Significance
Roots and shoots communicate with each other to synchronize and optimize plant growth and respond to environmental changes. Shoots and roots exchange signals to sense the status and respond to the needs of the other organ. Cytokinins, which are phytohormones that regulate various aspects of growth and development, are recognized as the most important signal transmitted from roots to shoots. Whereas the enzymes underlying cytokinin biosynthesis and the corresponding receptors have been identified, our knowledge of cytokinin transport is limited. In this study, we identified the Arabidopsis ATP-binding cassette transporter subfamily G14 as a major component in the transfer of cytokinins from roots to shoots and hence as a regulator of shoot development. This finding represents a major breakthrough in the field.
Keywords: ABC transporter, tZ-type cytokinin translocation, long-distance signal
Abstract
Cytokinins are phytohormones that induce cytokinesis and are essential for diverse developmental and physiological processes in plants. Cytokinins of the trans-zeatin type are mainly synthesized in root vasculature and transported to the shoot, where they regulate shoot growth. However, the mechanism of long-distance transport of cytokinin was hitherto unknown. Here, we report that the Arabidopsis ATP-binding cassette (ABC) transporter subfamily G14 (AtABCG14) is mainly expressed in roots and plays a major role in delivering cytokinins to the shoot. Loss of AtABCG14 expression resulted in severe shoot growth retardation, which was rescued by exogenous trans-zeatin application. Cytokinin content was decreased in the shoots of atabcg14 plants and increased in the roots, with consistent changes in the expression of cytokinin-responsive genes. Grafting of atabcg14 scions onto wild-type rootstocks restored shoot growth, whereas wild-type scions grafted onto atabcg14 rootstocks exhibited shoot growth retardation similar to that of atabcg14. Cytokinin concentrations in the xylem are reduced by ∼90% in the atabcg14 mutant. These results indicate that AtABCG14 is crucial for the translocation of cytokinin to the shoot. Our results provide molecular evidence for the long-distance transport of cytokinin and show that this transport is necessary for normal shoot development.
In plants, roots and shoots communicate to synchronize and optimize growth in response to environmental changes. The autotrophic shoot undergoes photosynthesis, and the products of photosynthesis are used as an energy source for root growth. The roots absorb water and nutrients from the soil and deliver these to the shoots. Thus, the growth of these two structures is coordinated; a change in root growth alters the growth of the shoot, and vice versa. The coordination requires communication mediated by signal molecules that move between the aboveground and belowground structures. The dominant signals transmitted between roots and shoots are (i) cytokinin, which is translocated both from roots to shoots and from shoots to roots (1, 2), and (ii) auxin, which moves from shoots to roots. Whereas the long-distance transport of auxin has been thoroughly investigated (3, 4), that of cytokinin is largely unexplored at the molecular level (1, 2, 5).
Cytokinins are a group of phytohormones that promote cytokinesis. They are involved in a myriad of developmental and physiological processes, including the maintenance of shoot and cambial meristem activities, which determine shoot size and structure (6). Several compounds that exhibit cytokinin activity have been characterized, including trans-zeatin (tZ), N6-(Δ2-isopentenyl) adenine (iP), and cis-zeatin (cZ). These are active at the site of synthesis (7), as well as in distant tissues, which they reach via long-distance transport. Recent studies revealed that tZ-type cytokinins are translocated from roots to shoots via xylem to regulate shoot growth (8). The iP-type cytokinins are the major form in phloem and are translocated from shoots to roots to maintain vascular patterning in the root meristem (1, 9). Hence, root-derived cytokinins coordinate shoot development, whereas shoot-derived cytokinins coordinate that of the root. Such cross-talk enables the plant to continuously adapt to an ever-changing environment. For instance, in nitrogen-starved maize roots resupplied with nitrate, synthesis of tZ-type cytokinin level increases in the root, which is then delivered to the xylem sap and results in an increase in cytokinin-responsive gene expression in the shoot (10). This sequence of events indicates the importance of long-distance cytokinin transport as a signal that modulates shoot growth.
The translocation of cytokinins was demonstrated in grafting experiments using a quadruple knockout mutant of cytokinin synthesis genes, adenosine phosphate isopentenyl transferase (IPT)1;3;5;7 (atipt1;3;5;7), and a double mutant of tZ-type cytokinin synthesis genes, cytochrome P450 monooxygenases, CYP735A1 and CYP735A2. These experiments revealed that wild-type rootstocks can complement the growth of the mutant shoot scions, indicating that tZ-type cytokinins synthesized in the wild-type root are delivered to the mutant shoot and stimulate its growth (8, 10). However, the molecular mechanism underlying the translocation was hitherto obscure. Here, we report that a member of the ABCG subfamily of ABC proteins, AtABCG14, is required for the root-to-shoot translocation of cytokinins.
Results
Search for Candidate Genes Important for Balanced Growth of Shoots and Roots.
Good candidates for transporters involved in the communication between the root and shoot are transporters that are colocalized with cytokinin biosynthesis genes in the root. To identify such candidates, we searched for genes with high transcript levels in root phloem companion cells (11), as this cell type expresses IPTs and is likely to be involved in cytokinin export. In addition, we searched for genes that are highly coexpressed with the most important cytokinin synthesis gene, IPT3 (Table S1), and genes induced by cytokinin treatment, both according to the literature (12) and the electronic fluorescent pictograph browser (Fig. S1A). AtABCG14 was the only gene that satisfied the three selection criteria. Induction of AtABCG14 by cytokinin treatment was confirmed by RT-PCR using RNA extracted from cytokinin-treated Arabidopsis seedlings (Fig. S1B). Thus, we hypothesized that AtABCG14 has a role in cytokinin transport. To test this hypothesis, we obtained the corresponding knockout mutant (SK_15918; Fig. S2E) and analyzed its phenotypes.
atabcg14 Exhibits Retarded Shoot Growth, Which Can Be Recovered by tZ Application.
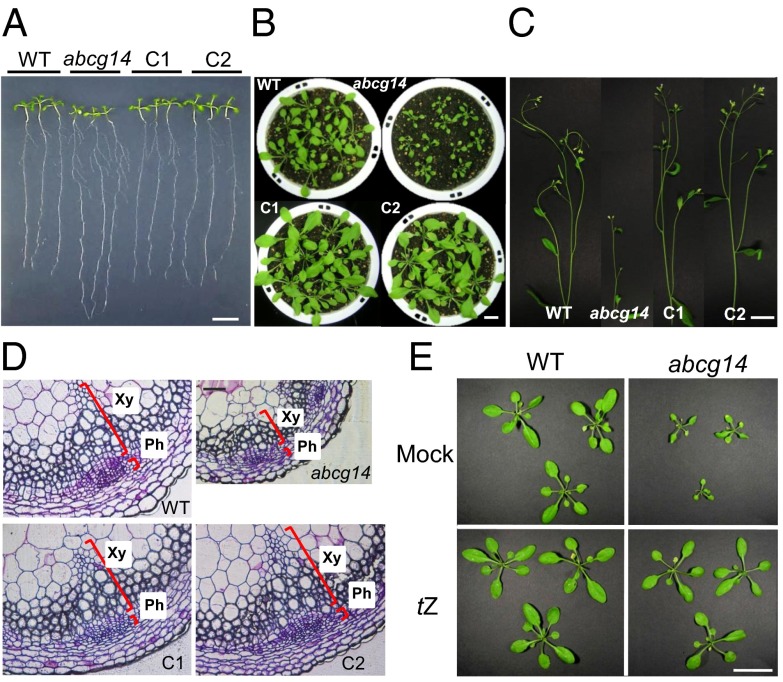
The shoot-to-root ratio of atabcg14 seedlings differed from that of the wild type (Fig. 1A and Fig. S2A). The leaves of atabcg14 plants grown on synthetic plant growth agar (MGRL-agar) medium (13) were smaller than those of the wild type, whereas the roots were longer. Mature atabcg14 mutant plants had considerably smaller rosette leaves (Fig. 1B) and shorter (Fig. 1C) and thinner (Fig. S2B) stems than the wild type. The number and size of xylem and phloem cells in the mutant was strikingly reduced (Fig. 1D and Fig. S2 C and D), and lignin content was decreased (Fig. S3). Furthermore, the mutant produced fewer siliques and seeds per plant than the wild type, but their seeds were larger (Fig. S2 F and G). The introduction of pAtABCG14::sGFP::AtABCG14 genomic DNA (gDNA) into atabcg14 restored the leaf size (Fig. 1B), shoot-to-root fresh weight ratio (Fig. S2A), and stem diameter (Fig. S2B) to those of the wild type.
Fig. 1.
Shoot growth retardation of atabcg14 and its recovery by tZ application. (A) Altered shoot-to-root ratio of atabcg14 knockout (abcg14) compared with the wild type. Plants were grown on MGRL-agar medium under short-day conditions (8 h/16 h, light/dark). (Scale bar, 1 cm.) (B and C) Leaves (B) and inflorescence stems (C) of 28-d-old wild-type (WT), abcg14, and complementation lines (C1 and C2). (Scale bar, 4 cm.) Note that abcg14 exhibits severely retarded shoot growth. Pictures were taken separately at the same time and composed into a single image. (D) Transverse sections of the inflorescence stems of wild-type, abcg14, and complementation lines. Sections were taken at the base of the stem of 35-d-old plants and stained with toluidine blue. Ph, phloem; Xy, xylem. (Scale bar, 80 µm.) (E) Leaves of wild-type and abcg14 plants sprayed with 0.1% DMSO (solvent control) solution (Mock, Upper) or 1 µM tZ (Lower) once a day for 21 d starting from 10 d after sowing. (Scale bar, 4 cm.)
The phenotypes of atabcg14 plants were similar to those described for cytokinin biosynthesis (14) and receptor Arabidopsis histidine kinase (AHK) mutants (15). To test our hypothesis that the dwarf leaf phenotype of the atabcg14 mutant is due to impaired cytokinin allocation to the shoot, we exogenously applied cytokinin to the leaves of atabcg14. Indeed, daily spraying with 1 µM tZ rescued the small leaf (Fig. 1E and Fig. S4A) and short stem (Fig. S4A) phenotypes. In contrast, a similar treatment with iP did not rescue the dwarf phenotypes of the mutant (Fig. S4).
AtABCG14 Is Expressed in the Vasculature and Localizes to the Plasma Membrane.
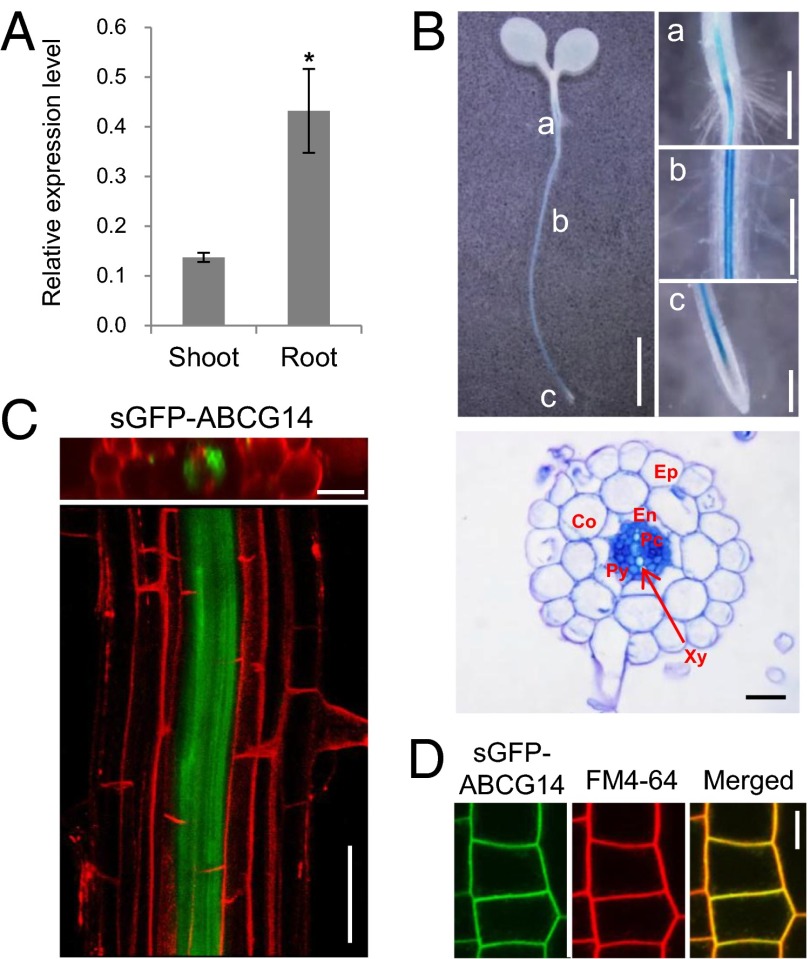
Quantitative real-time PCR indicated that AtABCG14 is expressed in both roots and shoots but that levels are higher in the roots (Fig. 2A). To analyze the cell-specific expression of AtABCG14, we generated Arabidopsis plants expressing a promoter–GUS fusion construct (pAtABCG14::GUS). Blue GUS staining was detected throughout the root, but not in the root tip (Fig. 2B, Upper). Root cross-sections revealed strong gene expression in almost all cells in the stele (i.e., phloem cells, procambial cells, and pericycle) (Fig. 2B, Lower), as reported previously (16). To further investigate gene expression, we analyzed transgenic plants expressing pAtABCG14::sGFP::AtABCG14 gDNA in the atabcg14 mutant background. Consistent with the GUS staining results, green fluorescence was detected in almost all stellar cells in the root (Fig. 2C). This expression pattern overlaps with that of the cytokinin-synthesizing genes IPT3 and CYP735A2 (8, 17, 18).
Fig. 2.
AtABCG14 was expressed in cells that produce tZ-type cytokinins and was localized in the plasma membrane. (A) Relative expression levels of AtABCG14 in shoots and roots. The expression level was normalized by that of the loading control, PP2A. Values are mean ± SEM (n = 4 each from two independent experiments). *P < 0.05 (Student t test) relative to expression level in shoot. (B) The tissue-specific expression pattern of AtABCG14. Five-day-old seedlings expressing pAtABCG14::GUS were used for GUS assays. A strong GUS signal was found mainly in the center of the root. a–c in the Upper Left panel indicate the root/hypocotyl junction, maturation zone, and root tip, respectively, and these are magnified in the Upper Right panels. [Scale bar, 2.5 mm (Upper Left), 5 mm (a), and 2 mm (b and c).] Cross-section of the maturation zone of a pAtABCG14::GUS-expressing root (Lower), showing high expression of AtABCG14 in the stele. (Scale bar, 20 µm.) Co, cortex; En, endodermis; Ep, epidermis; Pc, procambial cells; Py, pericycle; Xy, xylem. (C) Strong expression of AtABCG14 in the stele of roots. Confocal optical sections of a root stained with 15 μM propidium iodide for 10 min were stacked and rotated to view a cross-sectional image (Upper) or a longitudinal image (Lower) of the maturation zone of the root. The complementation line that expresses pAtABCG14::sGFP::AtABCG14 gDNA in the atabcg14 background shows high expression of AtABCG14 in the stele. [Scale bar, 25 µm (Upper) and 50 µm (Lower).] (D) Subcellular localization of AtABCG14 (ABCG14). Five-day-old transgenic plants expressing 35S promoter::sGFP::AtABCG14 gDNA were stained with FM4-64 for 10 min on ice, and the sGFP signal in the root epidermal cells was observed using confocal microscopy. (Scale bar, 10 µm.)
We then examined the intracellular localization of the transporter by expressing a 35S promoter-driven synthetic GFP (sGFP)–AtABCG14 fusion protein in the wild-type background. Green fluorescence was observed at the periphery of the root cells, and colocalized with the red fluorescence of FM4-64, which was applied to the root for 10 min on ice (Fig. 2D). FM4-64 applied in this manner did not enter the cells, but stained only the plasma membrane. Thus, the overlap of red and green fluorescence indicates that sGFP–AtABCG14 is targeted to the plasma membrane, consistent with the findings of a previous study (16). Together, these results suggest that AtABCG14 is present in the plasma membrane of cytokinin-synthesizing root cells.
atabcg14 Shoots Contain Reduced Levels of tZ-Type Cytokinins.
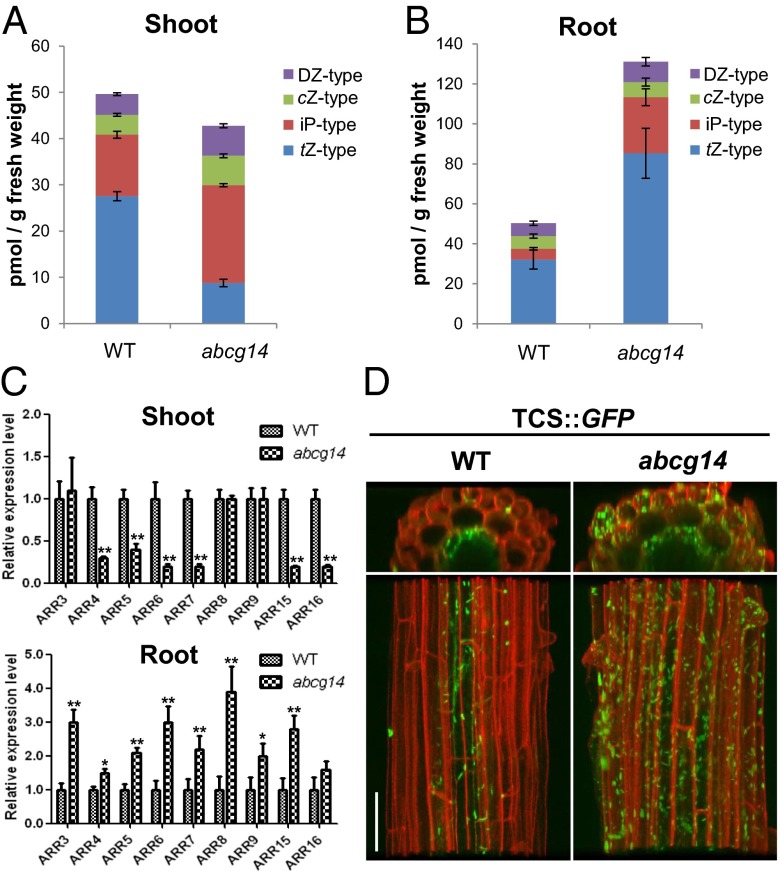
We hypothesized that if the root-to-shoot translocation of cytokinins is impaired in atabcg14, the cytokinin levels in the mutant shoot should be lower than those of the wild type. Indeed, quantification of cytokinins revealed that atabcg14 shoots contained lower levels of tZ-type cytokinins than the wild type (Fig. 3A and Fig. S5A). In contrast, atabcg14 roots had higher levels of tZ-type cytokinins than the wild type (Fig. 3B and Fig. S5B). Interestingly, iP-type cytokinin levels were higher in the atabcg14 mutant than in the wild type, in both roots and shoots (Fig. 3 A and B).
Fig. 3.
Cytokinin content and cytokinin response marker gene expression in shoots and roots of wild-type and atabcg14 plants. (A and B) Cytokinin concentrations in shoots (A) and roots (B). Shoots and roots of plants grown on MGRL-agar medium for 14 d were collected for cytokinin quantification. tZ, iP, cZ, and DZ type represent the concentration of each type of cytokinin present, and the individual cytokinins belonging to each type are described in Fig. S5. (C) Expression levels of type-A ARRs in shoots (Upper) and roots (Lower) of wild-type (WT) and atabcg14 (abcg14) plants. Note that the expression of ARRs is reduced relative to the wild type in abcg14 shoots, but increased in abcg14 roots. Values are mean ± SEM (n = 4 each from two independent experiments). *P < 0.05, **P < 0.01 (Student t test) relative to expression of the wild-type control. (D) TCS::GFP expression pattern in the roots of the wild type and abcg14. (Scale bar, 50 µm.)
We also compared the cytokinin signaling activity between shoots and roots by analyzing the expression levels of cytokinin response marker genes, immediate–early cytokinin-inducible type-A Arabidopsis response regulators (ARRs) (19). Transcript levels of six ARRs, including ARR7 and ARR15, which are important for shoot apical meristem function, were lower in the shoots of atabcg14 seedlings than in those of the wild type (Fig. 3C, Upper). This finding is in good agreement with the decrease in cytokinin levels observed in the shoots (Fig. 3A). In contrast, transcript levels of ARRs in the roots were higher in the mutant (Fig. 3C, Lower), and consistently, the expression of synthetic cytokinin response markers, TCS::GFP (20) and pARR5::GUS, was enhanced in atabcg14 roots (Fig. 3D and Fig. S5C).
Root-to-Shoot Cytokinin Translocation Is Reduced in atabcg14.
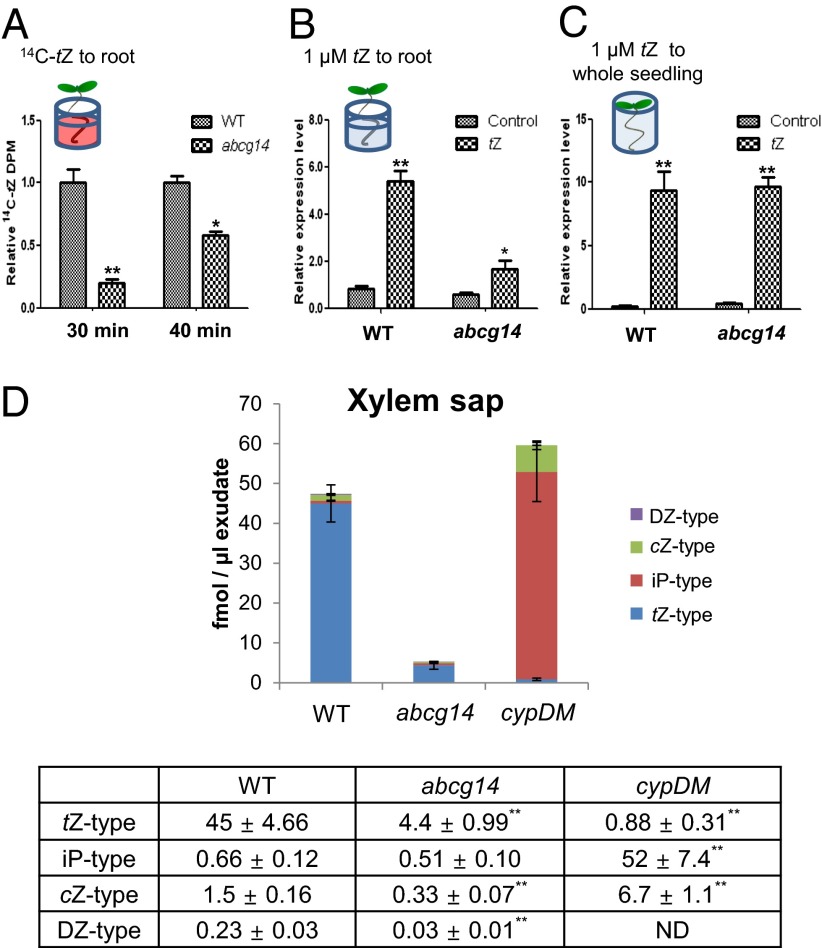
Next, we compared the capacity of the wild type and atabcg14 to transfer to the shoot cytokinin that had been exogenously applied to the roots. For this experiment, seedlings were grown for 5 d only, to minimize the effect of differences in leaf size between the wild type and atabcg14 mutant on translocation. The roots of whole seedlings were incubated in medium containing 14C-labeled tZ for 30 or 40 min. Much less radioactivity was detected in the shoots of atabcg14 than in those of the wild type (Fig. 4A), indicating that the translocation of exogenous cytokinin was reduced in atabcg14.
Fig. 4.
Translocation of exogenous and endogenous cytokinins was impaired in atabcg14. (A) Delayed root-to-shoot translocation of exogenously applied 14C-tZ in atabcg14 (abcg14). Only the roots of 5-d-old seedlings were treated with 4 μM of 14C-tZ and 3H-water for 30 min and 40 min. Then the shoots of each sample were harvested and radioactivity was measured. The dpm values of 14C-tZ were normalized by that of 3H-water and presented relative to the wild-type (WT) value, which was set to 1. Values are mean ± SEM (n = 3–4 each from two independent experiments). *P < 0.05, **P < 0.01 (Student t test) relative to the corresponding wild-type values. (B) ARR5 expression levels in shoots followed by tZ application to the root for 30 min. Exposure of the root to 1 µM tZ did not increase the transcription of ARR5 in abcg14 shoots as much as it did in wild-type shoots. Values are mean ± SEM (n = 3–4 each from three independent experiments). *P < 0.05, **P < 0.01 (Student t test) relative to the corresponding wild-type values. (C) ARR5 expression levels in shoots after direct tZ treatment. tZ, supplied by submerging 5-d-old whole plants in 1 µM tZ-containing medium for 30 min, strongly induced the transcription of ARR5 in both WT and abcg14 shoots. Values are mean ± SEM (n = 4 each from two independent experiments). **P < 0.01 (Student t test) relative to the mock-treated control. (D) Cytokinin concentration in the xylem sap. cypDM, cyp735a1 cyp735a2 double mutant; ND, none detected. Xylem exudates from 22-d-old plants were collected during the hour following decapitation. Values are mean ± SD of five samples. **P < 0.01 (Student t test) relative to the corresponding wild-type values (bottom table).
To confirm the AtABCG14-dependent translocation of cytokinin, only the roots were treated with tZ, and ARR5 induction in mutant and wild-type shoots was compared. The transcription level of ARR5 was much lower in mutant shoots than in those of the wild type (Fig. 4B), suggesting that AtABCG14-dependent translocation of cytokinins is required for a full cytokinin response. In contrast, when whole seedlings were immersed in cytokinin-containing medium, ARR5 was expressed at similar levels in the mutant and wild type (Fig. 4C).
Cytokinin Content in Xylem Sap Is Reduced in atabcg14.
To obtain direct proof that AtABCG14 is involved in delivery of cytokinins to the xylem, we analyzed the cytokinin content in the xylem exudate (xylem sap). The concentration of major root cytokinins—that is, the tZ-type—was reduced by more than 90%. The concentrations of the two minor cytokinins—that is, the dihydrozeatin (DZ) and cZ types, were also significantly reduced, whereas no significant change was observed for the iP-type cytokinins (Fig. 4D and Fig. S6). These results suggest that AtABCG14 is required for xylem loading of tZ-type cytokinins for root-to-shoot translocation. We also measured cytokinin contents in the xylem sap of the cyp735a1 cyp735a2 double mutant (8), which cannot synthesize tZ-type cytokinins. Although tZ-type cytokinin levels were low in this plant, the total amount of cytokinins was similar to that of the wild type, due to an increase in iP-type cytokinins (Fig. 4D).
Growth Recovery of atabcg14 Shoots Grafted onto Wild-Type Roots.
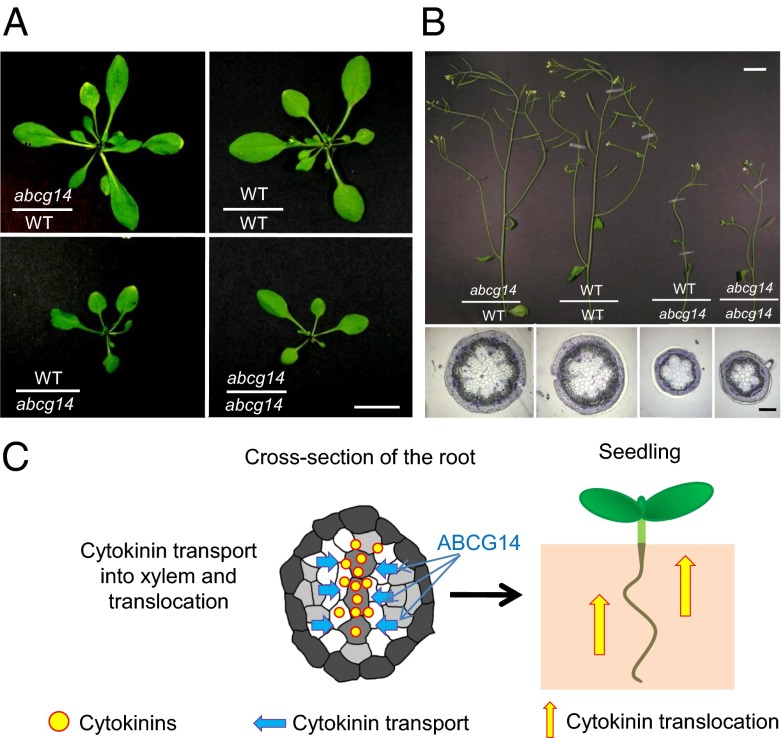
To examine whether the reduction in cytokinin translocation is the cause of the atabcg14 shoot phenotype, we reciprocally grafted atabcg14 shoot scions onto wild-type rootstocks (abcg14/WT) and wild-type shoot scions onto atabcg14 rootstocks (WT/abcg14). The grafted plants with the same genetic combinations exhibited consistent phenotypes. The growth defects of abcg14/WT restored to a level similar to that of the control grafting of WT/WT (Fig. 5A, Upper Right and Upper Left, and B), indicating that the roots of wild-type plants delivered sufficient cytokinin for normal development of the mutant shoot. In contrast, the WT/abcg14 graft exhibited a similar dwarf phenotype as the control grafting of abcg14/abcg14 (Fig. 5A, Lower Right and Lower Left, and B), indicating that atabcg14 roots could not transport sufficient cytokinins to support normal growth and development of the shoot. These results suggest that AtABCG14-mediated root-to-shoot translocation of cytokinins is crucial for shoot growth and development.
Fig. 5.
Growth recovery of atabcg14 shoots grafted onto wild-type roots. (A and B) Leaves (A) and inflorescence stems (B) of grafts between atabcg14 (abcg14) and the wild type (WT). Note that the leaves and stems of the graft between abcg14 scion and wild-type rootstock (abcg14/WT) grow normally, whereas those of the graft between wild-type scion and abcg14 rootstock (WT/abcg14) exhibit impaired growth. Cross-sections of inflorescence stems of each graft (B, Lower) also revealed that abcg14/WT developed normal vascular bundles, similar to those found in WT/WT, whereas WT/abcg14 did not. (C) Model of tZ-type cytokinin translocation mediated by AtABCG14. AtABCG14 is essential for the root-to-shoot translocation of cytokinins, which promotes shoot growth. [Scale bar, 0.5 cm (A), 2 cm (B, Upper), and 200 μm (B, Lower).]
Discussion
Cytokinins have long since been known to be transported from the root to the shoot via xylem (2). However, the molecular mechanism whereby cytokinin is transported into the xylem has been elusive. Our results strongly suggest that AtABCG14 is engaged in a critical step in cytokinin transport in roots—namely, it is involved in a step that loads cytokinin into the xylem of the root, facilitating its transfer to the shoot (Fig. 5C). The following lines of evidence presented in this work support this conclusion: (i) atabcg14 knockout plants phenocopied cytokinin biosynthesis (14) and receptor mutants (15) and were recovered by the exogenous application of cytokinin (Fig. 1); (ii) AtABCG14 was coexpressed with the cytokinin biosynthesis genes in the root (Fig. 2) (8, 17, 18); (iii) the levels of cytokinin and the expression of the cytokinin-responsive genes were decreased in the shoot and increased in the roots of the mutant (Fig. 3 and Fig. S5); (iv) exogenous cytokinin applied to the roots was transferred to the shoots of the knockout at a much slower rate than to those of the wild type (Fig. 4A); (v) cytokinin concentrations were extremely low in the xylem of the atabcg14 mutant (Fig. 4D and Fig. S6); (vi) grafting of the mutant shoot onto wild-type rootstock recovered the growth of the shoot (Fig. 5); and (vii) AtABCG14 was localized at the plasma membrane (Fig. 2D) and could therefore be involved in cellular efflux of cytokinin. These results imply that AtABCG14 functions as a cytokinin transporter that exports cytokinin to the apoplast. This notion prompted us to measure the transport activity of this protein directly. Because efflux transporter activity at the plasma membrane cannot be analyzed rigorously using a whole-cell system, we expressed AtABCG14 in budding yeast, isolated microsomes, and carried out transport assays. However, we could not detect any transport activity for tZ, either with the native form or with a codon-use adjusted form of AtABCG14, which produced the protein of the expected size. This negative result could be due to the fact that AtABCG14 requires an interacting partner and/or posttranscriptional modification that is absent in yeast. As a half-size ABC protein, AtABCG14 is only active when present as a homo- or heterodimer. Le Hir et al. (16) did not detect AtABCG14 homodimers, but observed that AtABCG14 forms heterodimers with AtABCG11. However, AtABCG11 is expressed at very low levels in roots and is not expressed in the vasculature (16). Thus, it is unlikely that the AtABCG11/AtABCG14 dimer functions as a cytokinin transporter for long-distance translocation. To identify the dimerization partner of AtABCG14, we observed phenotypes of 21 knockout plants among the 27 remaining half-size ABCG members, including two putative paralogs of AtABCG14, but could not find any with phenotypes similar to that of atabcg14 (Fig. S7). Thus, it remains to be determined whether, under conditions different from those tested by Le Hir et al. (16), ABCG14 forms homodimers or which other partners dimerize with AtABCG14 to form a functional unit.
Translocation of cytokinins from roots to shoots appears to be crucial for normal shoot growth, because atabcg14 rootstocks failed to support wild-type shoot growth (Fig. 5 A and B). This result is consistent with a previous report showing that tZ-type cytokinins are essential for shoot growth and that the enzymes that synthesize this type of cytokinin are mainly expressed in the root vasculature (8). However, our findings contradict those of other studies that indicate that root-derived cytokinins are not essential for shoot growth. This notion was based on two observations: (i) wild-type shoots grafted onto the roots of the quadruple cytokinin synthase knockout mutant, atipt1;3;5;7, exhibited normal growth (10), and (ii) when the roots of a double mutant of two genes involved in tZ-type cytokinin biosynthesis, cyp735a1 cyp735a2 (cypDM), were grafted onto wild-type shoots (WT/cypDM), shoot growth was similar to that of the wild type (8). Our analysis of xylem sap cytokinin contents provides a clue as to why the grafted plants exhibited different growth rates. The xylem sap of atabcg14 had very low levels of cytokinins, whereas that of cypDM had high levels of iP-type cytokinins and total cytokinin levels that did not differ from those of the wild type (Fig. 4D). It is therefore likely that the iP-type cytokinins in WT/cypDM are converted into tZ type in the wild-type shoot to support shoot growth, as CYP735A1 and A2 are also expressed in the shoot, albeit at low levels. Thus, as long as either the root or shoot can synthesize a type of cytokinin (as in WT/atipt1;3;5;7 or WT/cypDM), and can translocate it via the xylem, the shoot can grow normally. However, when transport of cytokinins from the root is severely blocked (as in WT/abcg14), growth is impaired in the shoot. Together, our results strongly suggest that the active translocation of tZ-type or iP-type cytokinins is necessary for shoot growth.
The deficiency in tZ-type cytokinins in the shoots of atabcg14 plants seems to have activated compensatory mechanisms to increase cytokinin levels (21); the expression of genes involved in cytokinin biosynthesis was up-regulated in atabcg14 (Fig. S8), and consequently, iP-type cytokinins were more abundant in atabcg14 shoots (Fig. 3). Intriguingly, atabcg14 roots exhibited increases in cytokinin biosynthesis (Fig. S8) and iP-type cytokinin levels (Fig. 3B), despite having high total cytokinin levels (Fig. 3B). These results suggest that a signal that heralds a cytokinin deficiency, originated in the atabcg14 shoot, is transmitted to the root.
The primary roots of atabcg14 were consistently longer than those of the wild type under short-day conditions (Fig. 1A), both when plants were grown on one-half Murashige and Skoog (MS) and MGRL media, and this phenotype was complemented by expression of AtABCG14 under its own promoter (Fig. 1A and Fig. S2A). A long primary root is a common phenotype of cytokinin-related mutants, such as the atipt1;3;5;7 quadruple mutant (14), and cytokinin oxidase/dehydrogenase overexpressing plants (22), which contain low levels of cytokinin. Although atabcg14 roots contained high cytokinin levels (Fig. 3B) and was enhanced in cytokinin signaling (Fig. S9 A and B), the expression of IAA3/Short Hypocoyl2 (SHY2), which is up-regulated by cytokinin and plays a major role in cytokinin-mediated cell differentiation in the root transition zone (23), was not increased at the root tip of the atabcg14 mutant (Fig. S9B). In addition, the number of root apical meristem cells was significantly higher in the mutant than in the wild type (Fig. S9C), indicating an increased cell division in the mutant root apical meristem. These results suggest that cytokinin content/signaling is not the sole determinant of root length and, furthermore, that complex crosstalk might exist between cytokinin and auxin. The expanded and enhanced cytokinin content/signaling in the root might influence root length by modulating auxin signaling. In support of this explanation, a high cytokinin concentration was reported to enhance auxin biosynthesis and/or auxin signal transduction (24, 25), which can result in an elongated root phenotype. More studies are necessary to decipher the reasons for the long primary root of the atabcg14 mutant.
The reduced lignin contents observed in the atabcg14 mutant (Fig. S3) raise the question of whether this transporter also acts as a monolignol transporter, such as the recently described AtABCG29 transporter that specifically exports coumaryl alcohol (26). However, in the case of AtABCG14, the effect on lignin seems to be indirect. The recovery of atabcg14 shoot grafted onto wild-type rootstock (abcg14/WT) suggests that the transporter has a role in long-distance signaling, rather than in the local supply of lignin precursors. Thus, it is very likely that the reduced cytokinin allocation to the shoot leads to the observed reduction in xylem development and, consequently, to the reduction in lignin content in atabcg14.
Besides being transported from the root to the shoot, cytokinins must be loaded into the phloem. It is tempting to speculate that members of the Arbidopsis purine permease (AtPUP) family or equilibrative nucleoside transporter (ENT) family, both of which have been reported to take up cytokinins, and other chemicals with similar structures (27–29), may be involved in this process. Because both AtPUP and ENT proteins are encoded by genes that belong to gene families, it is likely that a phenotype for single and even double mutants could not be observed, due to functional redundancy among family members (28). At the cellular level, cytokinins must be transported across the plasma membrane and finally into the endoplasmic reticulum, where, according to recent results, the cytokinin receptors are localized (30, 31). No clues are presently available as to the identity of the cytosolic exporter into the endoplasmic reticulum.
In conclusion, our results provide strong evidence that AtABCG14 is essential for loading cytokinins into the xylem (Fig. 5C). The translocation mediated by AtABCG14 has a great impact on shoot development, as well as on the coordinated growth of the shoot and root, by mediating the delivery of a potent growth-activating signal that is synthesized in the root to the shoot. Further work is needed to clarify how AtABCG14 contributes to cytokinin transport to the xylem in the root. Specifically, it will be interesting to (i) determine whether AtABCG14 directly transports cytokinins, (ii) identify the substrate specificity range, (iii) establish the identity of the dimerization partner, and (iv) test how AtABCG14 activity is regulated in response to environmental conditions that prompt changes in growth.
Materials and Methods
Plant Materials and Growth Conditions.
Wild-type, atabcg14 knockout (SK_15918), and transgenic seeds were surface-sterilized with NaOCl and placed in the dark at 4 °C for 2 d. After sowing the seeds on either MGRL (13) or on half-strength MS medium with 1% sucrose, plants were grown for the indicated periods of time in a controlled environment (22/18 °C; 16 h/8 h or 8 h/16 h light/dark). For further experiments, plants were transferred to soil and grown in a greenhouse (22/18 °C; 16 h/8 h light/dark).
Sectioning.
For stem sections, the base of the main stem was collected and fixed with 4% (vol/vol) formaldehyde and 4% (vol/vol) glutaraldehyde in 50 mM sodium phosphate buffer. Samples were dehydrated with a graded series of ethanol and then embedded in Technovit 7100 (Kulzer) for 24 h at 4 °C. Transverse sections (5 µm) were produced using a Leica RM2245 rotary microtome and stained with toluidine blue. Images were taken using a Zeiss Axiosckop2 microscope.
Exogenous Cytokinin Application.
Seeds were directly sowed on the soil. After 10 d of growth, the plants were sprayed with 1 µM of tZ, iP, or 0.1% DMSO solution (mock) every day for the following 21 d, and then photographed. The area of the fifth and sixth leaves was measured using ImageJ software (32).
Complementation.
The genomic DNA region of AtABCG14 was amplified and cloned into the pCR8/GW-TOPO vector (Invitrogen). The 2-kb promoter region of AtABCG14 was inserted into the SacII/SpeI site located immediately upstream of the AtABCG14 start codon. The GFP gene was inserted into the SpeI/ClaI site located between the promoter and AtABCG14 start codon. The completed construct, pAtABCG14::sGFP::AtABCG14 gDNA, was transferred into pMDC100 using LR clonase (Invitrogen) and introduced into the atabcg14 mutant by floral dipping (33).
Expression Pattern Analysis.
The fragment 2 kb upstream of the AtABCG14 start codon was amplified and inserted into pMDC163 (34). Wild-type Arabidopsis was transformed with the pAtABCG14::GUS construct using the Agrobacterium-mediated floral dip method (33). The transgenic plants were grown on half-strength MS medium for 5 d and then stained with GUS staining solution. Root sections were obtained as described previously (35). Briefly, roots were fixed in formalin–acetic–acid alcohol (FAA) solution [10% (vol/vol) formaldehyde, 5% (vol/vol) acetic acid, and 50% (vol/vol) ethanol] and embedded in 1% agarose. Samples were dehydrated, embedded, and sectioned as described above for stem sections.
Quantitative Real-Time PCR.
To analyze type-A ARR transcript levels, wild-type and atabcg14 plants were grown on MGRL-agar medium for 14 d, and shoots and roots were then separately sampled. Total RNA was extracted using TRIzol and treated with DNaseI (Takara) for 30 min. cDNA was generated from RNA purified by phenol:chloroform extraction, and relative transcript levels were measured using a SYBR Kit (Takara), according to the manufacturer’s instructions.
Micrografting.
Following a previously described protocol (36), the hypocotyls of 4- or 5-d-old seedlings of each genotype were cut using a sharp razor blade, aligned with their partners, and incubated under high-humidity conditions for 7 d. The seedlings that formed connections with their partners were transferred to soil, grown for 28 d, and then photographed. Fourteen plants were the product of atabcg14 shoot grafted onto wild-type rootstock (abcg14/WT), 12 plants of wild-type shoot grafted onto atabcg14 rootstock (WT/abcg14), six of wild-type shoot onto wild-type rootstock (WT/WT), and seven of atabcg14 shoot onto atabcg14 rootstock (abcg14/abcg14).
14C-tZ Translocation Assay.
Twenty 5-d-old seedlings per genotype were transferred to 96 wells, which contained 200 µl of half-strength MS (Mes pH 5.7–KOH) medium. Only the roots were submerged in medium. After 30 min of preincubation, 14C-tZ was added to the wells at a final concentration of 4 µM, and the plants were incubated for an additional 30 or 40 min. The shoots were harvested and their radioactivity was counted using Liquid Scintillation Counter.
Xylem Sap Collection.
After germination, seedlings were grown in the dark for 7 d, to allow the hypocotyls to elongate. Then, plants were transferred to 16 h/8 h light/dark cycles (100 µmol⋅m−2⋅s−1) at 22 °C and grown for 3 wk. The xylem exudate (xylem sap) was collected by cutting the hypocotyl with a razor blade. The exudate was then collected during a 1-h period, and the volumes of samples were measured. The exudate was immediately subjected to cytokinin quantification.
Cytokinin Quantification.
Cytokinins were extracted and determined from about 100 mg of fresh tissues as described previously using ultra-performance liquid chromatography–tandem mass spectrometry (AQUITY UPLC System/XEVO-TQS; Waters) with an octadecylsilyl column (AQUITY UPLC BEH C18, 1.7 µm, 2.1 × 100 mm; Waters) (37).
Note Added in Proof.
While this manuscript was under review, a similar function for AtABCG14 was reported by Zhang et al. (38).
Supplementary Material
Acknowledgments
We thank Ms. Nanae Ueda (RIKEN) for her technical assistance. We thank Dr. Bruno Müller for providing us with seeds of transgenic TCS::GFP–expressing plants and the TCS::GFP vector and Dr. Ykä Helariutta for seeds of transgenic pARR5::GUS–expressing plants. This research was supported by the Global Research Laboratory Program (to Y.L. and E.M.), a Swiss National foundation grant (to E.M.), and a Grant-in-Aid for Scientific Research on Innovative Areas (21114005) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to H.S.).
Footnotes
Conflict of interest statement: The authors are in the process of filing a patent based on the data presented in this paper.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321519111/-/DCSupplemental.
References
- 1.Hirose N, et al. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot. 2008;59(1):75–83. doi: 10.1093/jxb/erm157. [DOI] [PubMed] [Google Scholar]
- 2.Kudo T, Kiba T, Sakakibara H. Metabolism and long-distance translocation of cytokinins. J Integr Plant Biol. 2010;52(1):53–60. doi: 10.1111/j.1744-7909.2010.00898.x. [DOI] [PubMed] [Google Scholar]
- 3.Blakeslee JJ, Peer WA, Murphy AS. Auxin transport. Curr Opin Plant Biol. 2005;8(5):494–500. doi: 10.1016/j.pbi.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Morris DA, Friml J, Zazimalova E. (2004) in Plant Hormones: Biosynthesis, Signal Transduction, Action!, eds Davies PJ (Kluwer Academic, Dordrecht, The Netherlands), pp 437–470.
- 5.Sakakibara H. Cytokinins: Activity, biosynthesis, and translocation. Annu Rev Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- 6.Hwang I, Sheen J, Müller B. Cytokinin signaling networks. Annu Rev Plant Biol. 2012;63:353–380. doi: 10.1146/annurev-arplant-042811-105503. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Y. The role of local biosynthesis of auxin and cytokinin in plant development. Curr Opin Plant Biol. 2008;11(1):16–22. doi: 10.1016/j.pbi.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Kiba T, Takei K, Kojima M, Sakakibara H. Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev Cell. 2013;27(4):452–461. doi: 10.1016/j.devcel.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Bishopp A, et al. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr Biol. 2011;21(11):927–932. doi: 10.1016/j.cub.2011.04.049. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto-Kitano M, et al. Cytokinins are central regulators of cambial activity. Proc Natl Acad Sci USA. 2008;105(50):20027–20031. doi: 10.1073/pnas.0805619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318(5851):801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 12.Bhargava A, et al. Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol. 2013;162(1):272–294. doi: 10.1104/pp.113.217026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujiwara T, Hirai MY, Chino M, Komeda Y, Naito S. Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 1992;99(1):263–268. doi: 10.1104/pp.99.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyawaki K, et al. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA. 2006;103(44):16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riefler M, Novak O, Strnad M, Schmülling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell. 2006;18(1):40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Hir R, et al. ABCG9, ABCG11 and ABCG14 ABC transporters are required for vascular development in Arabidopsis. Plant J. 2013;76(5):811–824. doi: 10.1111/tpj.12334. [DOI] [PubMed] [Google Scholar]
- 17.Takei K, Yamaya T, Sakakibara H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J Biol Chem. 2004;279(40):41866–41872. doi: 10.1074/jbc.M406337200. [DOI] [PubMed] [Google Scholar]
- 18.Miyawaki K, Matsumoto-Kitano M, Kakimoto T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 2004;37(1):128–138. doi: 10.1046/j.1365-313x.2003.01945.x. [DOI] [PubMed] [Google Scholar]
- 19.To JP, et al. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16(3):658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453(7198):1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foo E, et al. Feedback regulation of xylem cytokinin content is conserved in pea and Arabidopsis. Plant Physiol. 2007;143(3):1418–1428. doi: 10.1104/pp.106.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner T, et al. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15(11):2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dello Ioio R, et al. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322(5906):1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- 24.Jones B, et al. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell. 2010;22(9):2956–2969. doi: 10.1105/tpc.110.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanstraelen M, Benková E. Hormonal interactions in the regulation of plant development. Annu Rev Cell Dev Biol. 2012;28:463–487. doi: 10.1146/annurev-cellbio-101011-155741. [DOI] [PubMed] [Google Scholar]
- 26.Alejandro S, et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr Biol. 2012;22(13):1207–1212. doi: 10.1016/j.cub.2012.04.064. [DOI] [PubMed] [Google Scholar]
- 27.Bürkle L, et al. Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J. 2003;34(1):13–26. doi: 10.1046/j.1365-313x.2003.01700.x. [DOI] [PubMed] [Google Scholar]
- 28.Cedzich A, Stransky H, Schulz B, Frommer WB. Characterization of cytokinin and adenine transport in Arabidopsis cell cultures. Plant Physiol. 2008;148(4):1857–1867. doi: 10.1104/pp.108.128454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirose N, Makita N, Yamaya T, Sakakibara H. Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinin transport. Plant Physiol. 2005;138(1):196–206. doi: 10.1104/pp.105.060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caesar K, et al. Evidence for the localization of the Arabidopsis cytokinin receptors AHK3 and AHK4 in the endoplasmic reticulum. J Exp Bot. 2011;62(15):5571–5580. doi: 10.1093/jxb/err238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wulfetange K, et al. The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol. 2011;156(4):1808–1818. doi: 10.1104/pp.111.180539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- 33.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 34.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133(2):462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heo J-O, et al. Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc Natl Acad Sci USA. 2011;108(5):2166–2171. doi: 10.1073/pnas.1012215108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turnbull CG, Booker JP, Leyser HM. Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 2002;32(2):255–262. doi: 10.1046/j.1365-313x.2002.01419.x. [DOI] [PubMed] [Google Scholar]
- 37.Kojima M, et al. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: An application for hormone profiling in Oryza sativa. Plant Cell Physiol. 2009;50(7):1201–1214. doi: 10.1093/pcp/pcp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang et al. (2014) Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat Commun, 10.1038/ncomms4274. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.