Abstract
Defects in intestinal barrier function characterized by an increase in intestinal permeability contribute to intestinal inflammation. Growing evidence has shown that an increase in intestinal permeability has a pathogenic role in diseases such as inflammatory bowel disease (IBD) and celiac disease, and functional bowel disorders such as irritable bowel syndrome. Therefore, clarification of the inflammatory responses, the defense pathway and the corresponding regulatory system is essential and may lead to the development of new therapies. MicroRNAs (miRNAs) are small (19-22 nt) noncoding RNA molecules that regulate genes at the post-transcriptional level by base-pairing to specific messenger RNAs for degradation to repress translation. Recent studies suggested that miRNAs are important in the immune response and mediate a critical role in multiple immune response-related disorders. Based on these discoveries, attention has been focused on understanding the role of miRNAs in regulating intestinal barrier dysfunction, especially in IBD. Here, we provide a review of the most recent state-of-the-art research on miRNAs in intestinal barrier dysfunction.
Keywords: MicroRNAs, Intestinal barrier dysfunction, Inflammatory bowel disease, Celiac disease, Therapeutic target
Core tip: This article summarizes the latest findings on the important roles of microRNAs (miRNAs) in regulating inflammation and autoimmune disorders in inflammatory bowel disease (IBD). Insight into miRNAs-21 as a novel biomarker is also provided, which shows that miRNAs-21 is a potential diagnostic and therapeutic target for IBD.
INTRODUCTION
The intestinal barrier plays an important role in absorbing nutrients and secreting waste[1]. In addition to its abilities to support paracellular transport, the intestinal barrier can also prevent luminal microbes and their products reaching the internal milieu. Tight junctions and their associated proteins, including claudins, occludin, and zonula occludens, are the most adhesive apical junctional complexes and act as a structural and functional barrier against paracellular permeation of luminal substances[2-4]. Breakdown or disruption of the epithelial barrier is thought to be an essential determinant in the predisposition to intestinal inflammation and a number of inflammatory disorders, such as Crohn’s disease[5], ulcerative colitis[6,7], celiac disease[8], and a series of infectious diarrheal syndromes[9,10]. The phenomenon of intestinal tight junction (TJ) barrier disruption has previously been reported, but the intracellular mechanism is still poorly understood. Of the essential factors relating to this issue, two are most critical. The first is to identify the early signaling event which triggers the immune response and inflammatory cascade which will help us to understand induction of the disease, and the second is to identify the regulatory system which will lead to the discovery of the therapeutic target. MicroRNAs (miRNAs), which are small noncoding RNAs, were recently discovered to have a promising role in the treatment of immune-related diseases[11,12]. By regulating the degradation of mRNAs at the post-transcriptional level, miRNAs can affect various signaling pathways, and may be good candidates for the treatment of immune-related diseases. More recently, investigations have focused on the role of miRNAs in intestinal-related diseases. These studies may not only provide novel insights into understanding the pathological and physiological process of intestinal barrier dysfunction, especially in inflammatory bowel disease (IBD), but have also suggested the therapeutic role played by miRNAs. The major purpose of this review was to examine current research on the role of miRNAs in the regulation of intestinal barrier function and their therapeutic potential.
miRNA BIOGENESIS AND FUNCTION
MiRNAs, non-coding small endogenous RNAs of 19-22 nucleotides in length, were first identified in C. elegans by Lee et al[13] in a study on the function of gene lin-14 as a sequence-specific regulator of gene expression[14]. MiRNAs which are encoded by eukaryotic nuclear DNA can target the 3’ untranslated region (3’UTR) of specific mRNAs, usually resulting in gene downregulation via translational repression or target degradation[15]. More than 1500 miRNAs have been found to be encoded in the human genome and over 60% of human genes are targeted by miRNAs[16-18]. miRNAs play an important role in many different types of human cells[19].
RNA polymerase II (Pol II) is associated with the miRNA promoter and induces primary miRNAs (pre-miRNAs) to be transcribed in the nucleus[20,21]. These pre-miRNAs have a capped structure and a poly(A) tail[20,22]. After being transported to the cytoplasm, the pre-miRNAs are further processed by the RNase III endonuclease, Dicer, in a complex with a trans-activator RNA binding protein into a double-stranded mature miRNA[23-25]. One strand of the mature miRNA is then incorporated into the RNA-induced silencing complex (RISC) and leads this complex to the untranslated region (3’UTR) of specific mRNAs, which causes repression of the corresponding protein[26-29]. In this way, miRNAs are thought to be fine-tune regulators in gene expression and disease control.
A number of biological processes are regulated by miRNAs, such as cell proliferation, apoptosis, differentiation, migration and cell cycle control[30-33]. miRNAs have also been reported to be involved in human diseases, including tumors[34-41], immune dysregulation[42-44], cardiovascular diseases[45-48], metabolic syndrome[49,50] and others[51].
EXPRESSION OF miRNAs IN INTESTINAL EPITHELIAL CELLS
In 2008, Wu et al[52] first reported miR-192 which was detected in the epithelial cells of colonic mucosa samples from healthy individuals, but not in patients with active UC using immunohistochemistry and in situ hybridization. These authors also found 11 differentially expressed miRNAs in active UC vs healthy samples and confirmed an inverse relationship between macrophage inflammatory peptide-2α (MIP-2α, previously shown to be involved in IBD[53]) and miR-192. Similarly, Bian et al[54] demonstrated that miR-150 was significantly increased in the epithelial cells of colonic mucosa in UC patients compared with controls, and suggested an inverse correlation between miR-150 and its target, c-Myb[55], a proto-oncogene involved in apoptosis. Consequently, these two pioneer studies have provided new insight into the pathogenesis of intestinal barrier dysfunction. A summary of the expression of miRNAs in IBD is shown in Figure 1.
Figure 1.
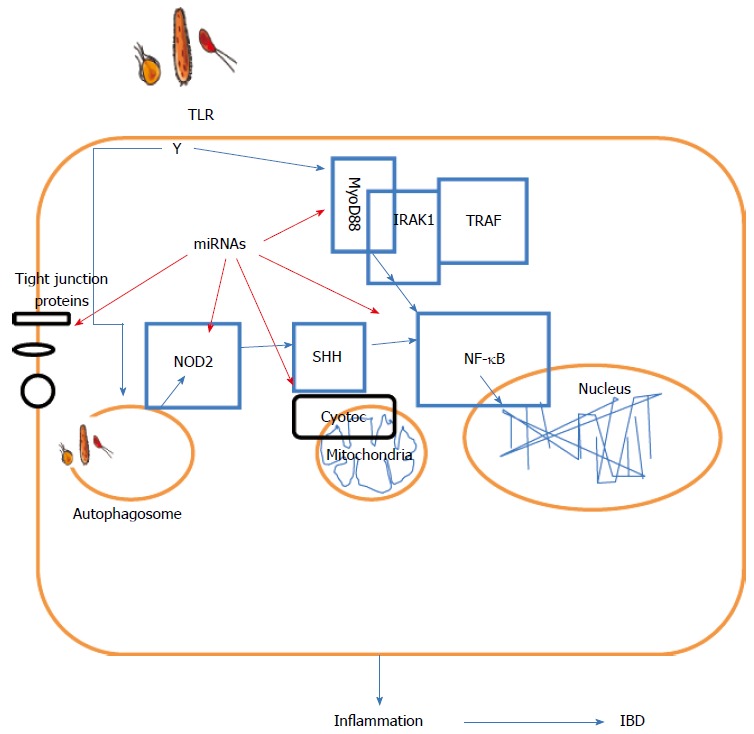
Summary of the role of microRNAs in inflammatory bowel disease. IBD: Inflammatory bowel disease. NF-κB: The nuclear factor NF-κB; TLR: Toll-like receptor; TRAF: TNF receptor-associated factor; NOD2: Nucleotide-binding oligomerization domain-containing protein 2.
ROLE OF miRNAs IN REGULATION OF TIGHT JUNCTION PROTEINS
Occludin and claudins are important transmembrane TJ proteins localized at the TJ strands and function in the TJ barrier[56-58]. Ye et al[59] demonstrated that miR-122a plays a central role in the regulation of intestinal TJ permeability by degrading the protein occludin. The regulation of claudins by miRNAs was reported in breast cancer[60,61] and HIV-associated neurological disorders[62], but not in the intestinal TJ barrier. Further studies on the relationship between claudins and miRNAs in the intestinal TJ barrier system are required. Zonula occludens 1 (ZO-1) is another major component of the TJ barrier which regulates intestinal permeability[63]. Tang et al[64] found miR-212 overexpression in colon biopsy samples from patients with alcoholic liver disease and in Caco-2 cells (a human intestinal epithelial cell line) treated with ethanol. Alcohol can induce miR-212 overexpression and leads to gut leakiness by down-regulating ZO-1 translation.
miRNAs AND INFLAMMATION
Many groups have demonstrated that miRNAs play pivotal roles in both adaptive and innate immunity[65]. miRNAs regulate the development of various immune cells as well as their immunological functions. miRNAs are also essential in B- and T-cell functions. A deficiency in Dicer, the key enzyme in processing miRNAs, results in inhibition of T cell development[65]. In addition, several miRNAs, including miR-155, miR-181a, miR-150 and the miR-17-92 cluster, are also involved in B- and T-cell regulation[66]. Forced overexpression of miR-150 blocks B cell development[67]. Innate immune responses provide the initial defense against pathogens. Pattern recognition receptors, such as Toll-like receptor (TLR), expressed on macrophages and dendritic cells (DCs), are regulated by miRNAs. miRNAs have also been shown to be important in regulation of the TLR signaling cascade[68,69]. MiR-146a expression can be induced by exposure to TLR ligands, such as lipopolysaccharide (LPS), peptidoglycan, and flagellin[70]. miR-146a then functions in a negative feedback mechanism in the TLR signaling cascade by decreasing the expression of TNF-receptor-associated factor-6 and IL-1 receptor associated kinase-1, two target genes of the TLR signaling cascade. Furthermore, loss of miR-155 in DCs impairs their antigen presenting capacity and costimulation activity. The target gene of miR-155 in DCs is SOCS1 which negatively regulates antigen presenting capacity in DCs. Therefore, deregulation of SOCS1 in the absence of miR-155 could account for impaired DC function[71]. In macrophages, the downregulation of miR-125b is required to ensure that the correct inflammatory response is produced[72]. The idea of miRNAs as regulators of IBD provides new insight into the development of appropriate therapies.
miRNAs AND AUTOIMMUNE RESPONSE IN IBD
Several studies have reported that miRNAs are involved in autoimmune diseases (AIDs)[42-44]. As miRNAs have been confirmed to play a role in immune cell development and have an impact on cell functions, it is reasonable to deduce that miRNAs are related to AIDs. Reports have shown that miRNAs take part in AIDs, such as rheumatoid arthritis[73], systemic lupus erythematosus[74], multiple sclerosis[75], primary biliary cirrhosis[76], inflammatory bowel disease[52], idiopathic thrombocytopenic purpura[77] and psoriasis[78].
IBD, including Crohn’s disease (CD) and ulcerative colitis (UC), is a chronic gastrointestinal inflammatory disorder whose pathophysiology has been extensively studied over the past several years, but is still poorly understood.
Wu and colleagues[52] studied mucosal tissues from healthy subjects and UC patients and found that miR-192 was predominantly expressed in the intestinal epithelial tissue of healthy subjects and was significantly decreased in UC patients. Furthermore, they also observed that the inflammatory protein, MIP-2, was mainly expressed in UC patients and was decreased in healthy subjects. The expression of MIP-2α had an inverse relationship with miR-192. Besides miR-192, they also identified several other differentially expressed miRNAs between UC patients and healthy subjects. MiR-21 was increased in UC patients compared with healthy subjects. miR-375, miR-422b and miR-23a were increased in healthy subjects and all three miRNAs had a similar level of expression in inactive UC[52]. A previous study also showed that TGF-β can induce miR-192[79]. This suggests that miR-192 may be the master regulator in the process of inflammation. These findings indicate that miRNAs are involved in the pathogenesis of IBD.
miRNAs AND MITOCHONRIAL STRESS IN IBD
Mitochondria are fundamental subcellular components that play a critical role in the maintenance of normal structure, function and survival of cells. Mitochondrial dysfunction is associated with metabolic diseases including insulin resistance, obesity, diabetes, and the cardiorenal metabolic syndrome[80-83]. Growing evidence suggests that miRNAs provide another layer of regulation with regard to mitochondrial function. MiR-338 can modulate mitochondrial function by targeting cytochrome c oxidase IV (COX IV) mRNA[84]. The miRNA-200 family is implicated in epithelial-to-mesenchymal transition which is accompanied by mitochondrial biogenesis and is involved in organ fibrosis and carcinoma progression[85]. Nishi et al[86] showed that miR-15b, miR-195 and miR-424 can down-regulate cellular ATP levels and affect mitochondrial integrity. In addition, miR-23a/b in hypertrophy acts in a compensatory mechanism to down-regulate mitochondrial glutaminase[87]. More recently, Yuan et al[88] suggested that prohibitin which can inhibit mitochondrial dysfunction may be a potential target in IBD. However, further studies are required to determine whether miRNAs can affect IBD by regulating mitochondrial function.
miRNAs AND AUTOPHAGY IN IBD
Autophagy is a unique cellular process of self-digestion, characterized by the engulfment of cytosolic macromolecules and organelles in a autophagosome, which are then transported to the lysosome for degradation[89,90]. Autophagy helps to recycle and store nutrients for stress conditions[89].
Recently, autophagy-related gene (ATG) 16L1 was reported to be involved in CD[91,92]. ATG 16L1 shares some sequence homology with yeast Apg16L and was originally identified in the protein complex ATG5-ATG12[93]. In autophagy, ATG16L plays an important role in autophagosome formation and functions as an E3-like enzyme to mediate lipidation[94]. Lu et al[95] recently provided evidence to show that miR106b and miR93 suppress autophagy-mediated removal of bacteria in epithelial cells by targeting ATG16L1. Furthermore, NOD2, an intracellular bacterial sensor of the nucleotide-binding and oligomerization domain (NOD)-like receptor family, can sense the presence of muramyl dipeptide, a component of the peptidoglycan cell wall from both Gram-positive and -negative bacteria. NOD2 activation results in pro-inflammatory and anti-bacterial molecule production dependent on cell signaling pathways mediated by RICK/RIP2, NF-κB and MAPKs. More recently, Ghorpade et al[96] found that miR-146a-mediated NOD2-SHH signaling regulated gut inflammation in a mouse model of IBD. In addition, Brest et al[97] demonstrated that the miR-196 family of miRNAs downregulates the CD protective variant (c.313C) of the immunity-related GTPase family M protein 1 gene in CD patients. Consequently, the control of intracellular replication of CD-associated adherent invasive E. coli by autophagy was lost due to a decrease in IRGM1[97]. By targeting the related gene, miRNAs may eventually contribute to the improvement of IBD.
NOVEL BIOMARKERES AND THERAPEUTIC TARGETS IN IBD
With regard to the involvement of miRNAs in the pathogenesis of IBD, it is vital to identify which miRNAs are consistently dysregulated in IBD and the target genes of the miRNAs. miR-21, the most investigated and well-described miRNA, also known as the “oncomiR”, has been shown to have potential clinical application[98]. According to published data[52,53,99,100], miR-21 is the only miRNA usually upregulated in inflamed tissue or serum in IBD patients. The expression of miR-21 is regulated by NF-κB which is a master gene in multiple immune diseases (including IBD)[101]. Thus, miR-21 has the potential as a biomarker. Iborra et al[102] recently conducted a study to establish the specific expression patterns of miRNAs in the serum and mucosa of IBD patients. They identified six and five differentially expressed miRNAs in the serum and mucosa of patients with active CD compared with those with inactive CD, respectively. Their study again suggested the utility of miRNAs as possible biomarkers. The actual role of miRNAs in IBD still need to be confirmed by functional studies, however, miRNAs have shown promise in the treatment of IBD.
CONCLUSION
Recently, increasing attention has been paid to the gene expression of miRNAs in IBD. Clinical trials have been carried out to test the therapeutic efficacy of miRNA-based therapies. “Miravirsen”, a specific inhibitor of miR-122, is now being evaluated in a phase II clinical trial[103]. Furthermore, a recent review[104] suggested the use of an miRNA inhibitor or synthetic miRNA targeting the PI3K and Ras/MAPK pathways in multiple myeloma (MM) treatment. MiR-29b is a promising target in MM treatment by multiple mechanisms, including the regulation of osteoclastic differentiation[105] and epigenetic regulation of the cell cycle[106,107]. Future studies will provide a basis for more clinical trials and shed light on miRNA-based therapies in IBD.
Footnotes
Supported by Grant for Key Clinical Discipline Construction of Shanghai Municipality, China, No. ZK2012B20; and Phase II Outstanding Young Medical Personnel Training Fund of Jinshan District Health Systems, Shanghai, China, No. JWKJ-RCYQ-201207
P- Reviewers: Caraglia M, de Magistris L S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Liu XM
References
- 1.Shen L, Su L, Turner JR. Mechanisms and functional implications of intestinal barrier defects. Dig Dis. 2009;27:443–449. doi: 10.1159/000233282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groschwitz KR, Hogan SP. Intestinal barrier function: molecular regulation and disease pathogenesis. J Allergy Clin Immunol. 2009;124:3–20; quiz 21-22. doi: 10.1016/j.jaci.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groschwitz KR, Ahrens R, Osterfeld H, Gurish MF, Han X, Abrink M, Finkelman FD, Pejler G, Hogan SP. Mast cells regulate homeostatic intestinal epithelial migration and barrier function by a chymase/Mcpt4-dependent mechanism. Proc Natl Acad Sci USA. 2009;106:22381–22386. doi: 10.1073/pnas.0906372106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr. 2011;141:769–776. doi: 10.3945/jn.110.135657. [DOI] [PubMed] [Google Scholar]
- 5.Sartor RB. Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 6.Clavel T, Haller D. Bacteria- and host-derived mechanisms to control intestinal epithelial cell homeostasis: implications for chronic inflammation. Inflamm Bowel Dis. 2007;13:1153–1164. doi: 10.1002/ibd.20174. [DOI] [PubMed] [Google Scholar]
- 7.Werner T, Haller D. Intestinal epithelial cell signalling and chronic inflammation: From the proteome to specific molecular mechanisms. Mutat Res. 2007;622:42–57. doi: 10.1016/j.mrfmmm.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Brandtzaeg P. Update on mucosal immunoglobulin A in gastrointestinal disease. Curr Opin Gastroenterol. 2010;26:554–563. doi: 10.1097/MOG.0b013e32833dccf8. [DOI] [PubMed] [Google Scholar]
- 9.Gecse K, Róka R, Ferrier L, Leveque M, Eutamene H, Cartier C, Ait-Belgnaoui A, Rosztóczy A, Izbéki F, Fioramonti J, et al. Increased faecal serine protease activity in diarrhoeic IBS patients: a colonic lumenal factor impairing colonic permeability and sensitivity. Gut. 2008;57:591–599. doi: 10.1136/gut.2007.140210. [DOI] [PubMed] [Google Scholar]
- 10.Gadewar S, Fasano A. Current concepts in the evaluation, diagnosis and management of acute infectious diarrhea. Curr Opin Pharmacol. 2005;5:559–565. doi: 10.1016/j.coph.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 14.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 17.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X, Ruan J, Wang G, Zhang W. Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 24.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 26.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 27.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 28.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 29.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 30.McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, Friedman JR, Kaestner KH. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;139:1654–1664, 1664.e1. doi: 10.1053/j.gastro.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–568. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 33.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 34.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mraz M, Pospisilova S, Malinova K, Slapak I, Mayer J. MicroRNAs in chronic lymphocytic leukemia pathogenesis and disease subtypes. Leuk Lymphoma. 2009;50:506–509. doi: 10.1080/10428190902763517. [DOI] [PubMed] [Google Scholar]
- 36.Zabolotneva AA, Zhavoronkov AA, Shegay PV, Gaifullin NM, Alekseev BY, Roumiantsev SA, Garazha AV, Kovalchuk O, Aravin A, Buzdin AA. A systematic experimental evaluation of microRNA markers of human bladder cancer. Front Genet. 2013;4:247. doi: 10.3389/fgene.2013.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandhu R, Rivenbark AG, Mackler RM, Livasy CA, Coleman WB. Dysregulation of microRNA expression drives aberrant DNA hypermethylation in basal-like breast cancer. Int J Oncol. 2014;44:563–572. doi: 10.3892/ijo.2013.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy R, De Sarkar N, Ghose S, Paul RR, Pal M, Bhattacharya C, Chowdhury SK, Ghosh S, Roy B. Genetic variations at microRNA and processing genes and risk of oral cancer. Tumour Biol. 2014;35:3409–3414. doi: 10.1007/s13277-013-1450-3. [DOI] [PubMed] [Google Scholar]
- 39.Sun F, Chen HG, Li W, Yang X, Wang X, Jiang R, Guo Z, Chen H, Huang J, Borowsky AD, et al. Androgen receptor splice variant AR3 promotes prostate cancer via modulating expression of autocrine/paracrine factors. J Biol Chem. 2014;289:1529–1539. doi: 10.1074/jbc.M113.492140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H, Gao G, Jiang L, Guo L, Lin M, Jiao X, Jia W, Huang J. Decreased expression of miR-218 is associated with poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2013;6:2904–2911. [PMC free article] [PubMed] [Google Scholar]
- 41.Lee HY, Mohammed KA, Kaye F, Sharma P, Moudgil BM, Clapp WL, Nasreen N. Targeted delivery of let-7a microRNA encapsulated ephrin-A1 conjugated liposomal nanoparticles inhibit tumor growth in lung cancer. Int J Nanomedicine. 2013;8:4481–4494. doi: 10.2147/IJN.S41782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–194. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonkoly E, Pivarcsi A. Advances in microRNAs: implications for immunity and inflammatory diseases. J Cell Mol Med. 2009;13:24–38. doi: 10.1111/j.1582-4934.2008.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 46.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 48.Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 49.Zhu H, Shyh-Chang N, Segrè AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maes OC, Chertkow HM, Wang E, Schipper HM. MicroRNA: Implications for Alzheimer Disease and other Human CNS Disorders. Curr Genomics. 2009;10:154–168. doi: 10.2174/138920209788185252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635.e24. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 53.Wu F, Dassopoulos T, Cope L, Maitra A, Brant SR, Harris ML, Bayless TM, Parmigiani G, Chakravarti S. Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13:807–821. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 54.Bian Z, Li L, Cui J, Zhang H, Liu Y, Zhang CY, Zen K. Role of miR-150-targeting c-Myb in colonic epithelial disruption during dextran sulphate sodium-induced murine experimental colitis and human ulcerative colitis. J Pathol. 2011;225:544–553. doi: 10.1002/path.2907. [DOI] [PubMed] [Google Scholar]
- 55.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 56.Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403–429. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- 57.Furuse M, Tsukita S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006;16:181–188. doi: 10.1016/j.tcb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Angelow S, Ahlstrom R, Yu AS. Biology of claudins. Am J Physiol Renal Physiol. 2008;295:F867–F876. doi: 10.1152/ajprenal.90264.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye D, Guo S, Al-Sadi R, Ma TY. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology. 2011;141:1323–1333. doi: 10.1053/j.gastro.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin W, Ren Q, Liu T, Huang Y, Wang J. MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett. 2013;587:1434–1439. doi: 10.1016/j.febslet.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 61.Li Q, Zhu F, Chen P. miR-7 and miR-218 epigenetically control tumor suppressor genes RASSF1A and Claudin-6 by targeting HoxB3 in breast cancer. Biochem Biophys Res Commun. 2012;424:28–33. doi: 10.1016/j.bbrc.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 62.Mishra R, Singh SK. HIV-1 Tat C modulates expression of miRNA-101 to suppress VE-cadherin in human brain microvascular endothelial cells. J Neurosci. 2013;33:5992–6000. doi: 10.1523/JNEUROSCI.4796-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Furuse M. Molecular basis of the core structure of tight junctions. Cold Spring Harb Perspect Biol. 2010;2:a002907. doi: 10.1101/cshperspect.a002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, Zhang LJ, Keshavarzian A. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res. 2008;32:355–364. doi: 10.1111/j.1530-0277.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 65.Lu LF, Liston A. MicroRNA in the immune system, microRNA as an immune system. Immunology. 2009;127:291–298. doi: 10.1111/j.1365-2567.2009.03092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 67.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci USA. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dunne A, O’Neill LA. Adaptor usage and Toll-like receptor signaling specificity. FEBS Lett. 2005;579:3330–3335. doi: 10.1016/j.febslet.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 69.Kracht M, Saklatvala J. Transcriptional and post-transcriptional control of gene expression in inflammation. Cytokine. 2002;20:91–106. doi: 10.1006/cyto.2002.0895. [DOI] [PubMed] [Google Scholar]
- 70.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA. 2009;106:2735–2740. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakamachi Y, Kawano S, Takenokuchi M, Nishimura K, Sakai Y, Chin T, Saura R, Kurosaka M, Kumagai S. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60:1294–1304. doi: 10.1002/art.24475. [DOI] [PubMed] [Google Scholar]
- 74.Nagata Y, Nakasa T, Mochizuki Y, Ishikawa M, Miyaki S, Shibuya H, Yamasaki K, Adachi N, Asahara H, Ochi M. Induction of apoptosis in the synovium of mice with autoantibody-mediated arthritis by the intraarticular injection of double-stranded MicroRNA-15a. Arthritis Rheum. 2009;60:2677–2683. doi: 10.1002/art.24762. [DOI] [PubMed] [Google Scholar]
- 75.Otaegui D, Baranzini SE, Armañanzas R, Calvo B, Muñoz-Culla M, Khankhanian P, Inza I, Lozano JA, Castillo-Triviño T, Asensio A, et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS One. 2009;4:e6309. doi: 10.1371/journal.pone.0006309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Padgett KA, Lan RY, Leung PC, Lleo A, Dawson K, Pfeiff J, Mao TK, Coppel RL, Ansari AA, Gershwin ME. Primary biliary cirrhosis is associated with altered hepatic microRNA expression. J Autoimmun. 2009;32:246–253. doi: 10.1016/j.jaut.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH, Xu ZM, Yin YB. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16:939–946. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 78.Sonkoly E, Wei T, Janson PC, Sääf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Godwin JG, Ge X, Stephan K, Jurisch A, Tullius SG, Iacomini J. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc Natl Acad Sci USA. 2010;107:14339–14344. doi: 10.1073/pnas.0912701107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ren J, Pulakat L, Whaley-Connell A, Sowers JR. Mitochondrial biogenesis in the metabolic syndrome and cardiovascular disease. J Mol Med (Berl) 2010;88:993–1001. doi: 10.1007/s00109-010-0663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim JA, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circ Res. 2008;102:401–414. doi: 10.1161/CIRCRESAHA.107.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Makino A, Scott BT, Dillmann WH. Mitochondrial fragmentation and superoxide anion production in coronary endothelial cells from a mouse model of type 1 diabetes. Diabetologia. 2010;53:1783–1794. doi: 10.1007/s00125-010-1770-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oba S, Kumano S, Suzuki E, Nishimatsu H, Takahashi M, Takamori H, Kasuya M, Ogawa Y, Sato K, Kimura K, et al. miR-200b precursor can ameliorate renal tubulointerstitial fibrosis. PLoS One. 2010;5:e13614. doi: 10.1371/journal.pone.0013614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nishi H, Ono K, Iwanaga Y, Horie T, Nagao K, Takemura G, Kinoshita M, Kuwabara Y, Mori RT, Hasegawa K, et al. MicroRNA-15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. J Biol Chem. 2010;285:4920–4930. doi: 10.1074/jbc.M109.082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yuan C, Chen WX, Zhu JS, Chen NW, Lu YM, Ou YX, Chen HQ. IL-10 treatment is associated with prohibitin expression in the Crohn’s disease intestinal fibrosis mouse model. Mediators Inflamm. 2013;2013:617145. doi: 10.1155/2013/617145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 2010;12:99–113. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS One. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 94.Noda T, Yoshimori T. Molecular basis of canonical and bactericidal autophagy. Int Immunol. 2009;21:1199–1204. doi: 10.1093/intimm/dxp088. [DOI] [PubMed] [Google Scholar]
- 95.Lu C, Chen J, Xu HG, Zhou X, He Q, Li YL, Jiang G, Shan Y, Xue B, Zhao RX, et al. MIR106B and MIR93 prevent removal of bacteria from epithelial cells by disrupting ATG16L1-mediated autophagy. Gastroenterology. 2014;146:188–199. doi: 10.1053/j.gastro.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ghorpade DS, Sinha AY, Holla S, Singh V, Balaji KN. NOD2-nitric oxide-responsive microRNA-146a activates Sonic hedgehog signaling to orchestrate inflammatory responses in murine model of inflammatory bowel disease. J Biol Chem. 2013;288:33037–33048. doi: 10.1074/jbc.M113.492496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hébuterne X, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 98.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 99.Fasseu M, Tréton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, Aparicio T, Daniel F, Soulé JC, Moreau R, et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 2010;5:pii: e13160. doi: 10.1371/journal.pone.0013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zahm AM, Thayu M, Hand NJ, Horner A, Leonard MB, Friedman JR. Circulating microRNA is a biomarker of pediatric Crohn disease. J Pediatr Gastroenterol Nutr. 2011;53:26–33. doi: 10.1097/MPG.0b013e31822200cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou R, Hu G, Gong AY, Chen XM. Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res. 2010;38:3222–3232. doi: 10.1093/nar/gkq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iborra M, Bernuzzi F, Correale C, Vetrano S, Fiorino G, Beltrán B, Marabita F, Locati M, Spinelli A, Nos P, et al. Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin Exp Immunol. 2013;173:250–258. doi: 10.1111/cei.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 104.Misso G, Zappavigna S, Castellano M, De Rosa G, Di Martino MT, Tagliaferri P, Tassone P, Caraglia M. Emerging pathways as individualized therapeutic target of multiple myeloma. Expert Opin Biol Ther. 2013;13 Suppl 1:S95–109. doi: 10.1517/14712598.2013.807338. [DOI] [PubMed] [Google Scholar]
- 105.Rossi M, Pitari MR, Amodio N, Di Martino MT, Conforti F, Leone E, Botta C, Paolino FM, Del Giudice T, Iuliano E, et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: implications for the treatment of multiple myeloma-related bone disease. J Cell Physiol. 2013;228:1506–1515. doi: 10.1002/jcp.24306. [DOI] [PubMed] [Google Scholar]
- 106.Amodio N, Leotta M, Bellizzi D, Di Martino MT, D’Aquila P, Lionetti M, Fabiani F, Leone E, Gullà AM, Passarino G, et al. DNA-demethylating and anti-tumor activity of synthetic miR-29b mimics in multiple myeloma. Oncotarget. 2012;3:1246–1258. doi: 10.18632/oncotarget.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Amodio N, Di Martino MT, Foresta U, Leone E, Lionetti M, Leotta M, Gullà AM, Pitari MR, Conforti F, Rossi M, et al. miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis. 2012;3:e436. doi: 10.1038/cddis.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]


