Abstract
Growing evidence indicates targeting PKCι may be effective in treating Hedgehog-dependent cancers. In this issue of Cancer Cell, Justilien and colleagues present the surprising finding that PKCι promotes Hedgehog ligand production and lung squamous cell carcinoma growth through SOX2, rather than the canonical transcription factor GLI.
Hedgehog (HH) signaling controls the morphogenesis of a panoply of organs by regulating the proliferation and differentiation of stem cells (Briscoe and Thérond, 2013). Because HH regulates key pluripotency and growth genes such as MYC, Cyclin D1, Nanog, and BMI1, it comes as no surprise that alteration of the HH pathway drives tumor growth. Although current HH pathway antagonists are quite effective in certain tumor contexts, recent advances in cancer-associated signaling pathways show a complex and incomplete picture as to the potential for “typical” pathway inhibitors (Atwood et al., 2012).
Tumorigenic HH signaling operates through cell-intrinsic mutations that cause inappropriate pathway activation or by autocrine/paracrine events where the tumor produces HH ligand to feed itself or adjacent growth factor-producing stroma (Figure 1). Before HH is competent to act, the ligand undergoes autoproteolytic cleavage and dual lipid modifications to convert to its active form (Briscoe and Thérond, 2013). HH acyl transferase (HHAT) partly controls production of active HH by adding a palmitoyl moiety to the amino-terminus of the ligand after cleavage, whereas a separate enzyme adds a cholesterol moiety to the C terminus. The dual lipid modified HH is transported to the cell surface where Dispatched1 (DISP1) regulates its release. After activation and secretion, HH binds to its target cell and inhibits the trans-membrane receptor Patched1, catalyzing the activation of signal transducer Smoothened (SMO) and translocation of GLI transcription factors to the nucleus to amplify oncogenic gene expression.
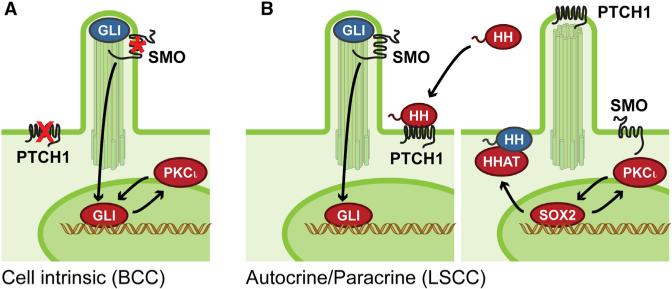
Figure 1. PKCι-Dependent Activation of the HH Pathway.
(A) PKCι phosphorylates and activates GLI1 to amplify target gene expression in basal cell carcinoma (BCC), a cell-intrinsic tumor with HH pathway mutations in PTCH1 or SMO. Blue denotes inactive protein, whereas red is active.
(B) PKCι promotes HHAT-mediated HH ligand production by phosphorylating and activating SOX2, which binds to the HHAT promoter in HH-driven autocrine/paracrine lung squamous cell carcinoma (LSCC) tumors.
Therapeutic inhibition of SMO effectively suppresses tumors driven by cell-intrinsic mutations, such as basal cell carcinoma (BCC) and medulloblastoma (Atwood et al., 2012). Vismodegib, a recently approved SMO antagonist, commands a high response rate in patients with basal cell nevus syndrome that predisposes them to develop hundreds of BCCs, whereas patients with locally advanced or metastatic BCCs show a lower response rate. Patients with more invasive paracrine-driven HH tumors, such as small cell lung or pancreatic cancer, show little to no responsiveness despite evidence of pathway inhibition. Why targeting the HH pathway would lead to such disparate outcomes remains unclear; however, identifying new targets that can act in both cell-intrinsic and autocrine/paracrine tumor proliferation may provide novel therapeutic strategies to treat both tumor types.
Atypical Protein Kinase C lambda/iota (PKCι) plays a central role in determining cell polarity, a fundamental property of cells and tissues that results from the differential distribution of cellular components (proteins, lipids, RNA, and organelles) by promoting asymmetric functions including oriented cell division, cell recognition, and cellular adhesion (Roignot et al., 2013). Apico-basolateral polarity stems from PKCι-dependent regulation of vesicle movements through asymmetric control of the actin cyto-skeleton, resulting in vectorial transport of nutrients and signals that play critical roles in the morphogenesis of most multicellular organisms. Loss of PKCι, or the polarity pathways it interacts with, can lead to abnormalities in cell polarity, epithelial-to-mesenchymal transition, and cancer invasiveness.
Apart from polarity signaling, a distinct role for PKCι as an oncogenic kinase in HH signaling has emerged. PKCι has been implicated in tumors with cell-intrinsic HH signaling, such as BCCs, where it phosphorylates the zinc finger domain of GLI1 to activate DNA binding and transcriptional activity (Figure 1A) (Atwood et al., 2013). Because PRKCI (the gene that encodes PKCι) is a GLI target gene, higher GLI1 activity produces more PKCι, which feeds back into the system to amplify HH pathway activation independently of SMO to feed tumor growth. BCCs that have become addicted to PKCι are vulnerable to pharmacological inhibition of kinase activity, which results in pathway suppression and the blockade of tumor growth. PKCι has also been implicated as an oncogene in non-small cell lung cancer (NSCLC), although the link between PKCι and HH signaling in this predominantly ligand-driven tumor remained unexplored until now (Regala et al., 2005).
In this issue of Cancer Cell, Justilien et al. (2014) show how PKCι maintains cancer stem cell-like properties of lung squamous cell carcinoma (LSCC), an NSCLC subtype (Justilien et al., 2014). LSCC oncospheres displayed increased PKCι activation, which correlated with activation of the HH pathway, as assessed by RNA sequencing from PKCι-deficient oncospheres and pharmacological agents that target either PKCι or SMO suppressed oncosphere growth. Interestingly, the expression of the HH component HHAT was also dependent on PKCι, suggesting a novel regulatory mechanism to control HH ligand production. Both HHAT and DISP1 have been shown to be required for the growth of NCSLC tumor cells and xenografts (Rodriguez-Blanco et al., 2013). Similarly, RNA knockdown of PKCι, GLI1, or HHAT in orthotopic tumors originating from LSCC oncospheres significantly reduced tumor take rate and decreased tumor size.
In addition, the authors found that PRKCI is frequently co-amplified and overexpressed in LSCC tumors on the 3q26 amplicon with SOX2, a master regulator of stem cell maintenance. Chromatin immunoprecipitation of SOX2 showed occupancy of the HHAT promoter and loss of SOX2 significantly decreased HHAT mRNA and protein levels suggesting a novel link between a stem cell gene and HH pathway activation that drives LSCC tumor growth. Surprisingly, SOX2 serves as a substrate for PKCι, with phosphorylation occurring adjacent to the HMG DNA-binding region. SOX2 phosphorylation enhanced binding to the HHAT promoter, increased expression of HH target genes, and augmented growth of oncospheres (Figure 1B). Whether PKCι-dependent phosphorylation of SOX2 controls stem cell maintenance or growth of cell-intrinsic HH pathway tumors such as BCCs remains to be determined.
This new work reinforces two new aspects of PRKCI biology. First, PKCι has now been implicated in two different HH-driven tumors, because NSCLC growth shows both cell-intrinsic and auto-crine/paracrine type signaling (Bermudez et al., 2013; Yuan et al., 2007). This has clinical importance in that targeting SMO appears only to be an effective therapy for cell-intrinsic, but not paracrine, HH-dependent tumors. By contrast, pharmacological inhibition of PKCι may prove useful to treat a variety of HH cancers, because targeting PKCι appears effective in paracrine-driven ovarian cancer (Wang et al., 2013). Second, although PKCι clearly regulates cortical trafficking in polarizing cells, PKCι also operates to regulate nuclear functions by controlling the affinity of specific transcription factors to chromatin. This report adds SOX2 to a growing list of transcription factors regulated by PKCι, which includes GLI1. Whether PKCι-dependent regulation of transcription factors plays similar roles in normal and tumor biology remains to be investigated. In any case, the emerging data implicating “atypical” PKCι in HH cancer biology suggests that PKCι involvement may be the rule rather than the exception.
ACKNOWLEDGMENTS
This work was supported by NIH grants 5ARO54780 and ARO46786.
REFERENCES
- Atwood SX, Chang ALS, Oro AE. J. Cell Biol. 2012;199:193–197. doi: 10.1083/jcb.201207140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood SX, Li M, Lee A, Tang JY, Oro AE. Nature. 2013;494:484–488. doi: 10.1038/nature11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez O, Hennen E, Koch I, Lindner M, Eickelberg O. PLoS ONE. 2013;8:e63226. doi: 10.1371/journal.pone.0063226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Thérond PP. Nat. Rev. Mol. Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- Justilien V, Walsh MP, Ali SA, Thompson EA, Murray NR, Fields AP. Cancer Cell. 2014;25:139–151. doi: 10.1016/j.ccr.2014.01.008. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regala RP, Weems C, Jamieson L, Khoor A, Edell ES, Lohse CM, Fields AP. Cancer Res. 2005;65:8905–8911. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Blanco J, Schilling NS, Tokhunts R, Giambelli C, Long J, Liang Fei D, Singh S, Black KE, Wang Z, Galimberti F, et al. Oncogene. 2013;32:2335–2345. doi: 10.1038/onc.2012.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignot J, Peng X, Mostov K. Cold Spring Harb. Perspect. Biol. 2013;5:5. doi: 10.1101/cshperspect.a013789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hill KS, Fields AP. Mol. Cancer Res. 2013;11:1624–1635. doi: 10.1158/1541-7786.MCR-13-0371-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Goetz JA, Singh S, Ogden SK, Petty WJ, Black CC, Memoli VA, Dmitrovsky E, Robbins DJ. Oncogene. 2007;26:1046–1055. doi: 10.1038/sj.onc.1209860. [DOI] [PubMed] [Google Scholar]