Abstract
The complete genomic sequence of Usutu virus (USUV, genus Flavivirus, family Flaviviridae) strain MB119/06, detected in a pool of Culex pipiens mosquitoes in northeastern Spain (Viladecans, Catalonia) in 2006, was determined and analyzed. The phylogenetic relationship with all other available complete USUV genome sequences was established. The Spanish sequence investigated showed the closest relationship to the USUV prototype strain SA AR 1776 isolated in South Africa in 1959 (96.9% nucleotide and 98.8% amino acid identities). Conserved structural elements and enzyme motifs of the putative polyprotein precursor were identified. Unique amino acid substitutions were recognized; however, their potential roles as virulence markers could not be verified. Comparisons of the polyprotein precursor sequences of USUV strains detected in mosquitoes, birds, and humans could not confirm the predicted role of unique amino acid substitutions in relation to virulence in humans. Phylogenetic analysis of a partial coding section of the NS5 protein gene region indicated that USUV strains circulating in Europe form three different genetic clusters. Broad and targeted surveys for USUV in mosquitoes could reveal further details of the geographic distribution and genetic diversity of the virus in Europe and in Africa.
Key Words: : Usutu virus, Flavivirus, Spain, Phylogenetic analysis, Virulence marker
Introduction
Usutu virus (USUV), a member of the mosquito-borne group of the genus Flavivirus, family Flaviviridae, was originally isolated from a pool of Culex univittatus mosquitoes in Ndumu, Natal, South Africa in 1959 (Woodall et al. 1964, McIntosh 1985). Subsequently, the virus was isolated from different species of mosquitoes and birds in Africa, and sporadic human infections were reported in central Africa (for review, see Nikolay et al. 2011). However, USUV in Africa received only scant attention, and it had never been associated with bird mortality. In 2001, a strain of USUV emerged in Austria and caused a significant die-off of wild birds, especially Eurasian blackbirds (Turdus merula) (Weissenböck et al. 2002). The virus managed to overwinter, became a resident pathogen, spread locally, and continued to cause wild bird mortality in the eastern part of Austria (Weissenböck et al. 2003, Chvala et al. 2007). Since 2004, however, USUV-associated bird mortality decreased and the antibody prevalence against USUV increased significantly, suggestive of the development of herd immunity (Meister et al. 2008, Buchebner et al. 2013). Although no USUV-related bird deaths were observed in Austria since 2006, the same virus strain was identified as the cause of sporadic or epizootic wild bird mortality in Hungary (Bakonyi et al. 2007), Italy (Mannarola et al. 2009), Switzerland (Steimetz et al. 2011), the Czech Republic (Hubálek et al. 2012), and Germany (Jöst et al. 2011, Becker et al. 2012).
Recent retrospective studies indicated that USUV was already present in Italy in 1996 and may have been the etiological agent of episodes of wild bird mortality since then (Weissenböck et al. 2013). Neuroinvasive human USUV infections were reported from immunocompromised patients in Italy (Cavrini et al. 2009, Pecorari et al. 2009). Genetic comparisons demonstrated very close relatedness between the USUV strains circulating in central Europe (Chvala et al. 2007, Hubálek et al. 2012, Gaibani et al. 2013, Nikolay et al. 2013, Weissenböck et al. 2013). The presence of USUV was also reported from the United Kingdom, on the basis of serological examinations of different species of birds (Buckley et al. 2003, Buckley et al. 2006); however, no associated bird mortality has been reported from the United Kingdom.
In the northeast region of Spain (Catalonia), USUV was detected in 2006 in a pool of Culex pipiens mosquitoes without associated bird deaths (Busquets et al. 2008). In Spain, the virus was subsequently detected in Cx. perexiguus in 2009 (Vázquez et al. 2011), and in two sick song thrushes (Turdus philomelos) in 2012 in southern Spain (Höfle et al. 2013). Phylogenetic comparisons of the Spanish strains, however, indicated significant differences between USUVs of mosquito and bird origin (Höfle et al. 2013).
Recently, the complete genome sequences of five USUV strains from central and western Africa were determined and analyzed (Nikolay et al. 2013). The study revealed that USUV strains isolated in Africa between 1969 and 2007 exhibited greater genetic diversity than USUVs detected in central Europe. One African strain (ArB1803, isolated from in the Central African Republic in 1969) showed considerable genetic differences compared to the other known USUVs; therefore, it was suggested to classify this divergent strain as a subtype of USUV (Nikolay et al. 2013).
The complete genome sequence of an USUV strain isolated from a human patient with neurological symptoms in Italy in 2009 has been determined recently (Gaibani et al. 2013). The authors identified amino acid substitutions in the envelope (E) protein and nonstructural protein 5 (NS5), as well as substitutions at the 3′-untranslated region (UTR), which, according to the authors, might be genetic markers for a human neuroinvasive phenotype. The complete genome sequence of another USUV strain (HB81P08), isolated from a human febrile patient in the Central African Republic in 1981, has been determined by Nikolay et al. (2013). Unique amino acid substitutions were identified in the NS2a, NS3, and NS5 protein regions, as well as nucleotide deletions in the 3′-UTR.
The aim of this study was to determine the complete genome sequence of the USUV strain detected in Cx. pipiens in 2006 in Spain (Busquets et al. 2008), and to compare it with other USUV sequences, particularly at the predicted putative virulence marker loci.
Materials and Methods
USUV RNA was demonstrated in a pool of three female Cx. pipiens mosquitoes, which were captured in mid-August 2006 in the center of the village of Viladecans, from the Delta del Llobregat, Catalonia, Spain (Busquets et al. 2008). The mosquitoes were stored in AVL buffer (Qiagen, Hilden, Germany) and were used for the determination of the complete genome sequence of this USUV strain designated MB119/06. Viral RNA was extracted from 140 μL of mosquito homogenate using the QIAamp viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The viral genome was amplified in continuous RT-PCR systems using overlapping oligonucleotide primer pairs and the QIAGEN OneStep RT-PCR Kit (Qiagen, Hilden, Germany). Amplification products were sequenced using the Sanger dideoxy method, as described earlier (Bakonyi et al. 2004). For a few gene regions, in which the originally designed primers did not result in specific PCR products, further primer pairs were designed based on adjacent specific sequences and used for amplification and sequencing of the missing regions. The additionally designed primers are listed in Table 1. After excluding primer regions, the sequences were compiled and aligned with all complete USUV genome sequences deposited in the GenBank database, using the Align Plus 4 (Scientific and Educational Software) and ClustalX (Thompson et al. 1997) programs. To specifically determine the genetic relationship of USUV strains circulating in Spain, an alignment of a partial NS5 protein coding region was also generated, in which the nucleotide sequences of USUV strains detected in Cx. perexiguus in Spain in 2009 (Vázquez et al. 2011) and in two song thrushes (T. philomelos) in Spain in 2012 (Höfle et al. 2013) were included. Accession numbers are listed in Table 2. Putative amino acid sequences of the polyprotein precursors were also aligned and analyzed.
Table 1.
Additional Specific Oligonucleotide Primers Used for Amplification and Sequencing of the Complete Genome Of USUV MB119/06
| Name | Sequence 5′ to 3′ | Product length (bp) |
|---|---|---|
| Usu 42f | TGGAGGATCGTGAGATTAAC | 1161 |
| UsuSp 1202r | GTTGGACAATTGGAGACAGT | |
| UsuSp 1326f | GACACTCATGGCAACTATTC | 567 |
| UsuSp 1892r | TCCGTACACATGCTATAGGT | |
| UsuSp 3651f | TCTACTCGTGCTGATTCTTG | 414 |
| UsuSp 4064r | CCGATGATGATGAGAGTGAT | |
| UsuSp 7249f | CTGATTACTGCGGCAGCTCT | 138 |
| UsuSp 7387r | TAGCCACCAAGCCATCTACC | |
| UsuSp 8499f | ACATGCCAATCAGGAGAAGA | 714 |
| UsuSp 9213r | TTCAACACCTCCTCCAGAAT |
Numbers in primer names indicate the 5′ end annealing positions.
f, forward (genomic) primer; r, reverse (complementary) primer.
Table 2.
Usutu Virus Nucleotide Sequences Included in the Phylogenetic Analyses
| Isolation origin | ||||||
|---|---|---|---|---|---|---|
| Code | Strain | Country | Year | Host | GenBank accession number | Reference |
| SA 1959 | SA Ar 1776 | South Africa | 1959 | Cx. neavei | AY453412 | Bakonyi et al. 2004 |
| CAR 1969 | ArB1803 | Central African Republic | 1969 | Cx. perfuscus | KC754958 | Nikolay et al. 2013 |
| SE 1974 | ArD19848 | Senegal | 1974 | Cx. perfuscus | KC754954 | Nikolay et al. 2013 |
| CAR 1981 | HB81P08 | Central African Republic | 1981 | Human | KC754955 | Nikolay et al. 2013 |
| SE 1993 | ArD101291 | Senegal | 1993 | Cx. univittatus | KC754956 | Nikolay et al. 2013 |
| AT 2001 | Vienna 2001 | Austria | 2001 | Blackbird | AY453411 | Bakonyi et al. 2004 |
| AT 2002 | Meise H | Austria | 2002 | Blue tit | JQ219843 | Becker et al. 2012 |
| HU 2005 | Budapest | Hungary | 2005 | Blackbird | EF206350 | Bakonyi et al. 2007 |
| SP 2006 | MB119/06 | Spain | 2006 | Cx. pipiens | KF573410 | This paper |
| SE 2007 | ArD192495 | Senegal | 2007 | Cx. neavei | KC754957 | Nikolay et al. 2013 |
| IT 2009A | Italia 2009 | Italy | 2009 | Blackbird | JF266698 | Savini et al. 2011 |
| IT 2009B | Bologna/09 | Italy | 2009 | Human | HM569263 | Gaibani et al. 2013 |
| SP 2009 | HU10279 09 | Spain | 2009 | Cx. perexiguus | HQ833022* | Vázquez et al. 2011 |
| GE 2011 | BH65/11-02-03 | Germany | 2011 | Blackbird | HE599647 | Becker et al. 2012 |
| SP 2012 | Flavi1 | Spain | 2012 | Song thrush | KC437386* | Höfle et al. 2013 |
NS5 gene region, partial coding section.
Phylogenetic reconstruction analyses were performed on the alignments employing the maximum likelihood (ML) method based on the Tamura–Nei model, neighbor-joining method, minimum evolution method, unweighted pair group method with arithmetic mean (UPGMA), and maximum parsimony method of the MEGA5 software package (Tamura et al. 2011). The stability of the trees was tested by bootstrap resampling analysis of 1000 replicates. The probable relationships were displayed in phylograms. Murray Valley encephalitis virus (MVEV; GenBank accession number NC_000943) was used as outgroup to root the trees.
Results
The complete genome sequence of USUV MB119/06 is composed of 11,064 nucleotides and contains an open reading frame between nucleotide positions 97 and 10,401, coding for a 3434-amino-acid-long putative polyprotein precursor. Conserved structural elements and putative enzyme motifs of the polyprotein precursor, described in Bakonyi et al. (2004) (12 Cys residues in the E and NS1 protein regions; putative integrin-binding domain, and receptor-binding domain for virus attachment to sulfated proteoglycans at the E protein region; catalytic triad, and substrate binding pocket of trypsin-like serine protease motif at the NS3 protein region; and RNA-dependent RNA polymerase motif at the NS5 protein region) were identified. Within the RNA helicase motif DEAH/DEAD at the carboxyl terminal of the NS3 protein region, an amino acid substitution was observed at H1791D. The putative N-glycosylation sites of the E protein, predicted by Nikolay et al. (2013), were also present in this Spanish USUV strain.
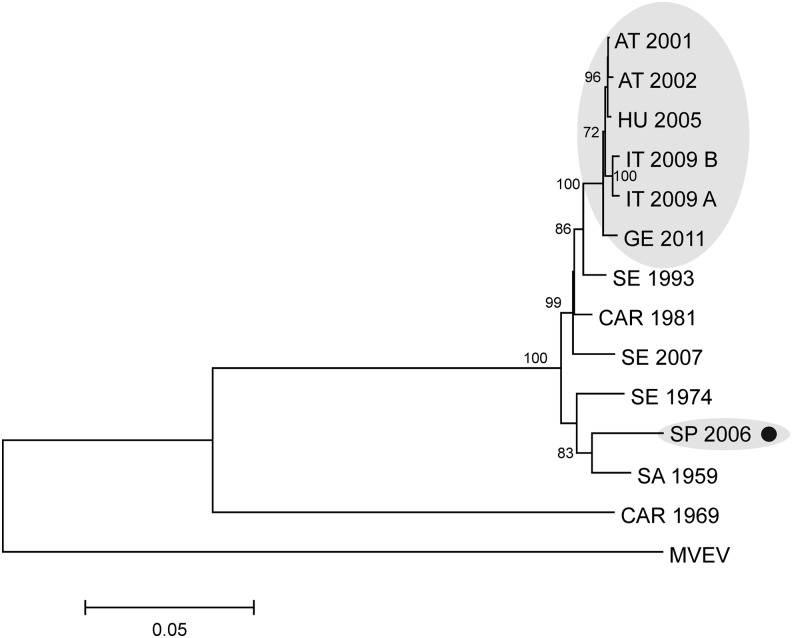
The genome sequence of USUV MB119/06 was aligned with 12 complete USUV genome sequences deposited in GenBank database. The nucleotide identity rates varied between 96.9% (strain SA Ar 1774, South Africa, 1959) and 78.3% (strain ArB1803, Central African Republic, 1969). Amino acid identity rates of the polypeptide precursor varied between 98.8% (strain Vienna_2001, Austria, 2001) and 94.4% (strain ArB1803, Central African Republic, 1969). The phylogenetic relationships between the investigated USUV genome sequences were inferred by five different statistical algorithms. Principally, similar topology trees were obtained; however, the bootstrap support values varied at particular junctions, depending on the applied method. One of the best resolution and bootstrap supports was obtained with the ML algorithm (Fig. 1). The USUV MB119/06 complete genome sequence (marked in red with a black dot in Fig. 1) showed the closest phlyogenetic relatedness to the SA Ar 1776 strain (isolated from Cx. neavei in South Africa in 1959). Another African isolate (ArD19848, isolated from Cx. perfuscus in Senegal in 1974) forms a monophyletic cluster with the above-mentioned two sequences. All other USUV sequences from Europe (marked with grey background, Fig. 1) and Africa cluster in distinct groups.
FIG. 1.
Phylogram demonstrating the genetic relationships of Usutu virus (USUV) complete genome nucleotide sequences. Details on the viruses are provided in Table 2. Murray Valley encephalitis virus (MVEV; GenBank accession number NC_000943) was used as outgroup to root the tree. The USUV MB119/06 sequence described in this article is marked with a black dot. European clusters are highlighted with grey background. Bootstrap values >70 are displayed at nodes. The horizontal bar on the left represents the genetic distance.
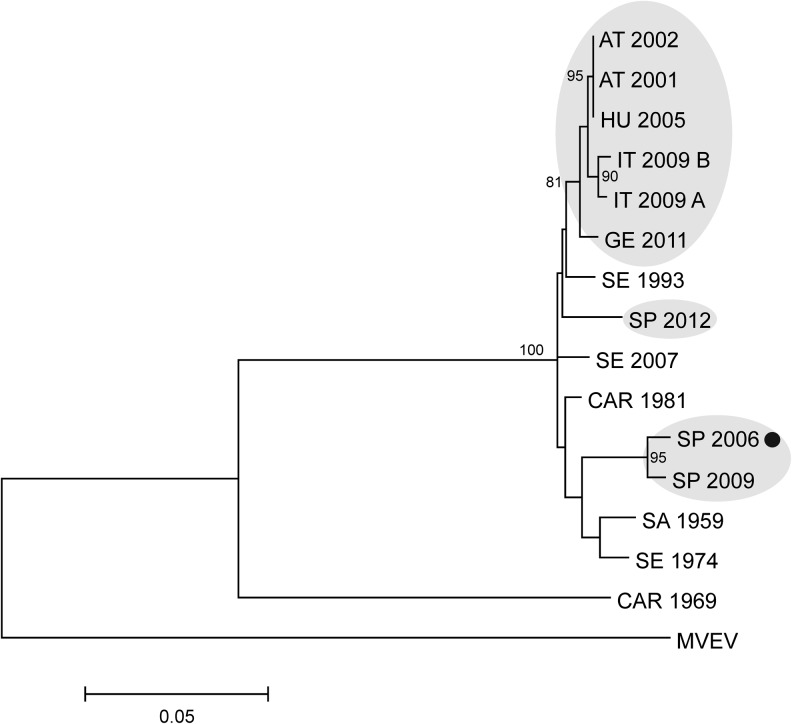
Partial nucleotide sequences of the NS5 protein-coding region of two further Spanish USUVs are also available in GenBank database (Vázquez et al. 2011, Höfle et al. 2013). Nucleotide sequences of the complete USUV genomes between nucleotide positions 8377 and 9305 were aligned with the above-mentioned partial sequences. The phylogram obtained with the ML method indicated the closest relationship of the MB119/06 USUV sequence with USUV HU10279 09, detected in Cx. perexiguus in Spain in 2009 (Vázquez et al. 2011) (Fig. 2). The other Spanish USUV (Flavi1, detected in two song thrushes in 2012; Höfle et al. 2013) formed a separated branch (Fig. 2, marked in pink), related to a sequence from Senegal, and sequences of central European USUVs (the latter marked in blue, Fig. 2). The ML phylogenetic reconstructions of the putative polypeptide precursor amino acid sequences indicated similar clustering of the viruses; however, generally lower bootstrap support values were obtained (tree not shown).
FIG. 2.
Phylogram demonstrating the genetic relationships of Usutu virus (USUV) nucleotide sequences in a partial NS5 (RNA-dependent RNA polymerase) gene region. Details on the viruses are provided in Table 2. Murray Valley encephalitis virus (MVEV; GenBank accession number NC_000943) was used as outgroup to root the tree. The USUV MB119/06 sequence described in this artcile is marked with a black dot. European clusters are highlighted with grey background. Bootstrap values >70 are displayed at nodes. The horizontal bar on the left represents the genetic distance.
In their analyses of sequences of African USUVs (Nikolay et al. 2013) and of a human isolate from Italy (Bologna/09, Gaibani et al. 2013), the authors predicted potential virulence markers and unique substitutions. Nikolay et al. (2013) identified amino acid substitutions A120Vand M2287I, specific to USUVs from Africa/Europe. The MB119/06 sequence is identical to the African viruses at these loci. The authors also characterized a human USUV isolate from Africa (strain HB81P08 isolated in the Central African Republic in 1981). Unique amino acid substitutions L1299S, H1977Y, and Q2702H were identified, as well as a deletion of the 3′-UTR between nucleotide positions 10494 and 10510. These mutations were found neither in the MB119/06 virus nor in the human isolate from Italy (Bologna/09). Gaibani et al. (2013) described two unique amino acid substitutions in the Bologna/09 strain. The S595G substitution in the E protein region is also present in the MB119/06 virus, but not in the African and in the central European strains. The D3425E substitution in the NS5 protein region is present only in the Italian sequence. Finally, the putative amino acid sequence of MB119/06 contains 24 unique amino acid substitutions compared to the other USUV sequences analysed (K101R, V/A112L, T153M, D/E172G, S273G, A454V, V563I, A636V, H967Y, V/T1067I, A1227V, A1236T, L1270F, I1460V, K1645R, H1771Y, H1791D, G1981S, V2009I, P2301H, N2355T, D2552E, H3055Y, and V3322I).
Discussion
The emergence and spread of USUV in central Europe has been investigated intensively and is well documented. The virus strain most likely emerged in Italy in 1996 or earlier (Weissenböck et al. 2013). The virus established itself in the region and circulated in avian hosts and mosquito vector species. The strain spread northward to Austria, Hungary, Switzerland, Germany, and the Czech Republic. Phylogenetic analyses indicate that presumably the same strain has been circulating and dispersing in the region (Chvala et al. 2007, Jöst et al. 2011, Becker et al. 2012, Hubálek et al. 2012, Gaibani et al. 2013, Nikolay et al. 2013, Weissenböck et al. 2013). It has obviously been pathogenic to particular species of birds (especially blackbirds, owls, and sparrows). Contrary to this, bird mortality was not or only exceptionally associated with USUV strains from Africa and Spain; and in the United Kingdom, only serological evidence indicated the presence of USUV both in migratory and sedentary bird populations (Buckley et al. 2003, Buckley et al. 2006). These observations point toward differences in the virulence of USUV strains.
The phylogenetic analysis of the complete genome sequence of the Spanish USUV strain MB119/06 confirmed the previous inferences on the genetic relatedness of the virus, on the basis of partial nucleotide sequence data (Busquets et al. 2008, Höfle et al. 2013). This virus is more closely related to the prototype USUV strain from South Africa than to the central European strain and to strains isolated in central and western Africa. A closely related USUV was detected in 2009 in southern Spain in Cx. perexiguus, indicating that this virus strain is circulating in the country, although it does not cause significant/detectable wild bird mortality. Because the principal vertebrate hosts of USUV are birds, migratory species of birds may play a role in the geographic spread of the virus. Because the bird migratory routes between Africa and western Europe are typically via Gibraltar and Spain, it is a reasonable scenario that USUVs from Spain (or from western Africa) were introduced to the United Kingdom. If these strains are not virulent to birds, they might covertly circulate in the populations. Targeted surveillance studies for USUV in mosquitoes in Western Europe could support this hypothesis.
Cases of USUV-associated avian neurological illness were recently reported in Spain (Höfle et al. 2013). On the basis of the phylogenetic analysis of NS5 partial sequences, the authors revealed that the USUV detected in song thrushes in 2012 is different from the USUVs detected in mosquitoes in Spain before. The authors hypothesize that this USUV was introduced to Spain by migrating birds from northern Europe. Our phylogenetic analysis involved novel USUV sequences from Africa described by Nikolay et al. (2013). The phylogram indicates that the USUV detected in 2012 in song thrushes in Spain is not monophyletic with the USUVs circulating in central Europe. Therefore, it is unlikely that the central European strain reached Spain. It seems more likely that a strain from central or western Africa was introduced. However, the thrush infections were detected in November, 2012, when southward bird migration is dominant in Spain.
Identification of virulence markers could play a key role for understanding USUV epidemiology. The analysis of increasing numbers of complete sequences might reveal candidate marker loci. Our study also identified unique amino acid substitutions in USUV strain MB119/06. The effects of these mutations on the virulence of USUV should further be investigated using, e.g., site-specific mutagenesis (Brault et al. 2007). Particularly important is the aspect of a possible human pathogenicity of USUV.
Within this study, we compared the complete genome sequences of the so-far available two human USUV isolates (HB81P08 from the Central African Republic, 1981, and Bologna/09 from Italy, 2009). Unique nucleotide or amino acid changes, present only in the USUVs of human origin, but not in the other analyzed sequences, were not found. Therefore, it is possible that the cumulative effect of multiple genetic markers influence the virulence of USUV in humans. However, the described human cases differ significantly in their clinical presentations. The patient in the Central African Republic suffered from a febrile illness with rash. These are frequent symptoms of flavivirus infections (e.g., in mild cases of West Nile, dengue, and yellow fever); therefore, it is quite possible that USUV may cause such a disease manifestation, although the exclusive role of USUV in the etiology of this case could not be confirmed. The human neuroinvasive USUV cases in Italy were diagnosed in immunocompromised patients (Cavrini et al. 2009, Pecorari et al. 2009). Impaired immune defense may allow the invasion of viruses, which are otherwise nonpathogenic for healthy people. Therefore, in these cases, the reactions of the host organisms might have played a greater role in the pathogenesis of the USUV infection, rather than an increased virulence of the virus involved.
Conclusions
The phylogenetic analysis of all currently available complete USUV sequences from Africa, Spain, and central Europe revealed a significant genetic diversity of the virus. The viruses circulating in Europe belong to (at least) three different genetic clusters. The USUVs detected in mosquitoes in Spain are genetically more closely related to African isolates than to central European ones. The viruses differ apparently in their virulence for avian hosts. Therefore, the geographical distribution of USUV is probably wider than so far identified because of covert cycles in bird hosts and mosquito vectors. Comparative genomic analyses revealed several differences among USUV strains. They might—or might not—act as virulence marker loci. The effects of these potential markers should be investigated further in controlled experiments. Considering the potential public health impact of USUV (conditional human pathogen), its interference in the serological diagnosis of pathogenic human flaviviruses due to cross-reactions, and its pathogenicity for several species of birds, a broader, targeted surveillance activity would be necessary to assess the geographical distribution and abundance of USUV in Europe and in Africa.
Acknowledgments
This study was partially supported by the European Union grants HEALTH.2010.2.3.3-3 Project 261391 EuroWestNile (www.eurowestnile.org/) and FP7-261504 EDENext (www.edenext.eu) and is catalogued by the EDENext Steering Committee as EDENext 186. The contents of this paper are the sole responsibility of the authors and do not necessarily reflect the views of the European Commission. The study was also partially sponsored by the TÁMOP–4.2.2.B-10/1 and TÁMOP–4.2.1.B-11/2/KMR-2011-0003 projects.
Author Disclosure Statement
No competing financial interest exists.
References
- Bakonyi T, Gould EA, Kolodziejek J, Weissenböck H, et al. Complete genome analysis and molecular characterization of Usutu virus that emerged in Austria in 2001: Comparison with the South African strain SAAR-1776 and other flaviviruses. Virology 2004; 328:301–310 [DOI] [PubMed] [Google Scholar]
- Bakonyi T, Erdélyi K, Ursu K, Ferenczi E, et al. Emergence of Usutu virus in Hungary. J Clin Microbiol 2007; 45:3870–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker N, Jöst H, Ziegler U, Eiden M, et al. Epizootic emergence of Usutu virus in wild and captive birds in Germany. PLoS One 2012; 7:e32604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault AC, Huang CY, Langevin SA, Kinney RM, et al. A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nat Genet 2007; 39:1162–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchebner N, Zenker W, Wenker C, Steinmetz HW, et al. Low Usutu virus seroprevalence in four zoological gardens in central Europe. BMC Vet Res 2013; 9:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley A, Dawson A, Moss SR, Hinsley SA, et al. Serological evidence of West Nile virus, Usutu virus and Sindbis virus infection of birds in the UK. J Gen Virol 2003; 84:2807–2817 [DOI] [PubMed] [Google Scholar]
- Buckley A, Dawson A, Gould EA. Detection of seroconversion to West Nile virus, Usutu virus and Sindbis virus in UK sentinel chickens. Virol J 2006; 3:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busquets N, Alba A, Allepuz A, Aranda C, et al. Usutu virus sequences in Culex pipiens (Diptera: Culicidae), Spain. Emerg Infect Dis 2008; 14:861–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavrini F, Gaibani P, Longo G, Pierro AM, et al. Usutu virus infection in a patient who underwent orthotropic liver transplantation, Italy, August–September 2009. Euro Surveill 2009; 14:pii.19448. [PubMed] [Google Scholar]
- Chvala S, Bakonyi T, Bukovsky C, Meister T, et al. Monitoring of Usutu virus activity and spread by using dead bird surveillance in Austria, 2003–2005. Vet Microbiol 2007; 122:237–245 [DOI] [PubMed] [Google Scholar]
- Gaibani P, Cavrini F, Gould EA, Rossini G, et al. Comparative genomic and phylogenetic analysis of the first usutu virus isolate from a human patient presenting with neurological symptoms. PLoS One 2013; 8:e64761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfle U, Gamino V, de Mera IG, Mangold AJ, et al. Usutu virus in migratory song thrushes, Spain. Emerg Infect Dis 2013; 19:1173–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubálek Z, Rudolf I, Capek M, Bakonyi T, et al. Usutu Virus in Blackbirds (Turdus merula), Czech Republic, 2011–2012. Transbound Emerg Dis 2012; Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Jöst H, Bialonski A, Maus D, Sambri V, et al. Isolation of usutu virus in Germany. J Am J Trop Med Hyg 2011; 85:551–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manarolla G, Bakonyi T, Gallazzi D, Crosta L, et al. Usutu virus in wild birds in northern Italy. Vet Microbiol 2009; 141:159–163 [DOI] [PubMed] [Google Scholar]
- McIntosh BM. Usutu (SA Ar 1776), nouvel arbovirus du groupe B. Int Catalogue Arboviruses 1985; 3:1059–1060 [Google Scholar]
- Meister T, Lussy H, Bakonyi T, Sikutová S, et al. Serological evidence of continuing high Usutu virus (Flaviviridae) activity and establishment of herd immunity in wild birds in Austria. Vet Microbiol 2008; 127:237–248 [DOI] [PubMed] [Google Scholar]
- Nikolay B, Diallo M, Boye CSB, Sall AA. Usutu virus in Africa. Vector Borne Zoonotic Dis 2011; 11:1417–1423 [DOI] [PubMed] [Google Scholar]
- Nikolay B, Dupressoir A, Firth C, Faye O, et al. Comparative full length genome sequence analysis of Usutu virus isolates from Africa. Virol J 2013; 10:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecorari M, Longo G, Gennari W, Grottola A, et al. First human case of Usutu virus neuroinvasive infection, Italy, August–September 2009. Euro Surveill 2009; 14:pii [PubMed] [Google Scholar]
- Savini G, Monaco F, Terregino C, Di Gennaro A, et al. Usutu virus in Italy: an emergence or a silent infection? Vet Microbiol 2011; 151:264–274 [DOI] [PubMed] [Google Scholar]
- Steinmetz HW, Bakonyi T, Weissenböck H, Hatt JM, et al. Emergence and establishment of Usutu virus infection in wild and captive avian species in and around Zurich, Switzerland—Genomic and pathologic comparison to other central European outbreaks. Vet Microbiol 2011; 148:207–212 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, et al. MEGA5: Molecular evolutionary genetics analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony methods. Mol Biol Evol 2011; 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, et al. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997; 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez A, Ruiz S, Herrero L, Moreno J, et al. West Nile and Usutu viruses in mosquitoes in Spain, 2008–2009. Am J Trop Med Hyg 2011; 85:178–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenböck H, Kolodziejek J, Url A, Lussy H, et al. Emergence of Usutu virus, an African mosquito-borne flavivirus of the Japanese encephalitis virus group, central Europe. Emerg Infect Dis 2002; 8:652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenböck H, Kolodziejek J, Fragner K, Kuhn R, et al. Usutu virus activity in Austria, 2001–2002. Microbes Infect 2003; 5:1132–1136 [DOI] [PubMed] [Google Scholar]
- Weissenböck H, Bakonyi T, Rossi G, Mani P, et al. Usutu virus, Italy, 1996. Emerg Infect Dis 2013; 19:274–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodall JP. The viruses isolated from arthropods at the East African Virus Research Institute in the 26 years ending December 1963. Proc E Afrc Acad 1964; II:141–146 [Google Scholar]