Abstract
AIM: To investigate the expression of key biomarkers in hepatoma cell lines, tumor cells from patients’ blood samples, and tumor tissues.
METHODS: We performed the biomarker tests in two steps. First, cells plated on coverslips were used to assess biomarkers, and fluorescence intensities were calculated using the NIH Image J software. The measured values were analyzed using the SPSS 19.0 software to make comparisons among eight cell lines. Second, eighty-four individual samples were used to assess the biomarkers’ expression. Negative enrichment of the blood samples was performed, and karyocytes were isolated and dropped onto pre-treated glass slides for further analysis by immunofluorescence staining. Fluorescence intensities were compared among hepatocellular carcinoma (HCC) patients, chronic HBV-infected patients, and healthy controls following methods similar to those used for cell lines. The relationships between the expression of biomarkers and clinical pathological parameters were analyzed by Spearman rank correlation tests. In addition, we studied the distinct biomarkers’ expression with three-dimensional laser confocal microscopy reconstructions, and Kaplan-Meier survival analysis was performed to understand the clinical significance of these biomarkers.
RESULTS: Microscopic examination and fluorescence intensity calculations indicated that cytokeratin 8/18/19 (CK) expression was significantly higher in six of the seven HCC cell lines examined than in the control cells, and the expression levels of asialoglycoprotein receptor (ASGPR) and glypican-3 (GPC3) were higher in all seven HCC cell lines than in the control. Cells obtained from HCC patients’ blood samples also displayed significantly higher expression levels of ASGPR, GPC3, and CK than cells from chronic HBV-infected patients or healthy controls; these proteins may be valuable surface biomarkers for identifying HCC circulating tumor cells isolated and enriched from the blood samples. The stem cell-like and epithelial-mesenchymal transition-related biomarkers could be detected on the karyocyte slides. ASGPR and GPC3 were expressed at high levels, and thus three-dimensional reconstructions were used to observe their expression in detail. This analysis indicated that GPC3 was localized in the cytoplasm and membrane, but that ASGPR had a polar localization. Survival analyses showed that expression of GPC3 and ASGPR is associated with a patient’s overall survival (OS).
CONCLUSION: ASGPR, GPC3, and CK may be valuable HCC biomarkers for CTC detection; the expression of ASGPR and GPC3 might be helpful for understanding patients’ OS.
Keywords: Hepatocellular carcinoma, Biomarker, Immunocytochemistry, Semiquantitative analysis, Three-dimensional reconstruction
Core tip: We report a novel workflow to detect potentially valuable biomarkers for hepatocellular carcinoma (HCC). We measured immunofluorescence intensity and performed statistical analyses to assess the expression of biomarkers in cell lines and patient blood samples. Furthermore, we determined the expression of biomarkers via three-dimensional reconstructions. These analyses indicated that asialoglycoprotein receptor (ASGPR), glypican-3 (GPC3), and cytokeratin (CK) may be valuable HCC biomarkers for detecting circulating tumor cells (CTCs). In addition, the expression of ASGPR and GPC3 might correlate with patients’ prognoses, and our CTC detection method can include epithelial cell adhesion molecule- and vimentin-positive tumor cells, and will thus supplement previous studies and potentially help predict future tumor recurrence and metastasis.
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in the world, ranking seventh and third in morbidity and mortality, respectively, for all cancers[1,2]. Although most of the burden lies in developing countries, where almost 85% of HCC cases occur, large increases in incidence, particularly in younger age groups, have been reported in the United States and Europe over the past two decades[2]. Currently, approximately 50% of the annual incidence occurs in China, making HCC the second leading cause of cancer-related deaths. This trend is likely to persist in the near future, given that 93 million hepatitis B virus (HBV) carriers and 980000 HBV patients live in China[3,4]. At present, hepatectomy is the main curative treatment, but postoperative risks can only be predicted by common clinical and pathological parameters, such as alpha fetoprotein (AFP) and tumor differentiation; prognosis remains poor due to the high incidence of recurrence and metastasis[5,6]. Considering that there are no reliable diagnostic and prognostic biomarkers for HCC patients, a variety of studies have been performed to identify new biomarkers, mainly using tumor sections or patients’ serum for the analyses[7-10].
In recent years, the number of circulating tumor cells (CTCs) has been reported to be associated with a poor prognosis[11-13]. The detection of CTCs may provide a method for assessing disease prognosis and a model for the biology of cancer metastasis. The CellSearch™ system (Veridex LLC, Warren, NJ, United States), based on the positive capture of epithelial cell adhesion molecule (EpCAM), has been approved by the United States Food and Drug Administration (FDA) for the detection of CTCs in patients with metastatic breast, prostate, and colorectal cancers[14]. In 2012, this system was approved by the China Food and Drug Administration for assessing patients with breast cancer[15]; however, few studies have examined HCC CTCs. We argued that the expression of EpCAM in HCC tissues was as low as 30%, and could be considered a cancer stem cell-like biomarker for HCC. The epithelial-mesenchymal transition (EMT), considered an initiation process for cancer metastasis, involves the loss of epithelial biomarkers such as EpCAM, which means that the CellSearch™ system may overlook HCC CTCs[16,17]. Although several studies have investigated a suitable method for identifying HCC CTCs[18,19], further efforts are required. Here, we analyzed the expression of potential cellular biomarkers for the identification of HCC CTCs in seven cell lines and in karyocytes isolated from HCC patients’ peripheral blood. We included HCC-related biomarkers, such as cytokeratin 8/18/19 (CK), glypican-3 (GPC3), and asialoglycoprotein receptor (ASGPR); stem cell-related biomarkers, such as EpCAM and CD133; and EMT-related biomarkers, such as vimentin. Cytokeratins are proteins in the intracytoplasmic cytoskeleton, and CK expression has been used as a biomarker in hepatoma histopathology[20,21]. GPC3 attached to the cell surface is overexpressed in most HCC foci and undetectable in normal livers and benign liver diseases[7]. ASGPR, as a membrane receptor, can specifically interact with the pre-S1 domain of HBV[22]. EpCAM was reported as a HCC stem cell-like biomarker, and CD133 is a common stem cell biomarker. EpCAM- and CD133-positive cells can potentially undergo self-renewal and differentiation[23,24]. Vimentin is a predominant mesenchymal marker in the EMT[25]. Given that normal epithelial cells and hepatocytes in the circulation of healthy adults are rare, any of these cells identified by CK, GPC3, or ASGPR are likely to be tumor cells[7,19,26]. Stem cell-related and EMT marker-positive tumor cells may be associated with recurrence and metastasis, which corresponds with the clinical significance of CTCs in several cancers[27-30].
The primary aim of this study was to assess biomarker expression in hepatoma cell lines and enriched patient blood cells using a semiquantitative cytopathological workflow. Secondly, the present study analyzes if exceptional biomarker expression in HCC patients could be valuable for determining cancer prognosis.
MATERIALS AND METHODS
Cell culture and preparation of cells plated on coverslips
Human hepatoma cell lines HepG2 (ATCC HB-8065), Hep3B (ATCC HB-8064), and SK-HEP-I (ATCC HTB-52) cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, United States), and Bel7402, Bel7404, SMMC7721, and the human hepatic cell line L02 cells were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, China. Huh7 cells were obtained from the Human Science Research Resources Bank (Osaka, Japan). HepG2, Hep3B, and SK-Hep-I cells were cultured in MEM (Hyclone, Logan, UT, United States) supplemented with 10% FBS (Hyclone, Logan, UT, United States), 1% 200 mmol glutamine, and 1% 100 mmol pyruvic acid sodium. Huh7 cells were cultured in Dulbecco’s modified Eagle medium (Hyclone, Logan, UT, United States) supplemented with 10% FBS. Bel7402, Bel7404, and SMMC7721 cells were cultured in RPMI-1640 (Hyclone, Logan, UT, United States) supplemented with 10% FBS. Cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2, and were harvested with trypsin before plating for experiments in tissue culture plates and on coverslips. The cells were seeded onto coverslips pretreated with 0.01% poly-L-lysine and fixed with 4% paraformaldehyde when the confluence reached approximately 60%-70%.
Blood sample preparation
Eighty-four individuals, including 62 HCC patients, 7 chronic HBV-infected patients, and 15 healthy individuals, were recruited in the present study. A total of 7.5 mL of peripheral blood was collected in BD vacutainer tubes containing acid citrate dextrose (Becton Dickinson, Franklin Lakes, NJ, United States). Written informed consent was obtained from each participant, and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki, as reflected in a priori approval by the Review Board at the Cancer Hospital affiliated with the Chinese Academy of Medical Sciences, Peking Union Medial College, and Navy General Hospital (Beijing, China). To avoid epithelial cell contamination during venous puncture, all samples were collected after discarding the first 2 mL of blood. Samples were processed within 24 h of collection. Diagnoses were pathologically confirmed using surgical specimens. The clinical characteristics of HCC patients are summarized in Table 1. HCC patients were classified according to the seventh edition of the cancer staging system published by the American Joint Committee on Cancer (AJCC) and the Union for International Cancer Control (UICC).
Table 1.
Clinical characteristics of individuals enrolled in the study n (%)
| Clinical characteristics | HCC number | Control number |
| Age at baseline, years | ||
| ≥ 60 | 20 (32.3) | 6 (27.3) |
| < 60 | 42 (67.7) | 16 (72.7) |
| Median | 55 | 51 |
| Range | 29-76 | 20-72 |
| Sex | ||
| Male | 53 (85.5) | 11 (50.0) |
| Female | 9 (14.5) | 11 (50.0) |
| KPS score | ||
| > 80 | 31 (50.0) | 22 (100.0) |
| 60-80 | 31 (50.0) | 0 (0.0) |
| Hepatitis infection | ||
| HBV positive | 59 (95.2) | 7 (31.2) |
| HBV negative | 3 (4.8) | 15 (68.2) |
| Tumor size (cm) | NA | |
| ≤ 3.0 | 21 (33.9) | |
| > 3.0 | 41 (66.1) | |
| Primary foci | NA | |
| Single | 54 (87.1) | |
| Multiple | 8 (12.9) | |
| Carcinoma cell embolus | NA | |
| Yes | 1 (1.6) | |
| No | 61 (98.4) | |
| AJCC stage at enrollment | NA | |
| Early (stage 1 + 2) | 55 (88.7) | |
| Late (stage 3 + 4) | 7 (11.3) |
HCC: Hepatocellular carcinoma; KPS: Karnofsky performance status; HBV: Hepatitis B virus; AJCC: American Joint Committee on Cancer; NA: Not applicable.
The karyocyte enrichment method was similar to previous descriptions[31]. Blood was transferred to a 50 mL centrifuge tube. The collecting tubes were rinsed twice with wash buffer (137 mmol/L NaCl, 2.7 mmol KCl, 10 mmol/L Na2HPO4, 2 mmol/L KH2PO4, 2 mmol/L EDTA, 0.5% BSA, pH = 7.4) to a combined volume of 45 mL. Blood samples were centrifuged at 1400 rpm for 5 min, and the supernatant was aspirated. Red blood cells (RBCs) were mixed with 37.5 mL of lysis buffer (155 mmol/L NH4Cl, 10 mmol/L KHCO3, 0.1 mmol/L EDTA), rotated for 8 min, and centrifuged at 1400 rpm for 5 min. The procedure was repeated twice. The resulting cell pellet was resuspended, washed, and incubated with CD45 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) at a proportion of 20 μL per 107 total white blood cells (WBCs) for 15 min. WBCs bound to microbeads were removed with an LS column in a MidiMACS™ separator (Miltenyi Biotec, Bergisch Gladbach, Germany). Supernatants were transferred into a new tube and centrifuged at 1400 rpm for 5 min. Cell pellets were fixed with 4% paraformaldehyde on SuperFrost Plus slides (Thermo Fisher Scientific, Pittsburgh, PA, United States), with immunofluorescence (IF) staining then being performed.
Immunofluorescence staining and microscopic examination
Cells on coverslips and karyocyte slides enriched from blood samples were blocked using 2% BSA (Sigma-Aldrich, St. Louis, MO, United States) for 45 min. Direct and indirect IF staining was performed at room temperature with the following HCC-related biomarkers (1:100 diluted in 2% BSA): anti-CK8/18/19-FITC (Miltenyi Biotec, Bergisch Gladbach, Germany), anti-GPC3 (Santa Cruz Biotechnology Inc, Dallas, TX, United States), anti-ASGPR (Sigma-Aldrich, St. Louis, MO, United States), anti-AFP (Zymed Laboratory Inc., South San Francisco, CA, United States), and anti-CD45-PE (Miltenyi Biotec, Bergisch Gladbach, Germany). EMT-related and stem cell-related biomarkers (1:100 diluted in 2% BSA) were also tested, including anti-vimentin (Thermo Fisher Scientific, Pittsburgh, PA, United States), anti-EpCAM-FITC (Miltenyi Biotec, Bergisch Gladbach, Germany), and anti-CD133-PE (Miltenyi Biotec, Bergisch Gladbach, Germany). Coverslips and slides were incubated for 60 min with primary antibodies for IF staining and with Alexa Fluor-labeled secondary antibody (Life Technologies, Grand Island, NY, United States) for another 60 min for indirect IF staining. They were then washed three times with 0.2% BSA for 3 min. Cells were mounted with medium containing the nuclear dye 4’,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, MO, United States). Blinded review of the fluorescent images was performed by three technicians using a Nikon 80i 3-color fluorescent microscope (Nikon Co, Tokyo, Japan) and a Leica TCS SP2 confocal microscope (Leica Co, Berlin, Germany). Fluorescence intensities were measured using the Image J software (NIH, Bethesda, MD, United States). Relative biomarker intensities on cells were assessed using the following formula that calculates how many times higher the biomarker staining intensity of the cell is than the background, which was based on the CellSearch™ model for HER2 staining of CTCs[32,33]:
Biomarker Relative Intensity = [Foreground Intensity (Cell staining)/Surface Area]/[Background Intensity/Surface Area]
The experiments were performed in triplicate. Representative images of 5 cells from each cell line and 3 cells from each patient were analyzed, and the mean intensity was calculated. Only cells from patients who met the following stringent criteria were analyzed: intact cells with diameters above 10 μm, biomarker-positive, CD45-negative, and DAPI-positive. The positive ratios of biomarker expression were calculated from the observation of 500 cells.
Immunohistochemical staining of liver cancer samples
Tissue arrays were composed of tumor samples and para-carcinoma liver tissues collected from 32 patients who were diagnosed with HCC and underwent surgical resection (Shanghai National Engineering Research Center for Biochip, Shanghai, China). From each paraffin block, 4-μm-thick sections were cut, cleared in xylene, and rehydrated. Slides were then moved from water to plastic slide holders, fully immersed in 10 mmol sodium citrate buffer (pH = 6.0), and heated for 20 min at 120 °C in a commercially available pressure cooker. After cooling to room temperature in the sodium citrate buffer, slides were treated with a solution of 3% hydrogen peroxide (H2O2) for 30 min at room temperature to abolish endogenous peroxidase activity. Sections were then incubated for 10 min in a moist chamber with non-immune goat serum diluted to 5% in PBS (pH = 7.4) to reduce non-specific background staining. Sections were then incubated overnight at 4 °C with the primary antibody (anti-GPC3 or anti-ASGPR) diluted to 1:100. Sections were subsequently incubated for 30 min with biotin-labeled secondary antibody (Santa Cruz Biotechnology Inc, Dallas, TX, United States) diluted to 1:600 in PBS, and then incubated in a streptavidin-biotin-peroxidase preformed complex (Zhongshan Jinqiao Biotechnology Co, Beijing, China) for 30 min. The immunologic reaction was visualized using 3,3′-diaminobenzidine substrate, and samples were counterstained with hematoxylin, dehydrated, and mounted with Canada balsam (Sigma-Aldrich, St. Louis, MO, United States). A negative control was performed by omitting the primary antibody.
The tumor expression of ASGPR and GPC3 was evaluated by two pathologists (HM and HZL) blinded to clinical data, and discrepancies were resolved by consensus. Images were visualized using an Olympus BX40 microscope (Olympus Co, Tokyo, Japan). Ten random fields per biomarker were selected, and the percentage of immunoreactive cells in a total of 1000 tumor cells was determined[34]. As described previously, the intensity of a biomarker and the percentage of positive cells were both graded with different scores. The product of the two scores was used to evaluate the biomarker’s staining[35,36]. An intensity score representing the average intensity of positive cells (0, none; 1, weak; 2, intermediate; 3, strong) was assigned. A proportion score representing the estimated proportion of positive-staining cells (0, 0-5% positive cells; 1, 6%-25%; 2, 26%-50%; 3, 51%-75%; 4, 76%-100%) was assigned. The proportion and intensity scores were multiplied to create an immunoreactivity score (IS). The IS was further divided as follows: 0-1 (−); 2-4 (+); 5-7 (++); > 8 (+++). “-” and “+” were considered low levels of immunoreactivity; “++” and “+++” were considered high levels of immunoreactivity.
Statistical analysis
Analyses were performed using SPSS version 19.0 (IBM, New York, NY, United States). The Mann-Whitney test was used to compare continuous variables between the two groups. Fluorescence intensities and clinical characteristics, such as age, AFP, ALT, AST, tumor size, differentiation, and AJCC stage, were subjected to Spearman rank correlation analysis. Survival analysis was performed using the Kaplan-Meier method. P values less than 0.05 were considered statistically significant, and P values less than 0.1 were considered potentially significant.
RESULTS
Expression of HCC-related biomarkers in cell lines
IF staining indicated that ASGPR (Figure 1A) and GPC3 (Figure 1B) were expressed in all liver cancer-related cell lines, and fluorescence intensity was significantly higher in the seven HCC cell lines than in L02, the control cells (Figure 1D). Confocal microscopic examination suggested that ASGPR is located in the nuclei or nucleoli of the Bel7404 and SMMC7721 cells. In addition, GPC3 expression was significantly stronger in cell lines derived from Asians than from Westerners (10.41 ± 6.67 vs 5.61 ± 2.89, P < 0.05). CK was expressed in all cell lines and located in the cytoplasm (Figure 1C). When the fluorescence intensities were compared, CK expression in six of the seven HCC cell lines was significantly higher than in the control cell line, L02 (Figure 1D). AFP was only expressed in Hep3B, HepG2, and SK-HEP-I cell lines.
Figure 1.
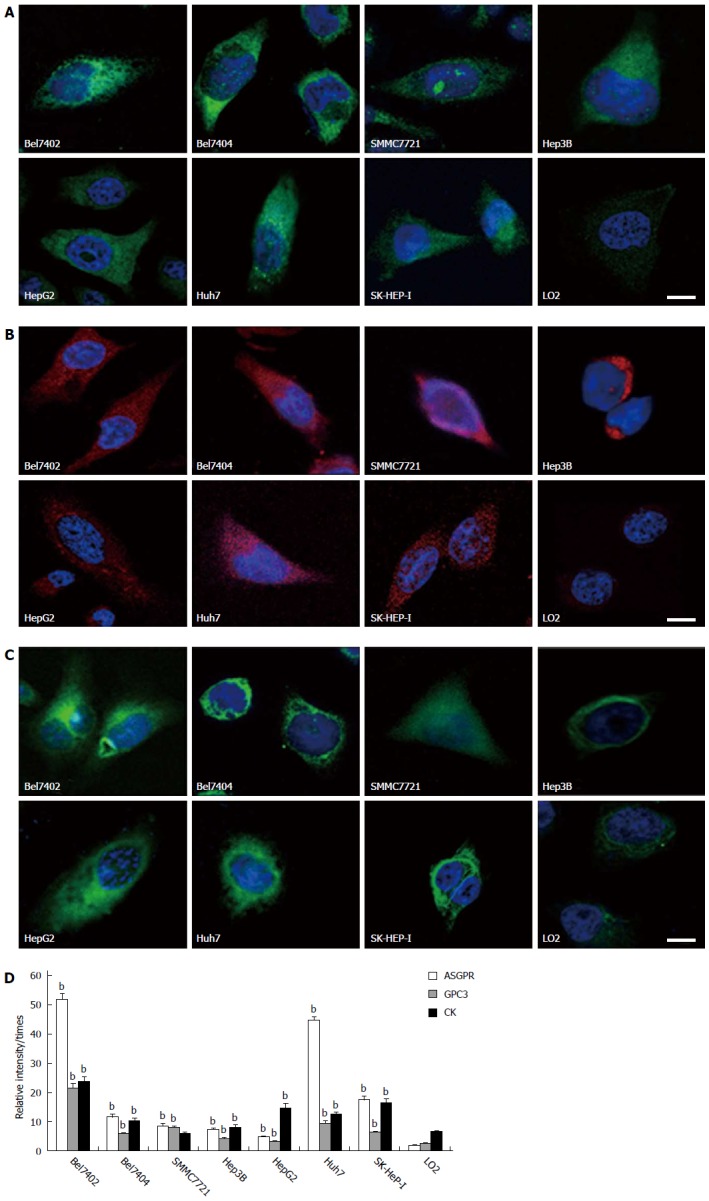
Asialoglycoprotein receptor, glypican-3 and cytokeratin expression, and fluorescence intensities in eight cell lines. A-C: Representative images of cells on the coverslips with DAPI-stained nuclei (blue) and asialoglycoprotein receptor (ASGPR) staining (green)/glypican-3 (GPC3) staining (red)/cytokeratin (CK) staining (green). The scale bar is 5 μm. D: The bar graph of fluorescence intensities in eight cell lines. Fluorescence intensities were measured using NIH Image J software. Biomarker relative intensities were calculated as the difference between the biomarker staining intensity of the cell and the background intensity. The comparisons between HCC cell lines and the control cell line were analyzed using the Mann-Whitney test (bP < 0.01 vs control).
Expression of EMT- and stem cell-related biomarkers in cell lines
IF staining indicated that there were considerably fewer cells expressing EpCAM than there were cells expressing the above three biomarkers (Figure 2). The cells expressing CD133 were even rarer than those expressing EpCAM. Table 2 presents the positive ratios in the cell lines tested. Vimentin was expressed at very low levels, and ratios in the cell lines determined by IF staining.
Figure 2.
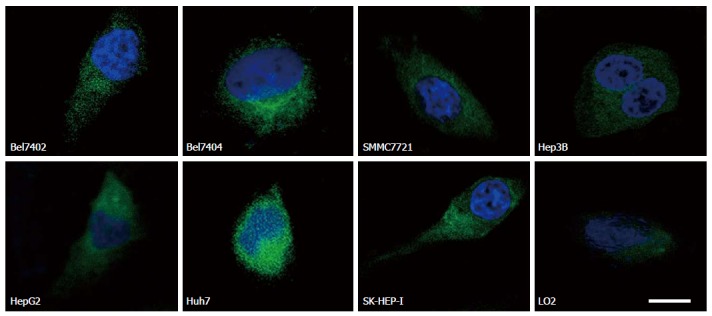
Epithelial cell adhesion molecule expression in eight cell lines. Representative images of cells on the coverslips with DAPI-stained nuclei (blue) and EpCAM staining (green). The scale bar is 5 μm.
Table 2.
Positive ratios of epithelial cell adhesion molecule/CD133 expression in hepatocellular carcinoma cells
| % | Bel7402 | Bel7404 | SMMC7721 | Huh7 | HepG2 | Hep3B | SK-Hep-I | L02 |
| EpCAM | 0.4 | 1.2 | 0.2 | 1 | 0.6 | 0.8 | 2 | 0 |
| CD133 | 0.4 | 0.4 | 0.2 | 0.2 | 0.2 | 0.4 | 0.6 | 0 |
EpCAM: Epithelial cell adhesion molecule.
Expression of biomarkers in blood samples
Based on the above results, three HCC-related biomarkers were further detected in patient samples. IF staining indicated that GPC3, ASGPR, and CK were expressed in most HCC patients (Figure 3), independent of HBV infection. Positive staining was rarely observed on the karyocyte slides from chronic HBV-infected patients or healthy controls.
Figure 3.
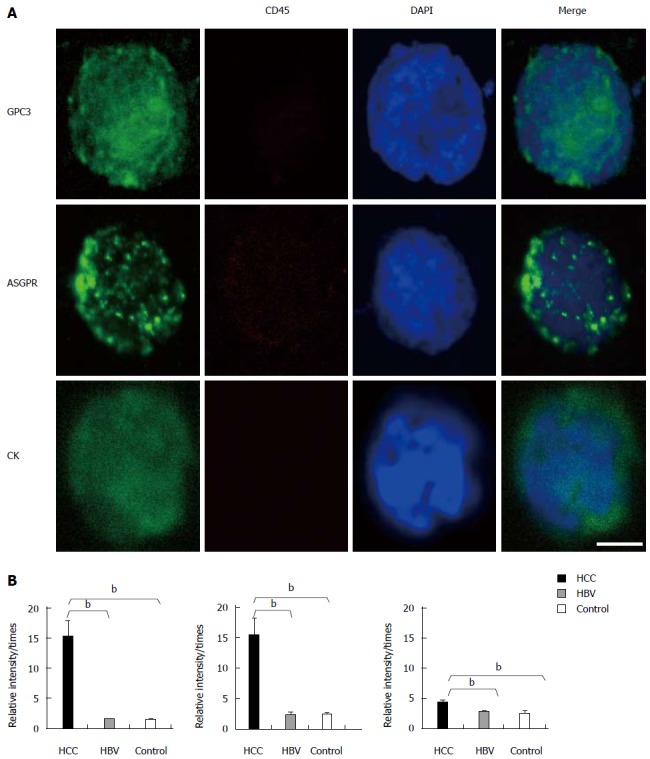
Representative images and fluorescence intensities of cells obtained from patients. A: Representative image of circulating tumor cells (CTCs) with DAPI-stained nuclei (blue), positive staining of ASGPR/GPC3/CK (green), and negative staining for CD45. The scale bar is 5 μm. A total of 7.5 mL of peripheral blood was collected from each individual, mixed with red blood cell lysis buffer, and incubated with CD45 microbeads to deplete the white blood cells. The remaining cell pellets were fixed on SuperFrost Plus slides for immunofluorescence staining; B: The bar graph of ASGPR/GPC3/CK (from left to right) fluorescence intensities in 30 HCC patients, 7 chronic HBV-infected patients, and 15 healthy controls. Fluorescence intensities were measured using NIH Image J software. Biomarker relative intensities were calculated as the difference between the biomarker staining intensity of the cell and the background intensity. The comparisons between HCC patients and healthy controls were analyzed by the Mann-Whitney test (bP < 0.01 vs control).
We compared the fluorescence intensities of three biomarkers between HCC patients, chronic HBV-infected patients, and healthy controls. The expression of the three biomarkers was significantly increased in HCC patients compared to chronic HBV-infected patients and healthy controls. The fluorescence intensities of GPC3 and ASGPR were considerably higher than that of CK. The concordance rates between a biomarker’s expression and pathological diagnosis were 90% for GPC3, 93.3% for ASGPR, and 63.3% for CK, whereas the concordance rate between AFP serum detection and pathological diagnosis was 46.7%. The positive and negative predictive values of these three biomarkers were 90% and 71.4% for GPC3, 93.1% and 75% for ASGPR, and 82.6% and 28.6% for CK, respectively.
To understand the clinical significance of the biomarkers’ expression levels, we used Spearman rank correlation analysis to assess the relationship between fluorescence intensities and clinical pathological parameters. No significant correlations were found.
Vimentin was positively expressed on some of the karyocyte slides and was accompanied by the expression of EpCAM (Figure 4). Because EpCAM is associated with stem cell-like properties in HCC patients, it was unnecessary to compare its fluorescence intensities in the manner performed for the HCC-related biomarkers discussed previously. EpCAM was expressed in 34.4% and vimentin was expressed in 24.1% of the cells on the karyocyte slides.
Figure 4.
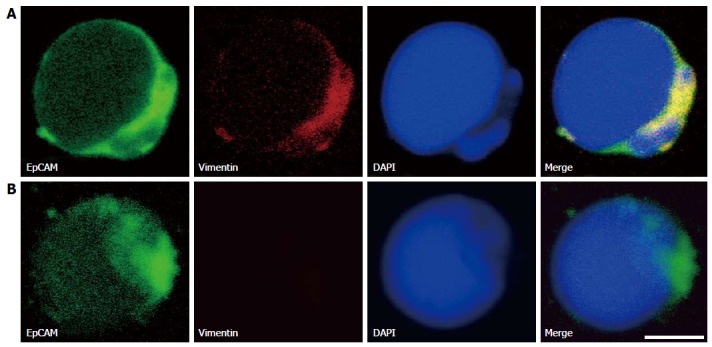
Representative images of epithelial-mesenchymal transition marker-related cells obtained from hepatocellular carcinoma patients. A: CTCs with dye 4’,6-diamidino-2-phenylindole, (DAPI) -stained nuclei (blue), positive staining of epithelial cell adhesion molecule (EpCAM) (green), and positive staining of vimentin (red). B: CTCs with DAPI-stained nuclei (blue), positive staining of EpCAM (green), and negative staining of vimentin. The scale bar is 5 μm.
Three-dimensional reconstruction for GPC3 and ASGPR
Three-dimensional reconstructions were generated to evaluate ASGPR and GPC3 localization. The cells were revolved around the X-, Y-, and Z-axes, which indicated that GPC3 was localized in the cytoplasm and on the membrane (mov 1), whereas ASGPR had a polar localization (mov 2).
Survival analysis for GPC3 and ASGPR
The expression of GPC3 and ASGPR is shown in Figure 5. Kaplan-Meier survival analyses indicated that ASGPR expression was significantly associated with the overall survival (OS) (P = 0.017), and GPC3 expression exhibited a trend toward significant association with OS (P = 0.068). The mean OS of patients with high and low levels of ASGPR was 24.1 and 48.5 mo, respectively. The mean OS of patients with high and low levels of GPC3 was 31.0 and 48.9 mo, respectively.
Figure 5.
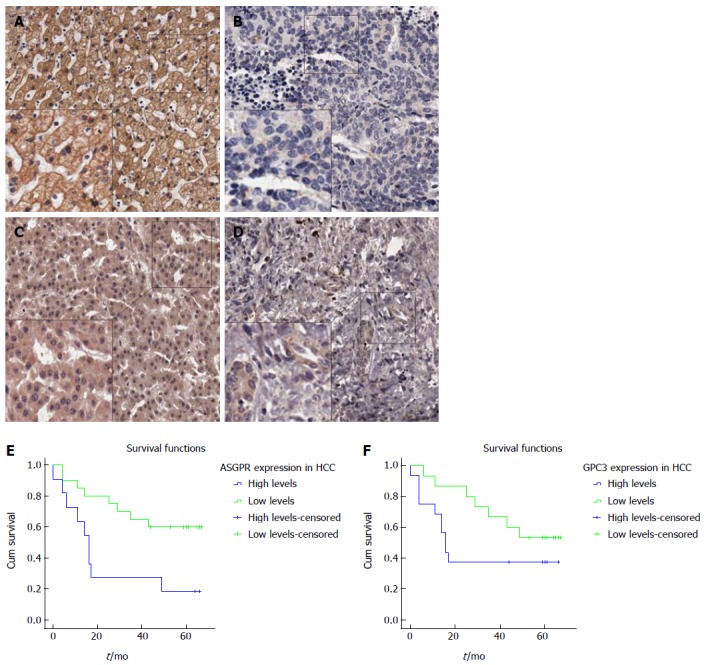
Representative images of asialoglycoprotein receptor/glypican-3 expression on tissue arrays, and Kaplan-Meier survival curves from follow-up studies. A-B: The expression of ASGPR in HCC (A) and paracarcinomatous (B) tissues. C-D: The expression of GPC3 in HCC (C) and matched paracarcinomatous (D) tissues (A-D: × 100; Inner × 200). The survival curves from six and a half years of follow-up indicate a significant difference between 32 HCC patients with high and low levels of ASGPR (E) and exhibit a trend toward significance between 32 HCC patients with high and low levels of GPC3 (F).
DISCUSSION
HCC ranks as the third most frequent cause of cancer-related death worldwide[2]. In China, it is the third most prevalent cancer, the second leading cause of cancer-related death, and new cases in the country account for 55% of those reported globally[4]. Currently serum AFP, a secretory protein, is widely used for detecting HCC patients and monitoring disease progression, but it has a sensitivity ranging from 39% to 97% and a specificity ranging from 76% to 95%, even when used to screen high-risk populations[37-39]. In a previous study, AFP was used to identify CTCs in patients with liver cancer[18]. However, given the shortcomings of AFP, there may be biomarkers that can detect CTCs with a higher sensitivity.
In this study, and based on our previous tests, we assessed the expression of potential key biomarkers in seven HCC cell lines and karyocytes isolated from patient blood samples. Cytokeratins are proteins that consist of keratin-containing intermediate filaments in the intracytoplasmic cytoskeleton, and their expression primarily depends on the type of epithelia, the moment of terminal differentiation, and the stage of development[40]. In many cases, cytokeratin expression in tumors and peripheral blood has prognostic significance for cancer patients, and the levels of CK8/18/19 expression in HCC have been used as biomarkers in histopathology[20,21]. In addition to conventional histopathology, CK8/18/19 have been used as diagnostic biomarkers in CTCs for breast, prostate, and colorectal cancers[11-13]. Expression in HCC cells obtained from peripheral blood was explored in this study in order to broaden knowledge regarding cytopathology. Positive results from cell lines and blood samples provide evidence for potential applications of biomarkers, although additional investigation may be needed in the future.
ASGPR is located on liver cells and was previously known as a hepatic galactose/N-acetylglucosamine (GlcNAc) receptor. Its primary physiological function is to bind, internalize, and subsequently clear glycoproteins containing terminal galactose or GlcNAc residues from circulation[41-43]. Several investigations have supported ASGPR as a multifunctional membrane receptor involved in the removal of apoptotic cells[44], a carrier of low-density lipoprotein (LDL)[45], and an entry point for hepatotropic viruses[46,47]. HBV DNA and virions can reportedly be transferred into hepatocytes via ASGPR[48]. Zhang et al[22] found that ASGPR on human hepatocytes can interact specifically and directly with the pre-S1 domain of HBV in vitro and in vivo[22]. The IF images of liver cancer cell lines and patient CTCs in this study indicated strong ASGPR expression, which suggested its potential use for identifying HCC cells in circulation, especially for HBV-related HCC.
GPC3 is a proteoglycan attached to the cell surface by a glycosylphosphatidylinositol (GPI) anchor[7,49]. GPC3 mRNA levels have been reported to be significantly elevated in most HCC foci compared with normal liver and non-malignant liver lesions, and they are elevated more frequently than AFP in HCC[50]. Similar results have been obtained at the protein level. Using immunohistochemical techniques, Capurro et al[7] found that GPC3 was specifically overexpressed in most HCC foci, but was undetectable in hepatocytes from normal liver and benign liver diseases. Staining was localized to the cell membrane and/or the cytoplasm. Zhang et al[8] found that 87.1% (128/147) of surgically excised HCC samples were GPC3 positive, and did not observed GPC3 expression in para-carcinoma or cirrhotic tissues. The detection of HCC cell lines and karyocytes in patients’ blood in our studies was consistent with the above results for both mRNA and protein expression, displaying positive results compared to the controls. Additionally, tumor cells can be identified by GPC3 in patients’ blood, in which AFP tests could be positive or negative. This result was aligned with a previous report that tested for GPC3 in HCC patients’ serum[51]. The GPC3 biomarker test results corresponded better to the pathological diagnosis in our study than the serum AFP test.
The results from Spearman rank correlation analysis (Table 3) indicated that there was no significant correlation between fluorescence intensities and clinical pathological parameters, which suggested that fluorescence intensity may be unaffected by clinical pathological parameters. From our analyses of the blood samples, the fluorescence intensities of the above three biomarkers were significantly increased in HCC patients compared to chronic HBV-infected patients and healthy controls, although the elevation of CK expression was not as pronounced as the other two biomarkers. This finding indicates that CK expression may be perturbed in chronic liver disease, whereas GPC3 and ASGPR could provide a better distinction between HCC and chronic liver disease. AFP was only expressed in the cell lines derived from Westerners in our study. AFP was reported to be expressed in 52.3% of HCC patients (23/44) for the identification of CTCs, whereas the three biomarkers that we considered were more commonly detected than positive changes in AFP in the serum[18]. In this study, survival analysis indicated that patients with high ASGPR expression levels had poorer OS, and a similar trend was observed for GPC3 expression. These results suggest that GPC3 and ASGPR may correlate with patient prognosis.
Table 3.
Spearman rank correlation analysis for fluorescence intensities
| P value | Age | AFP | ALT | AST | TBIL | Size | Differentiation | AJCC stage |
| ASGPR | 0.945 | 0.909 | 0.131 | 0.281 | 0.810 | 0.537 | 0.418 | 0.796 |
| GPC3 | 0.863 | 0.911 | 0.549 | 0.846 | 0.288 | 0.357 | 0.623 | 0.849 |
| CK | 0.889 | 0.173 | 0.573 | 0.771 | 0.970 | 0.173 | 0.974 | 0.425 |
ASGPR: Asialoglycoprotein receptor; GPC3: Glypican-3; CK: Cytokeratin 8/18/19; AFP: Alpha-fetoprotein; ALT: Alanine aminotransferase; AST: Aspartate aminotransferase; TBIL: Total bilirubin; AJCC: American Joint Committee on Cancer.
EMT is a process by which epithelial cells lose their cell polarity and cell-cell adhesion, and gain migratory and invasive properties to become mesenchymal cells[17]. This transition is considered one of the mechanisms underlying cancer metastasis and recurrence. EMT and MET form the initiation and completion of the invasion-metastasis cascade[27], and CTCs in the vessels act as “intermediates” between primary tumors and metastatic foci. We measured the expression of an EMT-related biomarker on the patients’ karyocyte slides to understand the biology of CTCs. Vimentin, a widely used biomarker for EMT, was expressed on some CTCs, whereas EpCAM, an epithelium-specific biomarker, was expressed at various levels. These results suggest that CTCs may be associated with cancer metastasis and recurrence via EMT mechanisms. Some recent evidence indicates that tumor cells undergoing EMT might gain stem cell-like properties, therefore giving rise to cancer stem cells (CSCs)[27,30], and EpCAM-positive HCC cells have been reported to have stem cell-like features, including self-renewal and differentiation capacities[23,29,52]. Previously, EpCAM was only known as an epithelial biomarker in many human cancers with an epithelium origin[53], and it has thus been used as a target to capture CTCs by the CellSearch™ system. Our results demonstrate that EpCAM was not widely expressed in the HCC cell lines, and the expression ratio on the karyocyte slides was consistent with histological reports[35], which implied that the CellSearch™ system may not be ideal for the detection of HCC CTCs[52]. The expression levels of vimentin and EpCAM were approximately 20% and 30%, respectively, of the total CTCs identified by GPC3, ASGPR, and CK, which indicates that our selected biomarkers could detect more tumor cells, including the EpCAM-positive and EMT-related cells.
Of the biomarkers detected, GPC3, ASGPR, and CK were all significantly expressed in HCC cell lines and karyocytes from patient blood samples, suggesting that these proteins could be used as valuable biomarkers of circulating HCC cells. Considering that GPC3 and ASGPR may be specific to HCC from tests on clinical samples, we applied three-dimensional reconstruction for further observation, revealing their localization. Additionally, the survival analysis indicated that GPC3 and ASGPR expression correlate with patient prognoses. To our knowledge, few studies have evaluated biomarker expression using a similar workflow. The results presented in this study improve the understanding of HCC biomarkers from a cytopathological perspective given that serum and tissues are widely used in other clinical examinations for biomarkers. The HCC-related biomarkers may help to identify tumor cells in the peripheral blood, which may be relevant to patient prognosis[11-13,54]. In addition, the expression of EMT-related biomarkers suggests that CTCs may help elucidate metastatic mechanisms, and the expression of EpCAM supports its novel function associated with stem cell-like properties. Although our study was limited by the small sample size of 84 individuals in the cohort, the biomarkers tested here may be used as capturing targets for HCC CTCs and may assist in the development of effective HCC therapies and individually targeted treatments based on biomarker expression and localization.
COMMENTS
Background
Hepatocellular carcinoma (HCC) is currently one of the most common malignant tumors in the world. Although the detection of circulating tumor cells (CTCs) will provide us with prognostic information for several tumors, it is difficult to identify HCC CTCs by cellular surface biomarkers, thus making it crucial to detect the expression of biomarkers in a novel workflow.
Research frontiers
The study of CTCs is an increasingly important field in several cancers, and may be relevant to metastasis and recurrence. However, the search for biomarkers for HCC CTC identification has not been unequivocally addressed. In this study, the authors demonstrate that asialoglycoprotein receptor (ASGPR), glypican-3 (GPC3), and cytokeratin (CK) can identify CTCs on karyocyte slides. The expression of GPC3 and ASGPR is clinically significant.
Innovations and breakthroughs
Recent reports have highlighted the importance of CTC detection. For HCC in particular, it is important to identify CTCs in the blood. This study is the first to detect several biomarkers of HCC in a semiquantitative immunocytochemical workflow. Furthermore, the results suggest that GPC3, ASGPR, and CK can identify CTCs on karyocyte slides and that GPC3 and ASGPR expression correlates to tumor prognosis.
Applications
By identifying HCC CTCs via the biomarkers that the authors detected, this study may represent a future strategy for monitoring CTC changes and assessing the therapeutic prognosis of HCC.
Terminology
CTCs are cells that have dislocated from a primary tumor to circulate in the blood, and they may be able to seed in distant organs to become a metastatic focus. Thus, they are thought to be relevant to metastasis and recurrence. However, potential biomarkers to identify HCC CTCs must be identified.
Peer review
The authors detected the expression levels of potential cellular biomarkers for HCC and demonstrated that GPC3, ASGPR, and CK may be valuable surface biomarkers for the identification of HCC CTCs. Furthermore, GPC3 and ASGPR can better distinguish HCC patients from chronic HBV patients and healthy controls, and their expression is clinically significant. In addition, the expression of EMT- and stem cell-related biomarkers might provide us with helpful information regarding metastasis and recurrence. The results are interesting and may represent a strategy for identifying HCC CTCs and developing effective targeted treatments.
Footnotes
Supported by Grants from the National High-tech R and D Program No. 2012AA020206, the Key Project for the Infectious Diseases No. 2012ZX10002-017 and No. 2013ZX10002009-001-004, the State Key Projects for Basic Research No. 2011CB910703, the National Natural Science Foundation No. 81372591, and No. 81321091 of China and the Center for Marine Medicine and Rescue of Tsinghua University
P- Reviewers: Bayliss J, Costantini S, Lin CW, Kim SS S- Editor: Qi Y L- Editor: Rutherford A E- Editor: Liu XM
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.International Agency for Research on Cancer. GLOBOCAN 2012: Estimated cancer incidence, mortality and prevalence worldwide in 2012. Available from: http://globocan.iarc.fr/factsheet.asp.
- 3.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273.e1. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong LZ, Zhao P. Report on cancers in China: Retrospective survey on malignant tumor in China. 1st ed. Beijing: People’s Medical Publishing House; 2010. pp. 37–38. [Google Scholar]
- 5.Ma WJ, Wang HY, Teng LS. Correlation analysis of preoperative serum alpha-fetoprotein (AFP) level and prognosis of hepatocellular carcinoma (HCC) after hepatectomy. World J Surg Oncol. 2013;11:212. doi: 10.1186/1477-7819-11-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah SA, Cleary SP, Wei AC, Yang I, Taylor BR, Hemming AW, Langer B, Grant DR, Greig PD, Gallinger S. Recurrence after liver resection for hepatocellular carcinoma: risk factors, treatment, and outcomes. Surgery. 2007;141:330–339. doi: 10.1016/j.surg.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 7.Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125:89–97. doi: 10.1016/s0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Liu H, Sun L, Li N, Ding H, Zheng J. Glypican-3 as a potential differential diagnosis marker for hepatocellular carcinoma: A tissue microarray-based study. Acta Histochem. 2012;114:547–552. doi: 10.1016/j.acthis.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Tomoda T, Nouso K, Miyahara K, Kobayashi S, Kinugasa H, Toyosawa J, Hagihara H, Kuwaki K, Onishi H, Nakamura S, et al. Prognotic impact of serum follistatin in patients with hepatocellular carcinoma. J Gastroenterol Hepatol. 2013;28:1391–1396. doi: 10.1111/jgh.12167. [DOI] [PubMed] [Google Scholar]
- 10.Yang H, Chen GD, Fang F, Liu Z, Lau SH, Zhang JF, Lau WY, Yang LY. Dickkopf-1: as a diagnostic and prognostic serum marker for early hepatocellular carcinoma. Int J Biol Markers. 2013;28:286–297. doi: 10.5301/jbm.5000015. [DOI] [PubMed] [Google Scholar]
- 11.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 12.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 13.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 14.Food and Drug Administration. 510(k) substantial equivalence determination. Available from: http://www.accessdata.fda.gov/cdrh_docs/reviews/K040898.pdf.
- 15.China Food and Drug Administration. Data search for instruments. Available from: http://app1.sfda.gov.cn/datasearch/face3/base.jsp.
- 16.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 17.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vona G, Estepa L, Béroud C, Damotte D, Capron F, Nalpas B, Mineur A, Franco D, Lacour B, Pol S, et al. Impact of cytomorphological detection of circulating tumor cells in patients with liver cancer. Hepatology. 2004;39:792–797. doi: 10.1002/hep.20091. [DOI] [PubMed] [Google Scholar]
- 19.Xu W, Cao L, Chen L, Li J, Zhang XF, Qian HH, Kang XY, Zhang Y, Liao J, Shi LH, et al. Isolation of circulating tumor cells in patients with hepatocellular carcinoma using a novel cell separation strategy. Clin Cancer Res. 2011;17:3783–3793. doi: 10.1158/1078-0432.CCR-10-0498. [DOI] [PubMed] [Google Scholar]
- 20.Kakehashi A, Kato A, Inoue M, Ishii N, Okazaki E, Wei M, Tachibana T, Wanibuchi H. Cytokeratin 8/18 as a new marker of mouse liver preneoplastic lesions. Toxicol Appl Pharmacol. 2010;242:47–55. doi: 10.1016/j.taap.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Tsuchiya K, Komuta M, Yasui Y, Tamaki N, Hosokawa T, Ueda K, Kuzuya T, Itakura J, Nakanishi H, Takahashi Y, et al. Expression of keratin 19 is related to high recurrence of hepatocellular carcinoma after radiofrequency ablation. Oncology. 2011;80:278–288. doi: 10.1159/000328448. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Lin SM, Chen TY, Liu M, Ye F, Chen YR, Shi L, He YL, Wu LX, Zheng SQ, et al. Asialoglycoprotein receptor interacts with the preS1 domain of hepatitis B virus in vivo and in vitro. Arch Virol. 2011;156:637–645. doi: 10.1007/s00705-010-0903-x. [DOI] [PubMed] [Google Scholar]
- 23.Terris B, Cavard C, Perret C. EpCAM, a new marker for cancer stem cells in hepatocellular carcinoma. J Hepatol. 2010;52:280–281. doi: 10.1016/j.jhep.2009.10.026. [DOI] [PubMed] [Google Scholar]
- 24.Yu X, Lin Y, Yan X, Tian Q, Li L, Lin EH. CD133, Stem Cells, and Cancer Stem Cells: Myth or Reality? Curr Colorectal Cancer Rep. 2011;7:253–259. doi: 10.1007/s11888-011-0106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Zhang B, Sun B, Wang X, Ban X, Sun T, Liu Z, Zhao X. A novel function for vimentin: the potential biomarker for predicting melanoma hematogenous metastasis. J Exp Clin Cancer Res. 2010;29:109. doi: 10.1186/1756-9966-29-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erdbruegger U, Haubitz M, Woywodt A. Circulating endothelial cells: a novel marker of endothelial damage. Clin Chim Acta. 2006;373:17–26. doi: 10.1016/j.cca.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 28.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaifi JT, Kunkel M, Zhu J, Dicker DT, Gusani N, Yang Z, Sarwani NE, Li G, Kimchi ET, Staveley-O’Carroll KF, et al. Circulating tumor cells are associated with diffuse spread in stage IV colorectal cancer patients. Cancer Biol Ther. 2013;14:1174–1181. doi: 10.4161/cbt.26884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tinhofer I, Saki M, Niehr F, Keilholz U, Budach V. Cancer stem cell characteristics of circulating tumor cells. Int J Radiat Biol. 2014:Epub ahead of print. doi: 10.3109/09553002.2014.886798. [DOI] [PubMed] [Google Scholar]
- 31.Witzig TE, Bossy B, Kimlinger T, Roche PC, Ingle JN, Grant C, Donohue J, Suman VJ, Harrington D, Torre-Bueno J, et al. Detection of circulating cytokeratin-positive cells in the blood of breast cancer patients using immunomagnetic enrichment and digital microscopy. Clin Cancer Res. 2002;8:1085–1091. [PubMed] [Google Scholar]
- 32.Riethdorf S, Müller V, Zhang L, Rau T, Loibl S, Komor M, Roller M, Huober J, Fehm T, Schrader I, et al. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin Cancer Res. 2010;16:2634–2645. doi: 10.1158/1078-0432.CCR-09-2042. [DOI] [PubMed] [Google Scholar]
- 33.Ignatiadis M, Rothé F, Chaboteaux C, Durbecq V, Rouas G, Criscitiello C, Metallo J, Kheddoumi N, Singhal SK, Michiels S, et al. HER2-positive circulating tumor cells in breast cancer. PLoS One. 2011;6:e15624. doi: 10.1371/journal.pone.0015624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao R, Bao S, Bai X, Blanchette C, Anderson RM, Dang T, Gishizky ML, Marks JR, Wang XF. Acquired expression of periostin by human breast cancers promotes tumor angiogenesis through up-regulation of vascular endothelial growth factor receptor 2 expression. Mol Cell Biol. 2004;24:3992–4003. doi: 10.1128/MCB.24.9.3992-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao XP, Pretlow TG, Rao JS, Pretlow TP. Beta-catenin expression is altered in human colonic aberrant crypt foci. Cancer Res. 2001;61:8085–8088. [PubMed] [Google Scholar]
- 36.Sun Y, Mi W, Cai J, Ying W, Liu F, Lu H, Qiao Y, Jia W, Bi X, Lu N, et al. Quantitative proteomic signature of liver cancer cells: tissue transglutaminase 2 could be a novel protein candidate of human hepatocellular carcinoma. J Proteome Res. 2008;7:3847–3859. doi: 10.1021/pr800153s. [DOI] [PubMed] [Google Scholar]
- 37.Gutman S, Kessler LG. The US Food and Drug Administration perspective on cancer biomarker development. Nat Rev Cancer. 2006;6:565–571. doi: 10.1038/nrc1911. [DOI] [PubMed] [Google Scholar]
- 38.Soresi M, Magliarisi C, Campagna P, Leto G, Bonfissuto G, Riili A, Carroccio A, Sesti R, Tripi S, Montalto G. Usefulness of alpha-fetoprotein in the diagnosis of hepatocellular carcinoma. Anticancer Res. 2003;23:1747–1753. [PubMed] [Google Scholar]
- 39.Sanai FM, Sobki S, Bzeizi KI, Shaikh SA, Alswat K, Al-Hamoudi W, Almadi M, Al Saif F, Abdo AA. Assessment of alpha-fetoprotein in the diagnosis of hepatocellular carcinoma in Middle Eastern patients. Dig Dis Sci. 2010;55:3568–3575. doi: 10.1007/s10620-010-1201-x. [DOI] [PubMed] [Google Scholar]
- 40.Franke WW, Schmid E, Osborn M, Weber K. Intermediate-sized filaments of human endothelial cells. J Cell Biol. 1979;81:570–580. doi: 10.1083/jcb.81.3.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spiess M. The asialoglycoprotein receptor: a model for endocytic transport receptors. Biochemistry. 1990;29:10009–10018. doi: 10.1021/bi00495a001. [DOI] [PubMed] [Google Scholar]
- 42.Ashwell G, Morell AG. The role of surface carbohydrates in the hepatic recognition and transport of circulating glycoproteins. Adv Enzymol Relat Areas Mol Biol. 1974;41:99–128. doi: 10.1002/9780470122860.ch3. [DOI] [PubMed] [Google Scholar]
- 43.Pricer WE, Hudgin RL, Ashwell G, Stockert RJ, Morell AG. A membrane receptor protein for asialoglycoproteins. Methods Enzymol. 1974;34:688–691. doi: 10.1016/s0076-6879(74)34090-6. [DOI] [PubMed] [Google Scholar]
- 44.Stockert RJ. The asialoglycoprotein receptor: relationships between structure, function, and expression. Physiol Rev. 1995;75:591–609. doi: 10.1152/physrev.1995.75.3.591. [DOI] [PubMed] [Google Scholar]
- 45.Windler E, Greeve J, Levkau B, Kolb-Bachofen V, Daerr W, Greten H. The human asialoglycoprotein receptor is a possible binding site for low-density lipoproteins and chylomicron remnants. Biochem J. 1991;276(Pt 1):79–87. doi: 10.1042/bj2760079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Bo XC, Ding XR, Dai JM, Zhang ML, Wang XH, Wang SQ. Antisense oligonucleotides targeted against asialoglycoprotein receptor 1 block human hepatitis B virus replication. J Viral Hepat. 2006;13:158–165. doi: 10.1111/j.1365-2893.2005.00666.x. [DOI] [PubMed] [Google Scholar]
- 47.Yang J, Wang F, Tian L, Su J, Zhu X, Lin L, Ding X, Wang X, Wang S. Fibronectin and asialoglyprotein receptor mediate hepatitis B surface antigen binding to the cell surface. Arch Virol. 2010;155:881–888. doi: 10.1007/s00705-010-0657-5. [DOI] [PubMed] [Google Scholar]
- 48.Treichel U, Meyer zum Büschenfelde KH, Stockert RJ, Poralla T, Gerken G. The asialoglycoprotein receptor mediates hepatic binding and uptake of natural hepatitis B virus particles derived from viraemic carriers. J Gen Virol. 1994;75(Pt 11):3021–3029. doi: 10.1099/0022-1317-75-11-3021. [DOI] [PubMed] [Google Scholar]
- 49.Stefaniuk P, Cianciara J, Wiercinska-Drapalo A. Present and future possibilities for early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2010;16:418–424. doi: 10.3748/wjg.v16.i4.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu ZW, Friess H, Wang L, Abou-Shady M, Zimmermann A, Lander AD, Korc M, Kleeff J, Büchler MW. Enhanced glypican-3 expression differentiates the majority of hepatocellular carcinomas from benign hepatic disorders. Gut. 2001;48:558–564. doi: 10.1136/gut.48.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H, Li P, Zhai Y, Qu CF, Zhang LJ, Tan YF, Li N, Ding HG. Diagnostic value of glypican-3 in serum and liver for primary hepatocellular carcinoma. World J Gastroenterol. 2010;16:4410–4415. doi: 10.3748/wjg.v16.i35.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun YF, Xu Y, Yang XR, Guo W, Zhang X, Qiu SJ, Shi RY, Hu B, Zhou J, Fan J. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology. 2013;57:1458–1468. doi: 10.1002/hep.26151. [DOI] [PubMed] [Google Scholar]
- 53.Litvinov SV, Balzar M, Winter MJ, Bakker HA, Briaire-de Bruijn IH, Prins F, Fleuren GJ, Warnaar SO. Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell interactions mediated by classic cadherins. J Cell Biol. 1997;139:1337–1348. doi: 10.1083/jcb.139.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK, Priest LJ, Greystoke A, Zhou C, Morris K, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]


