooperative/competitive behavior and selected clones, coming from their mixed mature biofilm, were compared through DNA fingerprinting and main virulence factors analysis.
Biofilms developed by mixed H. pylori strains were well-structured, with a higher amount of EPS matrix and viable cells than those detected by the parental strains. Finally, genetic analysis by both RAPD-PCR and cagA (EPIYA motifs)/vacA virulence genes of 45 clones showed a high number of recombinant clones together with the generation of more virulent strains. Thus, these recombinant clones might provide an advantage to the bacterial population by promoting the development of a more adhesive and stable biofilm. These data demonstrated that the biofilms developed by multiple H. pylori strains were more complex and structured than the ones associated with single strains. Such conditions might promote the genetic exchange favored by the protected environment and explain the development, in a single host, of more virulent and difficult to eradicate strains.
In an in vitro study, Cole et al[68] demonstrated the negative effect of mucin on H. pylori biofilm formation, suggesting that in the mucus-rich stomach, H. pylori planktonic growth is favored over biofilm formation. Moreover, these authors found, in the H. pylori luxS mutant, that biofilm formation was affected by overproduction of LuxS, as was observed in a Streptococcus mutans luxS mutant by Merritt et al[69]. In this study, Cole indicated the relative importance of the Quorum-Sensing gene, luxS, and also the cagE type IV secretion gene to the production of biofilms by H. pylori.
However, biofilm production and its characterization are strongly influenced by the different methods and media used for biofilm culture[70]. In a recent study by Bessa et al[24], the authors reported on the important influence of culture media on H. pylori growth, both in its free-living and biofilm growth modes. In particular, they suggested that the adherence and ability of H. pylori to form biofilm were not accomplished by the same mechanism in different media.
Finally, they demonstrated that sub-Minimal Inhibitory Concentration (MIC) values of amoxicillin and clarithromycin could increase biofilm biomass. The sub-MIC drug influence on H. pylori biofilm-forming capability may have clinical consequences, as during any antibiotic treatment focused to a particular infection, H. pylori bacteria can be exposed to sub-MICs of antibiotics, which constitute a condition that can stimulate the switching from planktonic to sessile cells forming biofilm, and consequently lead to recalcitrance to antibiotic treatment, and thus hampering eradication.
Similar results were obtained by Yonezawa et al[71] that displayed the increasing of biofilm biomass after various concentration of clarithromycin treatment. They also demonstrated that biofilm-forming capability in H. pylori affects the generation of clarithromycin resistance with the presence of a point mutation at positions 2142 and 2143 in the domain V loop of the 23 rRNA gene more frequently detected in sessile cells than their planktonic counterparts.
These conclusions strongly underline that biofilm formation can affect the generation of antibiotic resistance mutations in H. pylori.
The first evidence of an ex vivo H. pylori biofilm was raised by the Carron group[25,26]. They showed, via Scanning Electron Microscopy (SEM) analysis, the presence of dense, mature H. pylori biofilm detectable in urease H. pylori positive biopsy specimens that were absent in urease-negative controls. Of the patients who tested urease positive for H. pylori, the average percentage of total surface area covered by biofilms was 97.3%. Those testing negative had average surface area coverage of only 1.64%. This study demonstrated that, compared with controls, urease-positive specimens have significant biofilm formation, whereas urease-negative specimens have little to none. This was reflected in the significantly-increased biofilm surface density in urease positive specimens compared with urease-negative controls.
The dynamic behavior of H. pylori in the colonization of human gastric mucosa was investigated in patients previously treated for H. pylori infection by us[23]. In our study, biopsy samples were taken and analyzed for H. pylori detection by cultural, molecular, and ultra-structural methods. Viable H. pylori cells were isolated in 33% of performed cultures, whereas the expression of the glmM constitutive gene and the Quorum-Sensing related luxS gene were detected in 90% of the analyzed biopsies. In these positive cases, the analysis of glmM and luxS sequences confirmed H. pylori identity. The SEM analysis of biopsies coming from patients harboring culturable bacteria revealed a prevalent “S-shape” H. pylori morphotype co-existent with coccoid aggregated bacteria embedded in abundant matrix; samples coming from H. pylori positive patients showed clustered coccoid bacteria arranged in a microbial biofilm only through molecular method (Figure 2).
Figure 2.
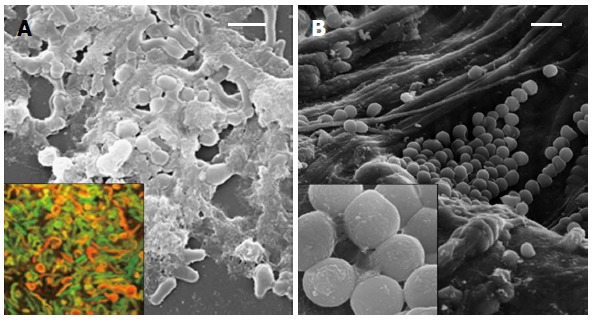
Biofilm of Helicobacter pylori. A: Scanning electron micrograph of mature biofilm on a polystyrene surface with rod shaped and coccoid cells embedded in an abundant matrix. Insert: Confocal laser scanning microscopy image of mature in vitro biofilm with viable (green) and dead (red) cells, live/dead staining; B: Clusters of coccoid Helicobacter pylori cells in the gastric mucosa also embedded in a matrix (insert). Bars represent 5 μm.
The undoubted clinical significance of coccoid H. pylori cells in epithelial gastric cells[72], also described in cases of adenocarcinoma[73], alone or grouped in clusters, underlines the need for planning of more efficacious testing protocols, such as RT-PCR methodology, to avoid underestimating H. pylori colonization by identifying camouflaged and protected clustered bacteria, and taking into account this serious microbial problem in medicine in the recommendation of therapeutic regimens.
CONCLUSION
H. pylori, more than other microorganisms, displays an amazing adaptive ability when confronted with stress conditions.
The viable coccoid morphotypes able to retain virulence factors and the aggregative behavior among H. pylori cells growing as a biofilm suggest a long-term survival of these bacterial communities outside and inside the host, enabling bacterial transmission with important clinical repercussions. In particular, these new living conditions, consisting of new self-organized populations, guarantee persistence, genetic variability, and antimicrobial resistance, as well as prolonging protection. For successful therapy, it may be essential not only to eliminate the bacillary forms but also to rapidly suppress and/or destroy the coccoid forms that are clustered in biofilm as well[74,75].
A recent study suggested a new effective treatment for the demolition of H. pylori biofilm which includes in the therapeutic regimen N-acetylcysteine (NAC), a mucolytic agent used in medical practice for the treatment of patients with chronic respiratory diseases[76]. In a clinical trial[77], the authors obtained a significantly higher percentage of H. pylori eradication (65% vs 20%) in patients with at least 4 treatment eradication failures by using NAC pretreatment prior to a culture guided antibiotic regimen. N-acetylcysteine may act by disrupting the biofilm agent and favoring the planktonic growth mode of H. pylori, thus overcoming the tolerance phenomenon described for bacterial biofilms[78].
Novel therapeutic regimens including plant extracts[79] or substances capable of inhibiting or destabilizing the formation of H. pylori biofilm should be explored to improve management of the infection.
Footnotes
P- Reviewers: Asahina K, Gharaee-Kermani M S- Editor: Ma YJ L- Editor: Rutherford A E- Editor: Liu XM
References
- 1.Höcker M, Hohenberger P. Helicobacter pylori virulence factors--one part of a big picture. Lancet. 2003;362:1231–1233. doi: 10.1016/S0140-6736(03)14547-3. [DOI] [PubMed] [Google Scholar]
- 2.Malfertheiner P. The intriguing relationship of Helicobacter pylori infection and acid secretion in peptic ulcer disease and gastric cancer. Dig Dis. 2011;29:459–464. doi: 10.1159/000332213. [DOI] [PubMed] [Google Scholar]
- 3.Calvet X, Ramírez Lázaro MJ, Lehours P, Mégraud F. Diagnosis and epidemiology of Helicobacter pylori infection. Helicobacter. 2013;18 Suppl 1:5–11. doi: 10.1111/hel.12071. [DOI] [PubMed] [Google Scholar]
- 4.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Biological agents. Volume 100 B. A review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum. 2012;100:1–441. [PMC free article] [PubMed] [Google Scholar]
- 5.Sasaki K, Tajiri Y, Sata M, Fujii Y, Matsubara F, Zhao M, Shimizu S, Toyonaga A, Tanikawa K. Helicobacter pylori in the natural environment. Scand J Infect Dis. 1999;31:275–279. doi: 10.1080/00365549950163572. [DOI] [PubMed] [Google Scholar]
- 6.Grübel P, Hoffman JS, Chong FK, Burstein NA, Mepani C, Cave DR. Vector potential of houseflies (Musca domestica) for Helicobacter pylori. J Clin Microbiol. 1997;35:1300–1303. doi: 10.1128/jcm.35.6.1300-1303.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsonnet J, Shmuely H, Haggerty T. Fecal and oral shedding of Helicobacter pylori from healthy infected adults. JAMA. 1999;282:2240–2245. doi: 10.1001/jama.282.23.2240. [DOI] [PubMed] [Google Scholar]
- 8.Kabir S. Review article: clinic-based testing for Helicobacter pylori infection by enzyme immunoassay of faeces, urine and saliva. Aliment Pharmacol Ther. 2003;17:1345–1354. doi: 10.1046/j.1365-2036.2003.01577.x. [DOI] [PubMed] [Google Scholar]
- 9.Boyanova L, Panov V, Yordanov D, Gergova G, Mitov I. Characterization of oral Helicobacter pylori strain by 4 methods. Diagn Microbiol Infect Dis. 2013;77:287–288. doi: 10.1016/j.diagmicrobio.2013.06.030. [DOI] [PubMed] [Google Scholar]
- 10.Sepúlveda E, Moreno J, Spencer ML, Quilodrán S, Brethauer U, Briceño C, García A. [Comparison of Helicobacter pylori in oral cavity and gastric mucosa according to virulence genotype (cagA and vacA m 1)] Rev Chilena Infectol. 2012;29:278–283. doi: 10.4067/S0716-10182012000300005. [DOI] [PubMed] [Google Scholar]
- 11.Hulten K, Han SW, Enroth H, Klein PD, Opekun AR, Gilman RH, Evans DG, Engstrand L, Graham DY, El-Zaatari FA. Helicobacter pylori in the drinking water in Peru. Gastroenterology. 1996;110:1031–1035. doi: 10.1053/gast.1996.v110.pm8612990. [DOI] [PubMed] [Google Scholar]
- 12.Baker KH, Hegarty JP. Presence of Helicobacter pylori in drinking water is associated with clinical infection. Scand J Infect Dis. 2001;33:744–746. doi: 10.1080/003655401317074536. [DOI] [PubMed] [Google Scholar]
- 13.Park SR, Mackay WG, Reid DC. Helicobacter sp. recovered from drinking water biofilm sampled from a water distribution system. Water Res. 2001;35:1624–1626. doi: 10.1016/s0043-1354(00)00582-0. [DOI] [PubMed] [Google Scholar]
- 14.Lu Y, Redlinger TE, Avitia R, Galindo A, Goodman K. Isolation and genotyping of Helicobacter pylori from untreated municipal wastewater. Appl Environ Microbiol. 2002;68:1436–1439. doi: 10.1128/AEM.68.3.1436-1439.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cellini L, Di Campli E, Grande R, Di Bartolomeo S, Prenna M, Pasquantonio MS, Pane L. Detection of Helicobacter pylori associated with zooplankton. Aquatic Microb Ecol. 2005;40:115–120. [Google Scholar]
- 16.Bahrami AR, Rahimi E, Ghasemian Safaei H. Detection of Helicobacter pylori in city water, dental units’ water, and bottled mineral water in Isfahan, Iran. ScientificWorldJournal. 2013;2013:280510. doi: 10.1155/2013/280510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellack NR, Koehoorn MW, MacNab YC, Morshed MG. A conceptual model of water’s role as a reservoir in Helicobacter pylori transmission: a review of the evidence. Epidemiol Infect. 2006;134:439–449. doi: 10.1017/S0950268806006005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKeown I, Orr P, Macdonald S, Kabani A, Brown R, Coghlan G, Dawood M, Embil J, Sargent M, Smart G, et al. Helicobacter pylori in the Canadian arctic: seroprevalence and detection in community water samples. Am J Gastroenterol. 1999;94:1823–1829. doi: 10.1111/j.1572-0241.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 19.Cellini L, Del Vecchio A, Di Candia M, Di Campli E, Favaro M, Donelli G. Detection of free and plankton-associated Helicobacter pylori in seawater. J Appl Microbiol. 2004;97:285–292. doi: 10.1111/j.1365-2672.2004.02307.x. [DOI] [PubMed] [Google Scholar]
- 20.Cellini L, Robuffo I, Di Campli E, Di Bartolomeo S, Taraborelli T, Dainelli B. Recovery of Helicobacter pylori ATCC43504 from a viable but not culturable state: regrowth or resuscitation? APMIS. 1998;106:571–579. [PubMed] [Google Scholar]
- 21.Andersen LP, Rasmussen L. Helicobacter pylori-coccoid forms and biofilm formation. FEMS Immunol Med Microbiol. 2009;56:112–115. doi: 10.1111/j.1574-695X.2009.00556.x. [DOI] [PubMed] [Google Scholar]
- 22.Cammarota G, Sanguinetti M, Gallo A, Posteraro B. Review article: biofilm formation by Helicobacter pylori as a target for eradication of resistant infection. Aliment Pharmacol Ther. 2012;36:222–230. doi: 10.1111/j.1365-2036.2012.05165.x. [DOI] [PubMed] [Google Scholar]
- 23.Cellini L, Grande R, Di Campli E, Traini T, Di Giulio M, Lannutti SN, Lattanzio R. Dynamic colonization of Helicobacter pylori in human gastric mucosa. Scand J Gastroenterol. 2008;43:178–185. doi: 10.1080/00365520701675965. [DOI] [PubMed] [Google Scholar]
- 24.Bessa LJ, Grande R, Di Iorio D, Di Giulio M, Di Campli E, Cellini L. Helicobacter pylori free-living and biofilm modes of growth: behavior in response to different culture media. APMIS. 2013;121:549–560. doi: 10.1111/apm.12020. [DOI] [PubMed] [Google Scholar]
- 25.Carron MA, Tran VR, Sugawa C, Coticchia JM. Identification of Helicobacter pylori biofilms in human gastric mucosa. J Gastrointest Surg. 2006;10:712–717. doi: 10.1016/j.gassur.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 26.Coticchia JM, Sugawa C, Tran VR, Gurrola J, Kowalski E, Carron MA. Presence and density of Helicobacter pylori biofilms in human gastric mucosa in patients with peptic ulcer disease. J Gastrointest Surg. 2006;10:883–889. doi: 10.1016/j.gassur.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Grande R, Di Campli E, Di Bartolomeo S, Verginelli F, Di Giulio M, Baffoni M, Bessa LJ, Cellini L. Helicobacter pylori biofilm: a protective environment for bacterial recombination. J Appl Microbiol. 2012;113:669–676. doi: 10.1111/j.1365-2672.2012.05351.x. [DOI] [PubMed] [Google Scholar]
- 28.Suerbaum S. Genetic variability within Helicobacter pylori. Int J Med Microbiol. 2000;290:175–181. doi: 10.1016/S1438-4221(00)80087-9. [DOI] [PubMed] [Google Scholar]
- 29.Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 31.Kersulyte D, Mukhopadhyay AK, Velapatiño B, Su W, Pan Z, Garcia C, Hernandez V, Valdez Y, Mistry RS, Gilman RH, et al. Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol. 2000;182:3210–3218. doi: 10.1128/jb.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cellini L, Di Campli E, Di Candia M, Marzio L. Molecular fingerprinting of Helicobacter pylori strains from duodenal ulcer patients. Lett Appl Microbiol. 2003;36:222–226. doi: 10.1046/j.1472-765x.2003.01295.x. [DOI] [PubMed] [Google Scholar]
- 33.Cellini L, Grande R, Di Campli E, Di Bartolomeo S, Capodicasa S, Marzio L. Analysis of genetic variability, antimicrobial susceptibility and virulence markers in Helicobacter pylori identified in Central Italy. Scand J Gastroenterol. 2006;41:280–287. doi: 10.1080/00365520510024223. [DOI] [PubMed] [Google Scholar]
- 34.Patra R, Chattopadhyay S, De R, Ghosh P, Ganguly M, Chowdhury A, Ramamurthy T, Nair GB, Mukhopadhyay AK. Multiple infection and microdiversity among Helicobacter pylori isolates in a single host in India. PLoS One. 2012;7:e43370. doi: 10.1371/journal.pone.0043370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu HS, Roberts N, Singleton FL, Attwell RW, Grimes DJ, Colwell RR. Survival and viability of nonculturableEscherichia coli andVibrio cholerae in the estuarine and marine environment. Microb Ecol. 1982;8:313–323. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]
- 36.Trevors JT. Viable but non-culturable (VBNC) bacteria: Gene expression in planktonic and biofilm cells. J Microbiol Methods. 2011;86:266–273. doi: 10.1016/j.mimet.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 37.Pinto D, Santos MA, Chambel L. Thirty years of viable but nonculturable state research: Unsolved molecular mechanisms. Crit Rev Microbiol. 2013:Epub ahead of print. doi: 10.3109/1040841X.2013.794127. [DOI] [PubMed] [Google Scholar]
- 38.Takeda Y. Vibrio parahaemolyticus, enterotoxigenic Escherichia coli, enterohemorrhagic Escherichia coli and Vibrio cholerae. Proc Jpn Acad Ser B Phys Biol Sci. 2011;87:1–12. doi: 10.2183/pjab.87.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Signoretto C, Burlacchini G, Lleò MM, Pruzzo C, Zampini M, Pane L, Franzini G, Canepari P. Adhesion of Enterococcus faecalis in the nonculturable state to plankton is the main mechanism responsible for persistence of this bacterium in both lake and seawater. Appl Environ Microbiol. 2004;70:6892–6896. doi: 10.1128/AEM.70.11.6892-6896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rahman I, Shahamat M, Kirchman PA, Russek-Cohen E, Colwell RR. Methionine uptake and cytopathogenicity of viable but nonculturable Shigella dysenteriae type 1. Appl Environ Microbiol. 1994;60:3573–3578. doi: 10.1128/aem.60.10.3573-3578.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lleò MM, Bonato B, Tafi MC, Signoretto C, Boaretti M, Canepari P. Resuscitation rate in different enterococcal species in the viable but non-culturable state. J Appl Microbiol. 2001;91:1095–1102. doi: 10.1046/j.1365-2672.2001.01476.x. [DOI] [PubMed] [Google Scholar]
- 42.Cellini L, Allocati N, Angelucci D, Iezzi T, Di Campli E, Marzio L, Dainelli B. Coccoid Helicobacter pylori not culturable in vitro reverts in mice. Microbiol Immunol. 1994;38:843–850. doi: 10.1111/j.1348-0421.1994.tb02136.x. [DOI] [PubMed] [Google Scholar]
- 43.Su X, Chen X, Hu J, Shen C, Ding L. Exploring the potential environmental functions of viable but non-culturable bacteria. World J Microbiol Biotechnol. 2013;29:2213–2218. doi: 10.1007/s11274-013-1390-5. [DOI] [PubMed] [Google Scholar]
- 44.Senoh M, Ghosh-Banerjee J, Ramamurthy T, Colwell RR, Miyoshi S, Nair GB, Takeda Y. Conversion of viable but nonculturable enteric bacteria to culturable by co-culture with eukaryotic cells. Microbiol Immunol. 2012;56:342–345. doi: 10.1111/j.1348-0421.2012.00440.x. [DOI] [PubMed] [Google Scholar]
- 45.Cellini L, Allocati N, Di Campli E, Dainelli B. Helicobacter pylori: a fickle germ. Microbiol Immunol. 1994;38:25–30. doi: 10.1111/j.1348-0421.1994.tb01740.x. [DOI] [PubMed] [Google Scholar]
- 46.Costa K, Bacher G, Allmaier G, Dominguez-Bello MG, Engstrand L, Falk P, de Pedro MA, García-del Portillo F. The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J Bacteriol. 1999;181:3710–3715. doi: 10.1128/jb.181.12.3710-3715.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaput C, Ecobichon C, Cayet N, Girardin SE, Werts C, Guadagnini S, Prévost MC, Mengin-Lecreulx D, Labigne A, Boneca IG. Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog. 2006;2:e97. doi: 10.1371/journal.ppat.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shahamat M, Mai U, Paszko-Kolva C, Kessel M, Colwell RR. Use of autoradiography to assess viability of Helicobacter pylori in water. Appl Environ Microbiol. 1993;59:1231–1235. doi: 10.1128/aem.59.4.1231-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams BL, Bates TC, Oliver JD. Survival of Helicobacter pylori in a natural freshwater environment. Appl Environ Microbiol. 2003;69:7462–7466. doi: 10.1128/AEM.69.12.7462-7466.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang KX, Wang XF. Cloning and sequencing of cagA gene fragment of Helicobacter pylori with coccoid form. World J Gastroenterol. 2004;10:3511–3513. doi: 10.3748/wjg.v10.i23.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mishra S, Singh V, Rao GR, Jain AK, Dixit VK, Gulati AK, Nath G. Detection of Helicobacter pylori in stool specimens: comparative evaluation of nested PCR and antigen detection. J Infect Dev Ctries. 2008;2:206–210. doi: 10.3855/jidc.264. [DOI] [PubMed] [Google Scholar]
- 52.Huston AL, Krieger-Brockett BB, Deming JW. Remarkably low temperature optima for extracellular enzyme activity from Arctic bacteria and sea ice. Environ Microbiol. 2000;2:383–388. doi: 10.1046/j.1462-2920.2000.00118.x. [DOI] [PubMed] [Google Scholar]
- 53.Datta S, Khan A, Nandy RK, Rehman M, Sinha S, Chattopadhyay S, Das SC, Nair GB. Environmental isolates of Aeromonas spp. harboring the cagA-like gene of Helicobacter pylori. Appl Environ Microbiol. 2003;69:4291–4295. doi: 10.1128/AEM.69.7.4291-4295.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watnick P, Kolter R. Biofilm, city of microbes. J Bacteriol. 2000;182:2675–2679. doi: 10.1128/jb.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wimpenny J, Manz W, Szewzyk U. Heterogeneity in biofilms. FEMS Microbiol Rev. 2000;24:661–671. doi: 10.1111/j.1574-6976.2000.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 56.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jefferson KK. What drives bacteria to produce a biofilm? FEMS Microbiol Lett. 2004;236:163–173. doi: 10.1016/j.femsle.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 58.Lewis K. Riddle of biofilm resistance. Antimicrob Agents Chemother. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 60.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 61.Stanley NR, Lazazzera BA. Environmental signals and regulatory pathways that influence biofilm formation. Mol Microbiol. 2004;52:917–924. doi: 10.1111/j.1365-2958.2004.04036.x. [DOI] [PubMed] [Google Scholar]
- 62.Azevedo NF, Pinto AR, Reis NM, Vieira MJ, Keevil CW. Shear stress, temperature, and inoculation concentration influence the adhesion of water-stressed Helicobacter pylori to stainless steel 304 and polypropylene. Appl Environ Microbiol. 2006;72:2936–2941. doi: 10.1128/AEM.72.4.2936-2941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li YH, Tian X. Quorum sensing and bacterial social interactions in biofilms. Sensors (Basel) 2012;12:2519–2538. doi: 10.3390/s120302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ehrlich GD, Ahmed A, Earl J, Hiller NL, Costerton JW, Stoodley P, Post JC, DeMeo P, Hu FZ. The distributed genome hypothesis as a rubric for understanding evolution in situ during chronic bacterial biofilm infectious processes. FEMS Immunol Med Microbiol. 2010;59:269–279. doi: 10.1111/j.1574-695X.2010.00704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol. 2012;65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 66.Stark RM, Gerwig GJ, Pitman RS, Potts LF, Williams NA, Greenman J, Weinzweig IP, Hirst TR, Millar MR. Biofilm formation by Helicobacter pylori. Lett Appl Microbiol. 1999;28:121–126. doi: 10.1046/j.1365-2672.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- 67.Grande R, Di Giulio M, Bessa LJ, Di Campli E, Baffoni M, Guarnieri S, Cellini L. Extracellular DNA in Helicobacter pylori biofilm: a backstairs rumour. J Appl Microbiol. 2011;110:490–498. doi: 10.1111/j.1365-2672.2010.04911.x. [DOI] [PubMed] [Google Scholar]
- 68.Cole SP, Harwood J, Lee R, She R, Guiney DG. Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol. 2004;186:3124–3132. doi: 10.1128/JB.186.10.3124-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Merritt J, Qi F, Goodman SD, Anderson MH, Shi W. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect Immun. 2003;71:1972–1979. doi: 10.1128/IAI.71.4.1972-1979.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams JC, McInnis KA, Testerman TL. Adherence of Helicobacter pylori to abiotic surfaces is influenced by serum. Appl Environ Microbiol. 2008;74:1255–1258. doi: 10.1128/AEM.01958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yonezawa H, Osaki T, Hanawa T, Kurata S, Ochiai K, Kamiya S. Impact of Helicobacter pylori biofilm formation on clarithromycin susceptibility and generation of resistance mutations. PLoS One. 2013;8:e73301. doi: 10.1371/journal.pone.0073301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu ZF, Chen CY, Tang W, Zhang JY, Gong YQ, Jia JH. Gene-expression profiles in gastric epithelial cells stimulated with spiral and coccoid Helicobacter pylori. J Med Microbiol. 2006;55:1009–1015. doi: 10.1099/jmm.0.46456-0. [DOI] [PubMed] [Google Scholar]
- 73.Chan WY, Hui PK, Leung KM, Chow J, Kwok F, Ng CS. Coccoid forms of Helicobacter pylori in the human stomach. Am J Clin Pathol. 1994;102:503–507. doi: 10.1093/ajcp/102.4.503. [DOI] [PubMed] [Google Scholar]
- 74.Berry V, Jennings K, Woodnutt G. Bactericidal and morphological effects of amoxicillin on Helicobacter pylori. Antimicrob Agents Chemother. 1995;39:1859–1861. doi: 10.1128/aac.39.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Figura N, Moretti E, Vaglio L, Langone F, Vernillo R, Vindigni C, Giordano N. Factors modulating the outcome of treatment for the eradication of Helicobacter pylori infection. New Microbiol. 2012;35:335–340. [PubMed] [Google Scholar]
- 76.Zuin R, Palamidese A, Negrin R, Catozzo L, Scarda A, Balbinot M. High-dose N-acetylcysteine in patients with exacerbations of chronic obstructive pulmonary disease. Clin Drug Investig. 2005;25:401–408. doi: 10.2165/00044011-200525060-00005. [DOI] [PubMed] [Google Scholar]
- 77.Cammarota G, Branca G, Ardito F, Sanguinetti M, Ianiro G, Cianci R, Torelli R, Masala G, Gasbarrini A, Fadda G, et al. Biofilm demolition and antibiotic treatment to eradicate resistant Helicobacter pylori: a clinical trial. Clin Gastroenterol Hepatol. 2010;8:817–820.e3. doi: 10.1016/j.cgh.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 78.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 79.Amin M, Anwar F, Naz F, Mehmood T, Saari N. Anti-Helicobacter pylori and urease inhibition activities of some traditional medicinal plants. Molecules. 2013;18:2135–2149. doi: 10.3390/molecules18022135. [DOI] [PMC free article] [PubMed] [Google Scholar]


