Abstract
Oral antiviral agents have been developed in the last two decades for the treatment of chronic hepatitis B (CHB). However, antiviral resistance remains an important challenge for long-term CHB therapy. All of the clinically available oral antiviral agents are nucleoside or nucleotide analogues that target the activity of viral reverse transcriptase (RT), and all are reported to have resistant mutations. Since the hepatitis B virus (HBV) RT, like other viral polymerases, lacks proofreading activity, the emergence of drug-resistance occurs readily under selective pressure from the administration of antiviral agents. The molecular diagnosis of drug-resistant HBV is based on sequence variations, and current diagnostic methods include sequencing, restriction fragment polymorphism analysis, and hybridization. Here, we will discuss the currently available molecular diagnosis tools, in vitro phenotypic assays for validation of drug-resistant HBV, and treatment options for drug-resistant HBV.
Keywords: Hepatitis B virus, Drug-resistance, Molecular diagnosis, Antiviral treatment, Chronic hepatitis B
Core tip: Although several antiviral agents have been developed in the last two decades for the treatment of chronic hepatitis B (CHB), antiviral resistance remains an important challenge for long-term CHB therapy. In this review, we discussed the currently available molecular diagnosis tools, in vitro phenotypic assays for validation of drug-resistant hepatitis B virus (HBV), and treatment options for drug-resistant HBV.
INTRODUCTION
Chronic hepatitis B (CHB) affects 240 million people worldwide and is a leading cause of liver-related morbidity and mortality[1-3]. The last two decades have seen the introduction of oral antiviral agents for the treatment of hepatitis B virus (HBV) infection[4-8]. Long-term antiviral therapy is needed in the majority of patients, and incomplete viral suppression and emergence of drug resistance is a major concern[9]. All of the clinically available HBV drugs are nucleoside or nucleotide analogues that inhibit the activity of viral reverse transcriptase (RT), and all drugs approved as anti-HBV agents are reported to have viral resistance due to specific mutations in the RT domain[10]. In treatment-naïve patients, the prevalence of such mutations is low and routine mutation analysis is not recommended[11]. The emergence of resistant strains is due to the selective pressure of the therapeutic regimen, although other factors, such as host immune response and therapy adherence, also play a role[12]. The development of antiviral resistance is one of the most important factors predicting the success or failure of CHB treatment[13]. The emergence of antiviral resistance results in the resumption of active viral replication that has been previously suppressed after the initiation of antiviral therapy, and can impair biochemical or histologic improvement[14]. Furthermore, increasing use of antiviral agents for CHB has led to a greater likelihood of antiviral resistance[12]. A recent prospective cohort study showed that antiviral drug resistance increased the risk of hepatocellular carcinoma in decompensated HBV-related cirrhotic patients, especially in those failed rescue therapy[15]. To avoid development of drug resistance, the current guidelines recommend tenofovir (TDF) and entecavir (ETV) as first-line antiviral agents[13,16-18].
In this review, we will discuss methods of diagnosis, phenotypic assays for drug resistance, and treatments for drug-resistant HBV strains.
MOLECULAR MECHANISM OF ANTIVIRAL RESISTANCE
The HBV RT, like other viral polymerases, lacks proofreading activity on the newly synthesized viral genome, resulting in the introduction of random mutations into progeny HBV DNA. The error rate of HBV polymerase is approximately 1 per 105 to 107 base syntheses due to the highly error-prone nature of the HBV RT[19]. Under selective pressure from the administration of antiviral agents, HBV quasispecies converge on a dominant HBV mutant that can escape selection pressure, producing a drug-resistant HBV strain.
Since the molecular mechanisms and clinical significance of resistance to HBV nucleoside/nucleotide analogue drugs have been extensively reviewed in literature[10,20-22], detailed explanations of the documented resistant mutants will not be reviewed here.
Lamivudine (LMV), a synthetic nucleoside analogue with activity against HBV and HIV, is sequentially phosphorylated to LMV triphosphate by cellular kinases and incorporated into the growing HBV DNA at the 3′-end by HBV polymerase, which induces premature chain termination. The primary resistance mutation to LMV is the rtM204I/V in the YMDD motif. This mutation is usually accompanied by a compensatory mutation including rtL180M, L80I/V, and V173L, which enhances the viral replication of replication-defective rtM204I/V mutants (Figure 1). LMV resistance does not confer cross-resistance to adefovir (ADV) or TDF[20,21].
Figure 1.
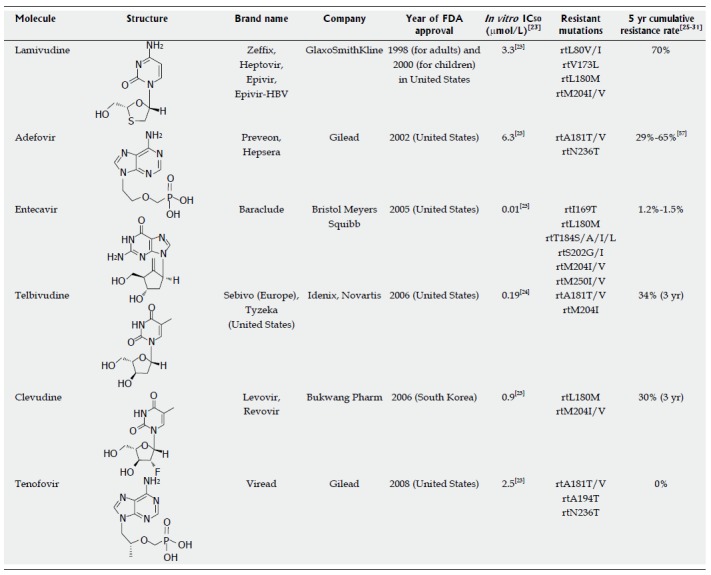
Approved anti-hepatitis B virus drugs and their resistant mutations. IC50 values are dependent on the duration of drug exposure to cells, the cells used, and the protocol used.
ADV dipivoxil, an analogue of adenosine monophosphate, can be easily phosphorylated by cellular kinases to the active metabolite ADV diphosphate, which inhibits HBV DNA polymerase by competing with the natural substrate deoxyadenosine triphosphate. The incorporation of ADV diphosphate into the growing viral DNA causes premature DNA chain termination similar to LMV. Genotypic analysis has revealed that ADV resistance is conferred by the rtN236T and/or rtA181T/V mutations. In vitro drug susceptibility assays showed that the rtN236T mutation does not affect sensitivity to LMV, telbivudine (LdT), or ETV; however, the rtA181T mutation was shown to decrease susceptibility to LMV (< 10-fold), ADV (2- to 8-fold), and TDF (2- to 3-fold)[32].
ETV, a guanosine nucleoside, is efficiently phosphorylated to the active triphosphate form. By competing with the natural substrate deoxyguanosine triphosphate, ETV triphosphate functionally inhibits the activities of HBV polymerase. ETV is the most potent among the currently available anti-HBV agents. The mutations associated with primary resistance to ETV are the most complex and have not been fully established in patients. Mutations associated with the emergence of ETV resistance have been mapped to the B domain (rtI169T, rtL180M, and rtS184S/A/I/L/G/C/M), C domain (rtM204I/V and rtS202G/I), and E domain (rtM250I/V). ETV resistance does not confer cross-resistance to ADV or TDF[22].
LdT, a synthetic thymidine nucleoside, is the unmodified L-isomer of the naturally occurring nucleoside, thymidine; therefore, phosphorylation to the active LdT triphosphate form by cellular kinases is easily accomplished. The LdT 5′-triphosphate eventually inhibits HBV DNA polymerase by competing with the natural substrate, thymidine 5′-triphosphate. The rtM204I substitution confers primary resistance to LdT treatment and frequently co-occurs with the rtL80I/V and rtL180M substitutions. An in vitro study revealed that LdT resistance does not confer cross-resistance to ADV, TDF, or ETV[33].
Clevudine (CLV), a pyrimidine analogue, inhibits HBV polymerase by competing with the natural substrate, thymidine. CLV inhibits the DNA-dependent DNA activity of HBV polymerase, as well as reverse transcription and priming. Since CLV is a fluorinated LdT, it has a similar resistance profile to LMV and LdT. Kwon et al[23] identified the rtM204I substitution as the most common mutation during viral breakthrough in four CLV-failure patients, whereas rtL229V was shown to be a compensatory mutation for the impaired replication of the rtM204I mutant. A quadruple mutant (rtL129M+rtV173L+rtM204I+rtH337N) conferred greater replicative ability and strong resistance to both CLV and LMV[23].
TDF disoproxil fumarate is a methyl derivative of ADV with activity against retroviruses, including HIV-1/2 and HBV. TDF demonstrates a mechanism of action and antiviral resistance pattern very similar to ADV. Like ADV, TDF is rapidly metabolized by cellular kinases into the active metabolite, TDF diphosphate. In turn, TDF diphosphate inhibits HBV DNA RT by competing with the natural substrate, deoxyadenosine triphosphate, causing the termination of the growing HBV DNA. In vitro drug susceptibility assay demonstrated that the rtA194T, rtA181T/V, and/or rtN236T mutations are associated with TDF resistance[22]. Resistance to TDF has been proven so far in in vitro studies.
MOLECULAR DIAGNOSIS OF DRUG-RESISTANT HBV
The molecular diagnosis of drug-resistant HBV is performed by genotypic assay, which determines the resistance-related mutations in the viral RT gene by comparing the patient-derived viral strain with wild-type. To date, several methods have been developed and clinically used to determine genotypic resistance. These methods include sequencing (PCR-based direct sequencing, cloning and sequencing, and ultra-deep pyrosequencing), restriction fragment length or mass polymorphism (RFLP or RFMP), and hybridization (DNA microarray and line-probe assay). Since the conventional methods for HBV genotyping have been extensively reviewed in literature[20,22], only those technologies that have received lesser attention elsewhere, such as ultra-deep pyrosequencing and RFMP, will be reviewed here in detail.
PCR-based direct or cloning sequencing
Currently, genotypic analysis is largely performed by sequencing-based assays. Only sequencing can provide all of the information on the mutations present in a viral genome. Therefore, this assay is useful to identify novel resistant mutations responsible for the insufficiency of new antiviral drugs[23]. However, limited by sensitivity, this assay can only detect the majority species present in the total viral population and is generally capable of detecting quasispecies comprising more than 20 percent of a viral population. This limitation can be overcome by multiple rounds of cloning followed by sequencing. This method is impractical for use in large cohort studies or clinical laboratories since the process is hard to standardize, labor-intensive, and time-consuming. However, cloning-based sequencing is the only method available to analyze the colocalization of mutations within the same HBV genome.
Ultra-deep pyrosequencing
Recently, ultra-deep pyrosequencing, a next-generation sequencing technology, was successfully applied to detect HBV quasispecies and also to identify drug-resistant mutants that present at very low concentrations in patient sera[34-37]. This is one of the most sensitive techniques capable of detecting minority virus populations that are less than 0.1% of the total, and that are typically missed by direct or cloning-based sequencing. However, application of this technology in determining drug resistance is limited by its high cost. The one advantage of this next-generation sequencing technique is the production of vast quantities of sequence data without prior information on the sequence of interest. Very recently, Rodriguez et al[37] generated approximately 480000 sequences (4010 ± 843 sequences per sample) from ADV-resistant patient sera by ultra-deep pyrosequencing. They found that the dynamics of ADV-resistant viral populations are very complex and more heterogeneous than expected. More importantly, the identified ADV-resistant variants (including rtA181 and rtN236) were already present as minor populations at baseline in most of the treatment-naïve patients who subsequently developed viral DNA breakthrough to ADV therapy.
Restriction fragment mass polymorphism
RFMP technology is based on detection of the mass difference in DNA fragments resulting from drug-resistant mutations in the RT gene[38]. Mass spectrometry generates precise information of the molecular mass of the analytes and enables quantitation of both strands of DNA in parallel using a fully automated procedure. Mass spectrometry directly assesses the nature of the PCR products, whereas other technologies indirectly measure the PCR products either through hybridization or by sequencing reactions.
The assay is based on the amplification and mass detection of oligonucleotides excised using a type IIS enzyme digestion and matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) as depicted in Figure 2. PCR is performed with primers designed to introduce a type IIS restriction endonuclease recognition sequence ahead of the polymorphism site. The use of a type IIS restriction enzyme means that this assay does not depend on the fortuitous occurrence of restriction sites, because these enzymes typically have cleavage sites distal to their recognition sites. The enzymatic cleavage of the reaction products leads to excision of multiple oligonucleotide fragments containing the mutated motifs, after which the masses of the resulting oligonucleotide fragments are examined by MALDI-TOF MS. Differences are observed as the presence, absence, or mass change of peaks corresponding to fragments affected by the existence of polymorphisms, including substitutions, deletions, and insertions in the RT gene[38]. In addition to its speed and high-throughput capacity, the RFMP assay is very sensitive and can detect drug-resistant mutants that constitute less than 1% of a total virus population, enabling distinctions between mixed-genotype samples[39]. A clear correlation has been observed between estimated peak heights and real proportions in mixed-genotype pools, indicating the RFMP assay enables better quantitative detection of mixed populations without the need for population-based cloning and subsequent sequencing. By combining the merits of unique assay chemistry and the established techniques of MALDI-TOF MS, the RFMP assay is able to screen for viral mutants in a robust, high-throughput manner and is known to be capable of simultaneously analyzing 384 samples in 3 h, which is almost 10 times faster than existing methods[40]. The improved sensitivity of the RFMP assay has allowed for its application in monitoring early intervention and prevention in antiviral therapy for HBV. An RFMP analysis of YMDD motifs within 740 consecutive samples collected from 116 hepatitis B patients demonstrated that YMDD mutants occur throughout the course of LMV therapy irrespective of the occurrence of viral DNA breakthrough. This indicates that the mere detection of YMDD mutants is not sufficient to predict viral DNA breakthrough, although the presence of YMDD mutants has been associated with a high incidence of viral DNA breakthrough, and a five-fold predominance of YMDD mutant-to-wild-type virus was significantly associated with viral DNA breakthrough. Periodic testing by RFMP assay was shown to be useful in detecting the predominance of YMDD mutants for monitoring drug resistance, enabling early intervention and prevention of viral breakthrough[40].
Figure 2.
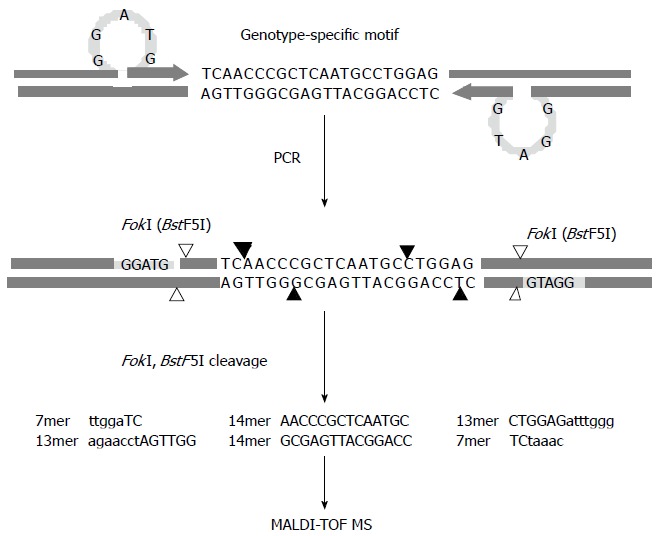
Schematic diagram of the restriction fragment mass polymorphism genotyping strategy. Polymerase chain reaction is performed with primers designed to introduce a type IIS restriction endonuclease recognition sequence (FokI) ahead of the genotype-specific motifs upon amplification. The enzymatic cleavage of the products leads to excision of multiple oligonucleotide fragments representing the motifs shown in capital letters, and then the masses of the resulting oligonucleotide fragments are examined by MALDI-TOF MS. Cleavage sites of FokI and BstF5I, an isoschizomer of FokI, are indicated by filled and blank arrows, respectively, and recognition sites for both restriction endonucleases are specified by the shaded bars.
Multiplex RFMP methods were developed to have practical advantages over existing methods, enabling the simultaneous detection of several resistant mutations within a large number of samples, and to address the dynamics and evolution of resistance and the relationship between viral genotypes and clinical outcomes[41,42]. This assay has proven to be reliable for clinical virus genotyping in concert with regular measurements of HBV viral load, for the early detection of LMV, ADV, ETV, and MDR mutations, and for the timely introduction of new salvage agents[43-45]. However, RFMP technology cannot provide information on the colocalization of mutations within the same HBV genome and requires access to a mass spectrometer.
Hybridization-based assays
Hybridization-based assays rely on affinity differences between amplified nucleotides harboring mutations and wild-type viral sequences. These techniques include the line-probe (LiPA) assay[46] and DNA microarray[47]. The commercially available LiPA assay (Innogenetics, Belgium) can detect single nucleotide mismatches and discriminate HBV resistance mutations within minor fractions constituting 5% or more of the total viral population[39,46]. DNA chip microarrays can simultaneously detect multiple resistant mutations with relatively low labor and cost[47].
Although hybridization techniques are generally reliable, the main limitations of all hybridization-based assays arise largely from their relatively low specificity and from the fact that they are qualitative rather than quantitative. Moreover, the design and optimization of new sets of specific probes are required for every mutant in order to detect a single nucleotide change because mutations in neighboring regions affect the sensitivity of the target sequence.
IN VITRO PHENOTYPIC ASSAY FOR VALIDATION OF DRUG-RESISTANT HBV
Only an in vitro phenotypic assay can confirm genotypic antiviral resistance. However, current methodology is labor-intensive and time consuming due to the need for construction of a replication-competent HBV replicon and the use of Southern blot analysis. Although the detailed methods for the validation of drug-resistant HBV may vary slightly based on approach, the basic concept is identical[48-50]. A typical approach to analyze phenotypic resistance is presented in Figure 3. The HBV genome is isolated from patient sera, and the RT mutation sequence is analyzed. To assess whether the mutations are colocalized within the same HBV genome and to select a specific RT mutant more easily, the PCR products are subcloned into a T-vector. After sequence analysis, the selected RT mutants are once again cloned into an HBV replicon such as an HBV 1.1mer, 1.2mer, 1.5mer, or dimer. For this procedure, the XhoI and NcoI restriction sites are preferentially used for insertion into both the T-vector and the replicon. The replicons are then transfected into Huh7 or HepG2 hepatocyte cell lines. Secreted HBeAg is considered to normalize transfection efficiency. Secreted HBsAg cannot be used as a control since the RT and surface genes overlap, and, thus, mutations in the RT gene can affect the antigenicity of HBsAg. Four to five days after treatment with antiviral drugs, cell lysates are subject to Southern blot analysis, which gives the most accurate result. Since drug potency varies, the preferred drug concentrations are indicated in Figure 3. After quantification of replication ability using a PhosphorImager imaging system, the IC50 (μmol/L) for each drug is calculated by interpolation. Finally, the fold resistance (R factor) is determined. By doing this, it is possible to determine the presence of drug-resistant mutations in the quasispecies contained in clinical isolates. To identify the sequence elements responsible for drug-resistance, artificial replicons that harbor conserved mutations in clinical isolates need to be constructed and tested for drug susceptibility. An example of the overall characterization of drug-resistant HBV was recently reported, including detailed methods[23]. Alternatively, real-time PCR can be used to measure replication ability, an approach that is suitable for automated and large-scale testing in a hospital setting[51].
Figure 3.
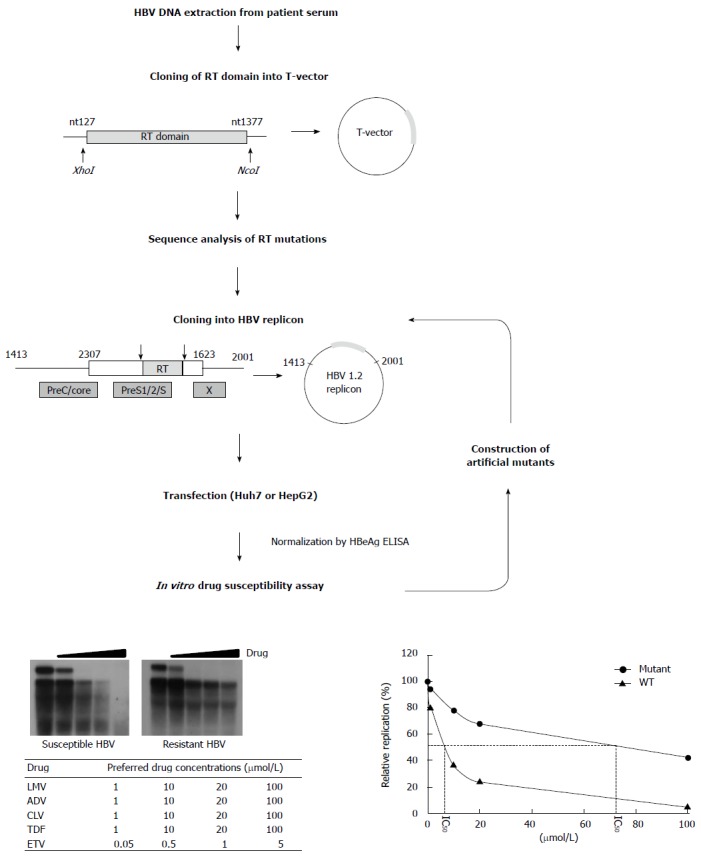
Scheme for in vitro phenotypic validation of drug-resistant hepatitis B virus. Hepatitis B virus (HBV) DNA is purified from patient serum, and the sequence of RT mutations is analyzed. After cloning into replication-competent HBV replicons, each mutant is transfected into hepatoma cell lines followed by Southern blot (or real time polymerase chain reaction) analysis. The IC50 (μmol/L) value is obtained by quantification of replication ability and curve-fitting. To characterize the specific mutation(s) conferring resistance to antiviral drugs, each artificial mutant must be constructed and individually tested.
TREATMENT
The development of drug resistance is associated with virological breakthrough, biochemical breakthrough, and, sometimes, hepatic decompensation[52]. Furthermore, resistance may also reverse histological improvement and oppose the reduction in disease progression among patients with advanced fibrosis and early cirrhosis[53]. LMV is not recommended as a first-line treatment due to high resistance development. Currently, the newer and potent drugs, TDF and ETV are recommended[13,16-18]. Several practice guidelines suggest strategies for treatment of CHB patients with resistance. The American Association for the Study of Liver Diseases guidelines were published in 2009[18]. The updated guidelines from the European Association for the Study of the Liver, the Asian Pacific Association for the Study of the Liver, and the Korean Association for the Study of the Liver were published in 2012[13,16,17]. The main principle of these guidelines is to choose antiviral agents without cross-resistance and to begin rescue therapy as soon as possible[13,16,17]. These guidelines are compared and summarized in Table 1.
Table 1.
Guidelines for treatment of resistance
| Resistant drug | AASLD 2009[18] | EASL 2012[16] | APASL 2012[17] | KASL 2012[13] |
| LMV | Add ADV or TDF Stop LMV, switch to Truvada (TDF + FTC) | Switch to TDF Add ADV (If TDF not available) | Add ADV Switch to TDF Switch to IFN-based therapy | Add ADV or TDF Stop LMV, switch to ADV or TDF + other nucleoside analogues Switch to TDF Stop LMV, consider to switch with Peg IFN |
| ADV | Add LMV Stop ADV, switch to Truvada Stop ADV, switch to or add ETV | NA-naïve; Switch to ETV or TDF Prior LMV-resistance; Switch to TDF + a nucleoside analogue | Add LMV, LdT, or ETV Switch to TDF Switch to IFN-based therapy | With prior LMV resistance, -Stop ADV, switch to TDF + other nucleoside analogues -Add ETV 1mg ADV as a first-line therapy -Stop ADV, switch to TDF + other nucleoside analogues -Add other nucleoside analogue, if rtA181T, add ETV |
| ETV | Switch to TDF or Truvada | Switch to TDF Add TDF Add ADV (If TDF not available) | Add TDF or ADV Switch to IFN-based therapy | Add nucleotide analogue |
| LdT | Same treatment as LMV | Switch to TDF Add TDF Add ADV (If TDF not available) | Same treatment as LMV | Same treatment as LMV |
| CLV | Same treatment as LMV | |||
| TDF | Add ETV, LdT, LMV or FTC (LMV-naïve; Switch to ETV/ prior LMV-resistance; Add ETV) | |||
| Multidrug-resistance | Combination of a nucleoside and a nucleotide (preferably TDF) | ETV + TDF Switch to IFN-based therapy | TDF + ETV 1 mg ADV + ETV 1 mg | |
| Comment | Update is required especially for multidrug-resistance | Recommendation about IFN-based therapy is limited | Recommendation about TDF resistance is limited | Recommendation about TDF resistance and IFN-based therapy is limited |
KASL: Korean Association for the Study of the Liver; AASLD: American Association for the Study of Liver Diseases; APASL: Asian Pacific Association for the Study of the Liver; EASL: European Association for the Study of the Liver; LMV: Lamivudine; ADV: Adefovir; LdT: Telbivudine; CLV: Clevudine; TDF: Tenofovir; FTC: Emtricitabine; ETV: Entecavir; IFN: Interferon.
LMV, LdT, and CLV resistance
LMV, LdT, and CLV are L-nucleoside analogues, and mutations at rtM204 are considered the primary cause of resistance to these agents[20,54,55]. rtM204I or rtM204V, with or without rtL180M mutations, are sensitive to ADV and TDF, but exhibit cross-resistance to ETV and show an eight-fold decrease in sensitivity. The rtA181T mutation has been detected in 5% of LMV-resistant patients. These mutants exhibit cross-resistance to ADV but remain sensitive to ETV[56].
Of the various forms of antiviral drug resistance, treatment for LMV resistance has been the most widely studied. The incidence of the rtM204I/V substitution has reportedly increased from 24% in 1 year to 70% in 5 years[25,54]. ADV monotherapy is not recommended due to the increased risk of ADV resistance, which has been shown to manifest in 18% of patients at 1 year, 25% at 2 years, and up to 65% after 5 years[57-59]. ADV add-on therapy is accepted as a first-line rescue therapy[60]. Many studies have shown that LMV-ADV combination therapy is superior to ADV monotherapy[61-63]. However, the LMV-resistant strain rtA181T has been reported to be continuously detected even after combination therapy with LMV-ADV, so caution is necessary to avoid the possibility of multidrug-resistant HBV[32,64,65]. LdT and ADV combination therapy, or a combination of ETV and ADV, are possible treatment options[66-68].
TDF has shown potent antiviral activity against LMV-resistant HBV and is reportedly superior to ADV monotherapy[69-71]. Therefore, treatment strategies that include TDF seem to be more effective than those involving ADV[13]. One study showed that switching to TDF is as effective as adding TDF to LMV[71]. However, there is one report of TDF resistance in an LMV-resistant CHB patient with HIV co-infection who received TDF monotherapy, so the efficacy of TDF monotherapy requires further evaluation[72]. One recent study also found that the reduction in HBV DNA levels was greater in a TDF-LMV combination therapy group than in TDF monotherapy, ADV monotherapy, or LMV-ADV combination groups[73].
ETV exhibits some cross-resistance with LMV, which has prompted the administration of 1.0 mg doses to LMV-resistant CHB patients[74]. However, ETV resistance was reportedly more frequent than in treatment-naïve patients. A 5-year cumulative genotypic resistance rate of 51% was reported, along with an accompanying virologic breakthrough of 43%[27]. A recent study found that LMV-ADV combination therapy showed superior antiviral efficacy over ETV 1.0 mg monotherapy in LMV-resistant CHB patients[75]. For these reasons, ETV 1.0 mg monotherapy is not recommended.
Peginterferon (PegIFN) is another option for the treatment of LMV-resistant CHB patients. A randomized control trial showed that 48 wk of PegIFN alfa-2a treatment for LMV-resistant CHB patients achieved undetectable HBV DNA levels in 10.6% of patients[76]. Another study showed similar efficacy of PegIFN alfa-2a in a comparison between treatment-naïve patients and LMV-resistant patients with HBeAg-positive CHB[77].
Few data related to LdT and CLV resistance are available. The 2-year risk of LdT resistance was shown to be 25.1% in HBeAg-positive patients and 10.8% in HBeAg-negative patients[78]. In the subgroup that had no genotypic resistance at year 2 and received LdT until year 3, an incremental 1.0% of HBeAg-positive, and 2.1% of HBeAg-negative patients developed genotypic resistance to LdT[79]. The cumulative resistance rate of LdT is 34% after 3 years[29]. In the case of LdT resistance, a switch to, or addition of TDF may be the preferred option[20]. The CLV resistance rate is approximately 20% and 30% in the 2nd and 3rd years of treatment, respectively[30,80]. According to clinical experience with LMV-resistant CHB patients, the general principles for treating LdT or CLV resistance are similar to those of LMV-resistance[13,17].
ADV and TDF resistance
ADV and TDF are nucleotide analogues. rtN236T and rtA181T/V are the primary mutations giving rise to ADV resistance[26,81]. The cumulative incidence of genotypic resistance to ADV is reported to be 0%, 3%, 11%, 18%, and 29% at the end of each successive year of therapy in HBeAg-negative patients[26]. As previously mentioned, ADV monotherapy in LMV-resistant patients leads to a 5-year cumulative ADV resistance of 65.6%[57]. No TDF resistance has been reported during treatment periods up to 3 years in length[82]. However, rtA181T/V and rtN236T mutations confer intermediate resistance to TDF[9]. Among patients who have the rtA181T/V and/or rtN236T substitutions, viral suppression by TDF is reduced[71,83]. rtA194T can decrease susceptibility to TDF by 10-fold in the presence of rtL180M+rtM204V mutations, according to a case study of a patient with HBV and HIV co-infection[72].
TDF significantly suppresses HBV replication in patients exhibiting LMV resistance who have failed to respond adequately to ADV, and in patients who are resistant to both LMV and ADV[83]. TDF alone or TDF plus emtricitabine (FTC) are similarly effective in ADV-treated CHB patients[84,85]. However, reduced sensitivity to TDF has been demonstrated in ADV-resistant HBV infections, indicating potential cross-resistance[71]. The addition of FTC led to a further decrease in serum HBV DNA levels in patients exhibiting ADV resistance and a suboptimal response to TDF therapy[86]. When LMV-TDF combination therapy was given to CHB patients who had previously failed to respond to LMV and subsequent ADV therapy, 64% achieved an undetectable level of HBV DNA after 96 wk of treatment[83].
ETV does not share cross resistance with ADV[56]. ETV has been shown to be effective against both rtA181T/V and rtN236T mutant HBV strains[32,43,87,88]. Switching to ETV monotherapy (1 mg daily) is initially effective in LMV-resistant patients (rtM204I/V), but the subsequent risk of ETV resistance is high[27]. Another study also showed increased ETV resistance risk with ETV monotherapy switching for ADV resistance[89]. In contrast, ADV-ETV combination therapy has been shown to be a better option[13,90]. The ETV-TDF combination can also be considered for multidrug-resistant HBV infections that include ADV resistance[13].
Although rtN236T mutants remain sensitive to LMV, the rtA181T/V mutant exhibits reduced susceptibility to LMV[56] Therefore, LMV-ADV combination therapy is recommended. For similar reasons, LdT or CLV monotherapy is not recommended for the rtA181T/V mutant in order to avoid cross-resistance[13,56], and add-on therapy is preferred[13,17]. One study showed that the LdT and ADV combination can be a good option for ADV resistance[91].
To date, there are no reports of resistance to TDF among patients with CHB monoinfection[31,92-94]. An in vitro study found that replication of the rtA194T mutant was suppressed effectively by ETV and intermediately by LdT[95]. The clinical impact of the rtA194T mutation is still unknown[92,96]. One recent report suggested that rtP177G and rtF249A mutants also reduce susceptibility to TDF[97]. In cases of confirmed TDF resistance, an add-on combination with a nucleoside analogue is preferred, while a switch to ETV may be sufficient if the patient had no prior LMV resistance[16].
ETV resistance
ETV is cyclopentene, a type of nucleoside analogue. ETV resistance develops via a two-hit mechanism, as previously described. Since this presents a high genetic barrier to ETV resistance, the resistance rate in treatment-naïve subjects is very low. In studies of the long-term follow-up of ETV treatment in CHB patients, the cumulative probability of ETV resistance was reportedly 1.2%-1.5% after 5 years of ETV treatment. However, a resistance rate as high as 51% has been reported after 5 years of treatment in LMV-refractory subjects[27,28,88].
ETV-resistant HBV maintains susceptibility to ADV, which could be considered an initial treatment option, and a clinical case has indicated that ADV can be effective in suppressing the ETV-resistant mutant[98,99]. Adding ADV to ETV would be more reasonable for reducing ADV resistance and improving antiviral efficacy[13,90,100]. Combination therapy of ADV plus LMV could be considered as another option, since a small study showed that the short-term efficacy of this combination was similar to that of combination therapy of ADV plus ETV[13]. Although TDF has not been fully evaluated in the treatment of ETV resistance, it is expected to be very effective since TDF does not show cross-resistance to ETV in vitro and has excellent potency[71]. Therefore, switching to or adding TDF may be preferred for addressing ETV resistance[20].
Multidrug resistance
Multidrug resistance (MDR) is defined as resistance to two or more classes of antiviral drugs[13]. The emergence of MDR is increasing and has raised serious concerns regarding antiviral therapy because it limits the selection of appropriate therapy[45]. Sequential monotherapy is associated with the development of MDR[22,98,101,102]. In these situations, pre-existing antiviral resistant mutations may reappear and become co-localized with newly developed resistant mutations in the same viral genome[98] The development of a triple-drug resistant (LMV, ADV, and ETV) HBV strain has been reported in LMV- and ADV-resistant patients after sequential ETV administration[103].
In patients with MDR, genotypic resistance testing is very useful, and a combination of a nucleoside and a nucleotide should be employed[16]. TDF-ETV combination therapy can be considered in case of resistance to both LMV and ADV[17,20]. A recent study showed that rescue therapy with TDF plus ETV achieved undetectable HBV DNA after a median of 6 mo in 51 (89.5%) of 57 patients, in whom nucleoside/nucleotide analogue therapy (LMV+ADV, ETV+ADV, or TDF+LMV) had failed and who had multidrug-resistant rtA181T/V or other MDR mutations[104].
If TDF is not available, combination therapy with ADV plus ETV is another option[13,90,100,105]. A recently published study showed that the ADV plus ETV combination is superior to the LMV-ADV combination or to ETV monotherapy for multidrug-resistant CHB patients[106]. On the other hand, the LMV plus ADV combination is usually insufficient for treatment due to low antiviral potency[45].
If resistant mutations to LMV, ETV, and ADV are detected in the same patient, combination therapy with TDF plus ETV may be the best option[13].
CONCLUSION AND PERSPECTIVES
Although ETV and TDF are powerful antiviral drugs with a high barrier to resistance, treatment failure due to resistance remains possible, especially among treatment-experienced CHB patients. The mechanisms of drug resistance have been evaluated, and many relevant diagnostic tools have been developed. Current CHB treatment guidelines suggest practical and effective strategies for resistance. However, there are still several treatment-related issues that need further evaluation. First, new diagnostic tools and in vitro phenotypic validation methods for drug-resistant HBV need to be developed that can identify mutations more efficiently, with greater accuracy, and with reduced cost. Second, the role of IFN must be evaluated further. A few studies suggest the possibility of PegIFN as a treatment option for LMV-resistant CHB patients. The efficacy of PegIFN in patients with resistance to other agents such as LdT, CLV, ADV, ETV, TDF, and MDR is not yet known. New IFN agents are also under development for hepatitis C virus treatment. These must also be studied for use in drug-resistant HBV infection. Third, the efficacy of new HBV agents in resistance treatment should be evaluated. For example, LB80830 is a new acyclic nucleotide phosphonate with chemistry similar to ADV and TDF. In a phase II, open-label, multicenter study among 65 LMV-resistant patients, a dose-dependent reduction in HBV DNA of up to -3.92 log copies/mL was observed at week 12 at the optimal dose of 150 mg daily[107]. These findings clearly need further evaluation. Finally, treatment strategies for MDR should be established. Most guidelines suggest combination therapies of nucleoside and nucleotide analogues, and the ETV plus TDF combination is preferred. However, clinical data on long-term efficacy is still lacking. Furthermore, there are patients who have experienced treatment failure without a known or confirmed genotypic resistance. No treatment guidelines have been suggested for these patients. If more sensitive diagnostic tools are developed, the novel combinations of drug-resistant mutations may be diagnosed in these patients, which can give more treatment options for these patients.
ACKNOWLEDGMENTS
We appreciate the valuable comments of Dr. SP Hong (GeneMatrix, South Korea).
Footnotes
Supported by Konkuk University
P- Reviewers: Abenavoli L, Al Olaby R, Ampuero J, Gatselis NK, Ikura Y, Komatsu H, Liula J, Tijera MFH, Wong GLH, Yonem O S- Editor: Gou SX L- Editor: A E- Editor: Zhang DN
References
- 1.Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11:97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- 2.Lai CL, Yuen MF. Chronic hepatitis B--new goals, new treatment. N Engl J Med. 2008;359:2488–2491. doi: 10.1056/NEJMe0808185. [DOI] [PubMed] [Google Scholar]
- 3.Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212–2219. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 4.Hadziyannis SJ, Tassopoulos NC, Heathcote EJ, Chang TT, Kitis G, Rizzetto M, Marcellin P, Lim SG, Goodman Z, Wulfsohn MS, et al. Adefovir dipivoxil for the treatment of hepatitis B e antigen-negative chronic hepatitis B. N Engl J Med. 2003;348:800–807. doi: 10.1056/NEJMoa021812. [DOI] [PubMed] [Google Scholar]
- 5.Lai CL, Chien RN, Leung NW, Chang TT, Guan R, Tai DI, Ng KY, Wu PC, Dent JC, Barber J, et al. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. N Engl J Med. 1998;339:61–68. doi: 10.1056/NEJM199807093390201. [DOI] [PubMed] [Google Scholar]
- 6.Lai CL, Gane E, Liaw YF, Hsu CW, Thongsawat S, Wang Y, Chen Y, Heathcote EJ, Rasenack J, Bzowej N, et al. Telbivudine versus lamivudine in patients with chronic hepatitis B. N Engl J Med. 2007;357:2576–2588. doi: 10.1056/NEJMoa066422. [DOI] [PubMed] [Google Scholar]
- 7.Lai CL, Shouval D, Lok AS, Chang TT, Cheinquer H, Goodman Z, DeHertogh D, Wilber R, Zink RC, Cross A, et al. Entecavir versus lamivudine for patients with HBeAg-negative chronic hepatitis B. N Engl J Med. 2006;354:1011–1020. doi: 10.1056/NEJMoa051287. [DOI] [PubMed] [Google Scholar]
- 8.Yuen MF, Lai CL. Treatment of chronic hepatitis B: Evolution over two decades. J Gastroenterol Hepatol. 2011;26 Suppl 1:138–143. doi: 10.1111/j.1440-1746.2010.06545.x. [DOI] [PubMed] [Google Scholar]
- 9.Zoulim F, Locarnini S. Management of treatment failure in chronic hepatitis B. J Hepatol. 2012;56 Suppl 1:S112–S122. doi: 10.1016/S0168-8278(12)60012-9. [DOI] [PubMed] [Google Scholar]
- 10.Kim KH, Kim ND, Seong BL. Discovery and development of anti-HBV agents and their resistance. Molecules. 2010;15:5878–5908. doi: 10.3390/molecules15095878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vutien P, Trinh HN, Garcia RT, Nguyen HA, Levitt BS, Nguyen K, da Silveira E, Daugherty T, Ahmed A, Garcia G, et al. Mutations in HBV DNA Polymerase Associated With Nucleos(t)ide Resistance Are Rare in Treatment-naive Patients. Clin Gastroenterol Hepatol. 2013:Epub ahead of print. doi: 10.1016/j.cgh.2013.11.036. [DOI] [PubMed] [Google Scholar]
- 12.Baxa DM, Thekdi AD, Golembieski A, Krishnan PV, Sharif O, Kizy A, Shetron-Rama L, Jovanovich J, Chappell BJ, Snow-Lampart A, et al. Evaluation of anti-HBV drug resistant mutations among patients with acute symptomatic hepatitis B in the United States. J Hepatol. 2013;58:212–216. doi: 10.1016/j.jhep.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Korean Association for the Study of the Liver. KASL Clinical Practice Guidelines: Management of chronic hepatitis B. Clin Mol Hepatol. 2012;18:109–162. doi: 10.3350/cmh.2012.18.2.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zoulim F, Durantel D, Deny P. Management and prevention of drug resistance in chronic hepatitis B. Liver Int. 2009;29 Suppl 1:108–115. doi: 10.1111/j.1478-3231.2008.01939.x. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Liu W, Chen YH, Fan CL, Dong PL, Wei FL, Li B, Chen DX, Ding HG. Antiviral drug resistance increases hepatocellular carcinoma: a prospective decompensated cirrhosis cohort study. World J Gastroenterol. 2013;19:8373–8381. doi: 10.3748/wjg.v19.i45.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.European Association For The Study Of The Liver. EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167–185. doi: 10.1016/j.jhep.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Liaw YF, Kao JH, Piratvisuth T, Chan HLY, Chien RN, Liu CJ, Gane E, Locarnini S, Lim SG, Han KH, et al. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2012 update. Hepatol Int. 2012;6:531–561. doi: 10.1007/s12072-012-9365-4. [DOI] [PubMed] [Google Scholar]
- 18.Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661–662. doi: 10.1002/hep.23190. [DOI] [PubMed] [Google Scholar]
- 19.Girones R, Miller RH. Mutation rate of the hepadnavirus genome. Virology. 1989;170:595–597. doi: 10.1016/0042-6822(89)90455-8. [DOI] [PubMed] [Google Scholar]
- 20.Lok AS, Zoulim F, Locarnini S, Bartholomeusz A, Ghany MG, Pawlotsky JM, Liaw YF, Mizokami M, Kuiken C. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology. 2007;46:254–265. doi: 10.1002/hep.21698. [DOI] [PubMed] [Google Scholar]
- 21.Ghany M, Liang TJ. Drug targets and molecular mechanisms of drug resistance in chronic hepatitis B. Gastroenterology. 2007;132:1574–1585. doi: 10.1053/j.gastro.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 22.Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593–608.e1-2. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]
- 23.Kwon SY, Park YK, Ahn SH, Cho ES, Choe WH, Lee CH, Kim BK, Ko SY, Choi HS, Park ES, et al. Identification and characterization of clevudine-resistant mutants of hepatitis B virus isolated from chronic hepatitis B patients. J Virol. 2010;84:4494–4503. doi: 10.1128/JVI.02066-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryant ML, Bridges EG, Placidi L, Faraj A, Loi AG, Pierra C, Dukhan D, Gosselin G, Imbach JL, Hernandez B, et al. Antiviral L-nucleosides specific for hepatitis B virus infection. Antimicrob Agents Chemother. 2001;45:229–235. doi: 10.1128/AAC.45.1.229-235.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao GB, Zhu M, Cui ZY, Wang BE, Yao JL, Zeng MD. A 7-year study of lamivudine therapy for hepatitis B virus e antigen-positive chronic hepatitis B patients in China. J Dig Dis. 2009;10:131–137. doi: 10.1111/j.1751-2980.2009.00375.x. [DOI] [PubMed] [Google Scholar]
- 26.Hadziyannis SJ, Tassopoulos NC, Heathcote EJ, Chang TT, Kitis G, Rizzetto M, Marcellin P, Lim SG, Goodman Z, Ma J, et al. Long-term therapy with adefovir dipivoxil for HBeAg-negative chronic hepatitis B for up to 5 years. Gastroenterology. 2006;131:1743–1751. doi: 10.1053/j.gastro.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 27.Tenney DJ, Rose RE, Baldick CJ, Pokornowski KA, Eggers BJ, Fang J, Wichroski MJ, Xu D, Yang J, Wilber RB, et al. Long-term monitoring shows hepatitis B virus resistance to entecavir in nucleoside-naïve patients is rare through 5 years of therapy. Hepatology. 2009;49:1503–1514. doi: 10.1002/hep.22841. [DOI] [PubMed] [Google Scholar]
- 28.Chang TT, Lai CL, Kew Yoon S, Lee SS, Coelho HS, Carrilho FJ, Poordad F, Halota W, Horsmans Y, Tsai N, et al. Entecavir treatment for up to 5 years in patients with hepatitis B e antigen-positive chronic hepatitis B. Hepatology. 2010;51:422–430. doi: 10.1002/hep.23327. [DOI] [PubMed] [Google Scholar]
- 29.Seto WK, Lai CL, Fung J, Wong DK, Yuen JC, Hung IF, Yuen MF. Significance of HBV DNA levels at 12 weeks of telbivudine treatment and the 3 years treatment outcome. J Hepatol. 2011;55:522–528. doi: 10.1016/j.jhep.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Choung BS, Kim IH, Jeon BJ, Lee S, Kim SH, Kim SW, Lee SO, Lee ST, Kim DG. Long-term treatment efficacy and safety of clevudine therapy in naïve patients with chronic hepatitis B. Gut Liver. 2012;6:486–492. doi: 10.5009/gnl.2012.6.4.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitrinos KM, Corsa A, Liu Y, Flaherty J, Snow-Lampart A, Marcellin P, Borroto-Esoda K, Miller MD. No detectable resistance to tenofovir disoproxil fumarate after 6 years of therapy in patients with chronic hepatitis B. Hepatology. 2014;59:434–442. doi: 10.1002/hep.26686. [DOI] [PubMed] [Google Scholar]
- 32.Villet S, Pichoud C, Billioud G, Barraud L, Durantel S, Trépo C, Zoulim F. Impact of hepatitis B virus rtA181V/T mutants on hepatitis B treatment failure. J Hepatol. 2008;48:747–755. doi: 10.1016/j.jhep.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 33.Seifer M, Patty A, Serra I, Li B, Standring DN. Telbivudine, a nucleoside analog inhibitor of HBV polymerase, has a different in vitro cross-resistance profile than the nucleotide analog inhibitors adefovir and tenofovir. Antiviral Res. 2009;81:147–155. doi: 10.1016/j.antiviral.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Ko SY, Oh HB, Park CW, Lee HC, Lee JE. Analysis of hepatitis B virus drug-resistant mutant haplotypes by ultra-deep pyrosequencing. Clin Microbiol Infect. 2012;18:E404–E411. doi: 10.1111/j.1469-0691.2012.03951.x. [DOI] [PubMed] [Google Scholar]
- 35.Solmone M, Vincenti D, Prosperi MC, Bruselles A, Ippolito G, Capobianchi MR. Use of massively parallel ultradeep pyrosequencing to characterize the genetic diversity of hepatitis B virus in drug-resistant and drug-naive patients and to detect minor variants in reverse transcriptase and hepatitis B S antigen. J Virol. 2009;83:1718–1726. doi: 10.1128/JVI.02011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margeridon-Thermet S, Shulman NS, Ahmed A, Shahriar R, Liu T, Wang C, Holmes SP, Babrzadeh F, Gharizadeh B, Hanczaruk B, et al. Ultra-deep pyrosequencing of hepatitis B virus quasispecies from nucleoside and nucleotide reverse-transcriptase inhibitor (NRTI)-treated patients and NRTI-naive patients. J Infect Dis. 2009;199:1275–1285. doi: 10.1086/597808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez C, Chevaliez S, Bensadoun P, Pawlotsky JM. Characterization of the dynamics of hepatitis B virus resistance to adefovir by ultra-deep pyrosequencing. Hepatology. 2013;58:890–901. doi: 10.1002/hep.26383. [DOI] [PubMed] [Google Scholar]
- 38.Hong SP, Kim NK, Hwang SG, Chung HJ, Kim S, Han JH, Kim HT, Rim KS, Kang MS, Yoo W, et al. Detection of hepatitis B virus YMDD variants using mass spectrometric analysis of oligonucleotide fragments. J Hepatol. 2004;40:837–844. doi: 10.1016/j.jhep.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Kim HS, Han KH, Ahn SH, Kim EO, Chang HY, Moon MS, Chung HJ, Yoo W, Kim SO, Hong SP. Evaluation of methods for monitoring drug resistance in chronic hepatitis B patients during lamivudine therapy based on mass spectrometry and reverse hybridization. Antivir Ther. 2005;10:441–449. [PubMed] [Google Scholar]
- 40.Lee CH, Kim SO, Byun KS, Moon MS, Kim EO, Yeon JE, Yoo W, Hong SP. Predominance of hepatitis B virus YMDD mutants is prognostic of viral DNA breakthrough. Gastroenterology. 2006;130:1144–1152. doi: 10.1053/j.gastro.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Han KH, Hong SP, Choi SH, Shin SK, Cho SW, Ahn SH, Hahn JS, Kim SO. Comparison of multiplex restriction fragment mass polymorphism and sequencing analyses for detecting entecavir resistance in chronic hepatitis B. Antivir Ther. 2011;16:77–87. doi: 10.3851/IMP1702. [DOI] [PubMed] [Google Scholar]
- 42.Choe WH, Hong SP, Kim BK, Ko SY, Jung YK, Kim JH, Yeon JE, Byun KS, Kim KH, Ji SI, et al. Evolution of hepatitis B virus mutation during entecavir rescue therapy in patients with antiviral resistance to lamivudine and adefovir. Antivir Ther. 2009;14:985–993. doi: 10.3851/IMP1417. [DOI] [PubMed] [Google Scholar]
- 43.Shim JH, Suh DJ, Kim KM, Lim YS, Lee HC, Chung YH, Lee YS. Efficacy of entecavir in patients with chronic hepatitis B resistant to both lamivudine and adefovir or to lamivudine alone. Hepatology. 2009;50:1064–1071. doi: 10.1002/hep.23145. [DOI] [PubMed] [Google Scholar]
- 44.Woo HY, Park H, Kim BI, Jeon WK, Cho YK, Kim YJ. Comparison of mass spectrometric analysis and TRUGENE HBV genotyping for monitoring lamivudine resistance in chronic hepatitis B patients. Antivir Ther. 2007;12:7–13. [PubMed] [Google Scholar]
- 45.Kim SS, Cho SW, Kim SO, Hong SP, Cheong JY. Multidrug-resistant hepatitis B virus resulting from sequential monotherapy with lamivudine, adefovir, and entecavir: clonal evolution during lamivudine plus adefovir therapy. J Med Virol. 2013;85:55–64. doi: 10.1002/jmv.23440. [DOI] [PubMed] [Google Scholar]
- 46.Stuyver L, Van Geyt C, De Gendt S, Van Reybroeck G, Zoulim F, Leroux-Roels G, Rossau R. Line probe assay for monitoring drug resistance in hepatitis B virus-infected patients during antiviral therapy. J Clin Microbiol. 2000;38:702–707. doi: 10.1128/jcm.38.2.702-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jang H, Cho M, Heo J, Kim H, Jun H, Shin W, Cho B, Park H, Kim C. Oligonucleotide chip for detection of Lamivudine-resistant hepatitis B virus. J Clin Microbiol. 2004;42:4181–4188. doi: 10.1128/JCM.42.9.4181-4188.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Günther S, Li BC, Miska S, Krüger DH, Meisel H, Will H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol. 1995;69:5437–5444. doi: 10.1128/jvi.69.9.5437-5444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durantel D, Carrouée-Durantel S, Werle-Lapostolle B, Brunelle MN, Pichoud C, Trépo C, Zoulim F. A new strategy for studying in vitro the drug susceptibility of clinical isolates of human hepatitis B virus. Hepatology. 2004;40:855–864. doi: 10.1002/hep.20388. [DOI] [PubMed] [Google Scholar]
- 50.Yang H, Westland C, Xiong S, Delaney WE. In vitro antiviral susceptibility of full-length clinical hepatitis B virus isolates cloned with a novel expression vector. Antiviral Res. 2004;61:27–36. doi: 10.1016/j.antiviral.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 51.Ronsin C, Pillet A, Bali C, Denoyel GA. Evaluation of the COBAS AmpliPrep-total nucleic acid isolation-COBAS TaqMan hepatitis B virus (HBV) quantitative test and comparison to the VERSANT HBV DNA 3.0 assay. J Clin Microbiol. 2006;44:1390–1399. doi: 10.1128/JCM.44.4.1390-1399.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liaw YF, Chien RN, Yeh CT, Tsai SL, Chu CM. Acute exacerbation and hepatitis B virus clearance after emergence of YMDD motif mutation during lamivudine therapy. Hepatology. 1999;30:567–572. doi: 10.1002/hep.510300221. [DOI] [PubMed] [Google Scholar]
- 53.Liaw YF, Sung JJ, Chow WC, Farrell G, Lee CZ, Yuen H, Tanwandee T, Tao QM, Shue K, Keene ON, Dixon JS, Gray DF, Sabbat J. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N Engl J Med. 2004;351:1521–1531. doi: 10.1056/NEJMoa033364. [DOI] [PubMed] [Google Scholar]
- 54.Lai CL, Dienstag J, Schiff E, Leung NW, Atkins M, Hunt C, Brown N, Woessner M, Boehme R, Condreay L. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin Infect Dis. 2003;36:687–696. doi: 10.1086/368083. [DOI] [PubMed] [Google Scholar]
- 55.Lok AS, Lai CL, Leung N, Yao GB, Cui ZY, Schiff ER, Dienstag JL, Heathcote EJ, Little NR, Griffiths DA, et al. Long-term safety of lamivudine treatment in patients with chronic hepatitis B. Gastroenterology. 2003;125:1714–1722. doi: 10.1053/j.gastro.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 56.Qi X, Xiong S, Yang H, Miller M, Delaney WE. In vitro susceptibility of adefovir-associated hepatitis B virus polymerase mutations to other antiviral agents. Antivir Ther. 2007;12:355–362. [PubMed] [Google Scholar]
- 57.Lee JM, Park JY, Kim do Y, Nguyen T, Hong SP, Kim SO, Chon CY, Han KH, Ahn SH. Long-term adefovir dipivoxil monotherapy for up to 5 years in lamivudine-resistant chronic hepatitis B. Antivir Ther. 2010;15:235–241. doi: 10.3851/IMP1510. [DOI] [PubMed] [Google Scholar]
- 58.Lee YS, Suh DJ, Lim YS, Jung SW, Kim KM, Lee HC, Chung YH, Lee YS, Yoo W, Kim SO. Increased risk of adefovir resistance in patients with lamivudine-resistant chronic hepatitis B after 48 weeks of adefovir dipivoxil monotherapy. Hepatology. 2006;43:1385–1391. doi: 10.1002/hep.21189. [DOI] [PubMed] [Google Scholar]
- 59.Yeon JE, Yoo W, Hong SP, Chang YJ, Yu SK, Kim JH, Seo YS, Chung HJ, Moon MS, Kim SO, et al. Resistance to adefovir dipivoxil in lamivudine resistant chronic hepatitis B patients treated with adefovir dipivoxil. Gut. 2006;55:1488–1495. doi: 10.1136/gut.2005.077099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim JH, Ko SY, Choe WH, Kwon SY, Lee CH. Lamivudine plus adefovir combination therapy for lamivudine resistance in hepatitis-B-related hepatocellular carcinoma patients. Clin Mol Hepatol. 2013;19:273–279. doi: 10.3350/cmh.2013.19.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rapti I, Dimou E, Mitsoula P, Hadziyannis SJ. Adding-on versus switching-to adefovir therapy in lamivudine-resistant HBeAg-negative chronic hepatitis B. Hepatology. 2007;45:307–313. doi: 10.1002/hep.21534. [DOI] [PubMed] [Google Scholar]
- 62.Yatsuji H, Suzuki F, Sezaki H, Akuta N, Suzuki Y, Kawamura Y, Hosaka T, Kobayashi M, Saitoh S, Arase Y, et al. Low risk of adefovir resistance in lamivudine-resistant chronic hepatitis B patients treated with adefovir plus lamivudine combination therapy: two-year follow-up. J Hepatol. 2008;48:923–931. doi: 10.1016/j.jhep.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 63.Yun TJ, Jung JY, Kim CH, Um SH, An H, Seo YS, Kim JD, Yim HJ, Keum B, Kim YS, et al. Treatment strategies using adefovir dipivoxil for individuals with lamivudine-resistant chronic hepatitis B. World J Gastroenterol. 2012;18:6987–6995. doi: 10.3748/wjg.v18.i47.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lampertico P, Viganò M, Manenti E, Iavarone M, Sablon E, Colombo M. Low resistance to adefovir combined with lamivudine: a 3-year study of 145 lamivudine-resistant hepatitis B patients. Gastroenterology. 2007;133:1445–1451. doi: 10.1053/j.gastro.2007.08.079. [DOI] [PubMed] [Google Scholar]
- 65.Gaia S, Barbon V, Smedile A, Olivero A, Carenzi S, Lagget M, Alessandria C, Rizzetto M, Marzano A. Lamivudine-resistant chronic hepatitis B: an observational study on adefovir in monotherapy or in combination with lamivudine. J Hepatol. 2008;48:540–547. doi: 10.1016/j.jhep.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 66.Lin MT, Chou YP, Hu TH, Yu HC, Hsu YC, Tsai MC, Tseng PL, Chang KC, Yen YH, Chiu KW. Telbivudine and adefovir combination therapy for patients with chronic lamivudine-resistant hepatitis B virus infections. Arch Virol. 2014;159:29–37. doi: 10.1007/s00705-013-1786-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ha M, Zhang G, Diao S, Lin M, Wu J, Sun L, She H, Shen L, Huang C, Shen W, et al. Rescue therapy for lamivudine-resistant chronic hepatitis B: adefovir monotherapy, adefovir plus lamivudine or entecavir combination therapy. Intern Med. 2012;51:1509–1515. doi: 10.2169/internalmedicine.51.7329. [DOI] [PubMed] [Google Scholar]
- 68.Lim YS, Lee JY, Lee D, Shim JH, Lee HC, Lee YS, Suh DJ. Randomized trial of entecavir plus adefovir in patients with lamivudine-resistant chronic hepatitis B who show suboptimal response to lamivudine plus adefovir. Antimicrob Agents Chemother. 2012;56:2941–2947. doi: 10.1128/AAC.00338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Bömmel F, Wünsche T, Mauss S, Reinke P, Bergk A, Schürmann D, Wiedenmann B, Berg T. Comparison of adefovir and tenofovir in the treatment of lamivudine-resistant hepatitis B virus infection. Hepatology. 2004;40:1421–1425. doi: 10.1002/hep.20464. [DOI] [PubMed] [Google Scholar]
- 70.van Bömmel F, Zöllner B, Sarrazin C, Spengler U, Hüppe D, Möller B, Feucht HH, Wiedenmann B, Berg T. Tenofovir for patients with lamivudine-resistant hepatitis B virus (HBV) infection and high HBV DNA level during adefovir therapy. Hepatology. 2006;44:318–325. doi: 10.1002/hep.21253. [DOI] [PubMed] [Google Scholar]
- 71.van Bömmel F, de Man RA, Wedemeyer H, Deterding K, Petersen J, Buggisch P, Erhardt A, Hüppe D, Stein K, Trojan J, et al. Long-term efficacy of tenofovir monotherapy for hepatitis B virus-monoinfected patients after failure of nucleoside/nucleotide analogues. Hepatology. 2010;51:73–80. doi: 10.1002/hep.23246. [DOI] [PubMed] [Google Scholar]
- 72.Sheldon J, Camino N, Rodés B, Bartholomeusz A, Kuiper M, Tacke F, Núñez M, Mauss S, Lutz T, Klausen G, et al. Selection of hepatitis B virus polymerase mutations in HIV-coinfected patients treated with tenofovir. Antivir Ther. 2005;10:727–734. [PubMed] [Google Scholar]
- 73.Hann HW, Chae HB, Dunn SR. Tenofovir (TDF) has stronger antiviral effect than adefovir (ADV) against lamivudine (LAM)-resistant hepatitis B virus (HBV) Hepatol Int. 2008;2:244–249. doi: 10.1007/s12072-008-9045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang TT, Gish RG, Hadziyannis SJ, Cianciara J, Rizzetto M, Schiff ER, Pastore G, Bacon BR, Poynard T, Joshi S, et al. A dose-ranging study of the efficacy and tolerability of entecavir in Lamivudine-refractory chronic hepatitis B patients. Gastroenterology. 2005;129:1198–1209. doi: 10.1053/j.gastro.2005.06.055. [DOI] [PubMed] [Google Scholar]
- 75.Ryu HJ, Lee JM, Ahn SH, Kim do Y, Lee MH, Han KH, Chon CY, Park JY. Efficacy of adefovir add-on lamivudine rescue therapy compared with switching to entecavir monotherapy in patients with lamivudine-resistant chronic hepatitis B. J Med Virol. 2010;82:1835–1842. doi: 10.1002/jmv.21898. [DOI] [PubMed] [Google Scholar]
- 76.Sun J, Hou JL, Xie Q, Li XH, Zhang JM, Wang YM, Wang H, Lai JY, Chen SJ, Jia JD, et al. Randomised clinical trial: efficacy of peginterferon alfa-2a in HBeAg positive chronic hepatitis B patients with lamivudine resistance. Aliment Pharmacol Ther. 2011;34:424–431. doi: 10.1111/j.1365-2036.2011.04750.x. [DOI] [PubMed] [Google Scholar]
- 77.Suh DJ, Lee HC, Byun KS, Cho M, Kweon YO, Tak WY, Chon CY, Koh KC, Lee YS. Efficacy and safety of pegylated interferon-α2a in patients with lamivudine-resistant HBeAg-positive chronic hepatitis B. Antivir Ther. 2013;18:765–773. doi: 10.3851/IMP2664. [DOI] [PubMed] [Google Scholar]
- 78.Liaw YF, Gane E, Leung N, Zeuzem S, Wang Y, Lai CL, Heathcote EJ, Manns M, Bzowej N, Niu J, et al. 2-Year GLOBE trial results: telbivudine Is superior to lamivudine in patients with chronic hepatitis B. Gastroenterology. 2009;136:486–495. doi: 10.1053/j.gastro.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 79.Gane EJ, Wang Y, Liaw YF, Hou J, Thongsawat S, Wan M, Moon YM, Jia J, Chao YC, Niu J, et al. Efficacy and safety of prolonged 3-year telbivudine treatment in patients with chronic hepatitis B. Liver Int. 2011;31:676–684. doi: 10.1111/j.1478-3231.2011.02490.x. [DOI] [PubMed] [Google Scholar]
- 80.Gwak GY, Eo S, Shin S, Choi M, Lee J, Koh K, Paik S, Yoo B. A comparison of clevudine and entecavir for treatment-naïve patients with chronic hepatitis B: results after 2 years of treatment. Hepatol Int. 2013;7:106–110. doi: 10.1007/s12072-012-9368-1. [DOI] [PubMed] [Google Scholar]
- 81.Villeneuve JP, Durantel D, Durantel S, Westland C, Xiong S, Brosgart CL, Gibbs CS, Parvaz P, Werle B, Trépo C, et al. Selection of a hepatitis B virus strain resistant to adefovir in a liver transplantation patient. J Hepatol. 2003;39:1085–1089. doi: 10.1016/j.jhep.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 82.Heathcote EJ, Marcellin P, Buti M, Gane E, De Man RA, Krastev Z, Germanidis G, Lee SS, Flisiak R, Kaita K, et al. Three-year efficacy and safety of tenofovir disoproxil fumarate treatment for chronic hepatitis B. Gastroenterology. 2011;140:132–143. doi: 10.1053/j.gastro.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 83.Patterson SJ, George J, Strasser SI, Lee AU, Sievert W, Nicoll AJ, Desmond PV, Roberts SK, Locarnini S, Bowden S, et al. Tenofovir disoproxil fumarate rescue therapy following failure of both lamivudine and adefovir dipivoxil in chronic hepatitis B. Gut. 2011;60:247–254. doi: 10.1136/gut.2010.223206. [DOI] [PubMed] [Google Scholar]
- 84.Berg T, Marcellin P, Zoulim F, Moller B, Trinh H, Chan S, Suarez E, Lavocat F, Snow-Lampart A, Frederick D, et al. Tenofovir is effective alone or with emtricitabine in adefovir-treated patients with chronic-hepatitis B virus infection. Gastroenterology. 2010;139:1207–1217. doi: 10.1053/j.gastro.2010.06.053. [DOI] [PubMed] [Google Scholar]
- 85.Svarovskaia ES, Curtis M, Zhu Y, Borroto-Esoda K, Miller MD, Berg T, Lavocat F, Zoulim F, Kitrinos KM. Hepatitis B virus wild-type and rtN236T populations show similar early HBV DNA decline in adefovir refractory patients on a tenofovir-based regimen. J Viral Hepat. 2013;20:131–140. doi: 10.1111/j.1365-2893.2012.01638.x. [DOI] [PubMed] [Google Scholar]
- 86.Tan J, Degertekin B, Wong SN, Husain M, Oberhelman K, Lok AS. Tenofovir monotherapy is effective in hepatitis B patients with antiviral treatment failure to adefovir in the absence of adefovir-resistant mutations. J Hepatol. 2008;48:391–398. doi: 10.1016/j.jhep.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 87.Fung SK, Andreone P, Han SH, Rajender Reddy K, Regev A, Keeffe EB, Hussain M, Cursaro C, Richtmyer P, Marrero JA, et al. Adefovir-resistant hepatitis B can be associated with viral rebound and hepatic decompensation. J Hepatol. 2005;43:937–943. doi: 10.1016/j.jhep.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 88.Reijnders JG, Deterding K, Petersen J, Zoulim F, Santantonio T, Buti M, van Bömmel F, Hansen BE, Wedemeyer H, Janssen HL. Antiviral effect of entecavir in chronic hepatitis B: influence of prior exposure to nucleos(t)ide analogues. J Hepatol. 2010;52:493–500. doi: 10.1016/j.jhep.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 89.Chen CH, Wang JH, Lu SN, Hu TH, Hung CH, Chang MH, Changchien CS, Lee CM. Treatment response and evolution of HBV resistance during lamivudine plus adefovir or entecavir therapy in patients with adefovir-resistant mutants. Antivir Ther. 2012;17:701–709. doi: 10.3851/IMP2074. [DOI] [PubMed] [Google Scholar]
- 90.Yang HJ, Lee JH, Kim YJ, Yoon JH, Lee HS. Antiviral efficacy of combination therapy with entecavir and adefovir for entecavir/lamivudine-resistant hepatitis B virus with or without adefovir resistance. J Med Virol. 2012;84:424–430. doi: 10.1002/jmv.23229. [DOI] [PubMed] [Google Scholar]
- 91.Lu JJ, Liu K, Ma YJ, Wang J, Chen EQ, Tang H. Efficacy and safety of telbivudine plus adefovir dipivoxil combination therapy and entecavir monotherapy for HBeAg-positive chronic hepatitis B patients with resistance to adefovir dipivoxil. J Viral Hepat. 2013;20 Suppl 1:40–45. doi: 10.1111/jvh.12062. [DOI] [PubMed] [Google Scholar]
- 92.Seto WK, Yuen MF, Fung J, Lai CL. Tenofovir disoproxil fumarate for the treatment of chronic hepatitis B monoinfection. Hepatol Int. 2011:Epub ahead of print. doi: 10.1007/s12072-011-9282-y. [DOI] [PubMed] [Google Scholar]
- 93.Marcellin P, Heathcote EJ, Buti M, Gane E, de Man RA, Krastev Z, Germanidis G, Lee SS, Flisiak R, Kaita K, et al. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N Engl J Med. 2008;359:2442–2455. doi: 10.1056/NEJMoa0802878. [DOI] [PubMed] [Google Scholar]
- 94.Snow-Lampart A, Chappell B, Curtis M, Zhu Y, Myrick F, Schawalder J, Kitrinos K, Svarovskaia ES, Miller MD, Sorbel J, et al. No resistance to tenofovir disoproxil fumarate detected after up to 144 weeks of therapy in patients monoinfected with chronic hepatitis B virus. Hepatology. 2011;53:763–773. doi: 10.1002/hep.24078. [DOI] [PubMed] [Google Scholar]
- 95.Amini-Bavil-Olyaee S, Herbers U, Sheldon J, Luedde T, Trautwein C, Tacke F. The rtA194T polymerase mutation impacts viral replication and susceptibility to tenofovir in hepatitis B e antigen-positive and hepatitis B e antigen-negative hepatitis B virus strains. Hepatology. 2009;49:1158–1165. doi: 10.1002/hep.22790. [DOI] [PubMed] [Google Scholar]
- 96.Dupouey J, Gerolami R, Solas C, Colson P. Hepatitis B virus variant with the a194t substitution within reverse transcriptase before and under adefovir and tenofovir therapy. Clin Res Hepatol Gastroenterol. 2012;36:e26–e28. doi: 10.1016/j.clinre.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 97.Qin B, Budeus B, Cao L, Wu C, Wang Y, Zhang X, Rayner S, Hoffmann D, Lu M, Chen X. The amino acid substitutions rtP177G and rtF249A in the reverse transcriptase domain of hepatitis B virus polymerase reduce the susceptibility to tenofovir. Antiviral Res. 2013;97:93–100. doi: 10.1016/j.antiviral.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 98.Villet S, Ollivet A, Pichoud C, Barraud L, Villeneuve JP, Trépo C, Zoulim F. Stepwise process for the development of entecavir resistance in a chronic hepatitis B virus infected patient. J Hepatol. 2007;46:531–538. doi: 10.1016/j.jhep.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 99.Yatsuji H, Hiraga N, Mori N, Hatakeyama T, Tsuge M, Imamura M, Takahashi S, Fujimoto Y, Ochi H, Abe H, et al. Successful treatment of an entecavir-resistant hepatitis B virus variant. J Med Virol. 2007;79:1811–1817. doi: 10.1002/jmv.20981. [DOI] [PubMed] [Google Scholar]
- 100.Chae HB, Kim MJ, Seo EG, Choi YH, Lee HS, Han JH, Yoon SM, Park SM, Youn SJ. High efficacy of adefovir and entecavir combination therapy in patients with nucleoside-refractory hepatitis B. Korean J Hepatol. 2012;18:75–83. doi: 10.3350/kjhep.2012.18.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Villet S, Pichoud C, Villeneuve JP, Trépo C, Zoulim F. Selection of a multiple drug-resistant hepatitis B virus strain in a liver-transplanted patient. Gastroenterology. 2006;131:1253–1261. doi: 10.1053/j.gastro.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 102.Shaw T, Bartholomeusz A, Locarnini S. HBV drug resistance: mechanisms, detection and interpretation. J Hepatol. 2006;44:593–606. doi: 10.1016/j.jhep.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 103.Liu Y, Wang C, Zhong Y, Chen L, Li X, Ji D, Wang H, Xin S, Zoulim F, Xu D. Evolution and suppression of HBV strains with multidrug resistance to lamivudine, adefovir dipivoxil and entecavir in a patient with chronic hepatitis B. Antivir Ther. 2010;15:1185–1190. doi: 10.3851/IMP1679. [DOI] [PubMed] [Google Scholar]
- 104.Petersen J, Ratziu V, Buti M, Janssen HL, Brown A, Lampertico P, Schollmeyer J, Zoulim F, Wedemeyer H, Sterneck M, et al. Entecavir plus tenofovir combination as rescue therapy in pre-treated chronic hepatitis B patients: an international multicenter cohort study. J Hepatol. 2012;56:520–526. doi: 10.1016/j.jhep.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 105.Lim YS, Lee JY, Lee D, Shim JH, Lee HC, Lee YS, Suh DJ. Randomized trial of the virologic response during up to two years of entecavir-adefovir combination therapy in multiple-drug-refractory chronic hepatitis B virus patients. Antimicrob Agents Chemother. 2013;57:3369–3374. doi: 10.1128/AAC.00587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park MS, Kim BK, Kim KS, Kim JK, Kim SU, Park JY, Kim do Y, Baartarkhuu O, Han KH, Chon CY, et al. Antiviral efficacies of currently available rescue therapies for multidrug-resistant chronic hepatitis B. Clin Mol Hepatol. 2013;19:29–35. doi: 10.3350/cmh.2013.19.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yuen MF, Han KH, Um SH, Yoon SK, Kim HR, Kim J, Kim CR, Lai CL. Antiviral activity and safety of LB80380 in hepatitis B e antigen-positive chronic hepatitis B patients with lamivudine-resistant disease. Hepatology. 2010;51:767–776. doi: 10.1002/hep.23462. [DOI] [PubMed] [Google Scholar]


