Significance
Assembly of actin filaments near the plasma membrane drives extension of the cell edge as it migrates. Cells recruit a protein machine called the Arp2/3 complex to the leading edge to initiate actin filament branches that grow and push on the membrane. Equally important, capping protein (CP) binds to and terminates polymerization of the fast-growing end of the filament. Like other aspects of actin assembly, CP is subject to regulation. Specifically, the protein V-1/myotrophin inactivates CP, while capping protein Arp2/3 myosin I linker (CARMIL) proteins moderately inhibit CP. We describe how CARMIL and V-1 cooperate in a cycle of reactions to regulate CP and promote assembly of the actin network by the Arp2/3 complex.
Keywords: cell migration, VASP
Abstract
Although capping protein (CP) terminates actin filament elongation, it promotes Arp2/3-dependent actin network assembly and accelerates actin-based motility both in vitro and in vivo. In vitro, capping protein Arp2/3 myosin I linker (CARMIL) antagonizes CP by reducing its affinity for the barbed end and by uncapping CP-capped filaments, whereas the protein V-1/myotrophin sequesters CP in an inactive complex. Previous work showed that CARMIL can readily retrieve CP from the CP:V-1 complex, thereby converting inactive CP into a version with moderate affinity for the barbed end. Here we further clarify the mechanism of this exchange reaction, and we demonstrate that the CP:CARMIL complex created by complex exchange slows the rate of barbed-end elongation by rapidly associating with, and dissociating from, the barbed end. Importantly, the cellular concentrations of V-1 and CP determined here argue that most CP is sequestered by V-1 at steady state in vivo. Finally, we show that CARMIL is recruited to the plasma membrane and only at cell edges undergoing active protrusion. Assuming that CARMIL is active only at this location, our data argue that a large pool of freely diffusing, inactive CP (CP:V-1) feeds, via CARMIL-driven complex exchange, the formation of weak-capping complexes (CP:CARMIL) at the plasma membrane of protruding edges. In vivo, therefore, CARMIL should promote Arp2/3-dependent actin network assembly at the leading edge by promoting barbed-end capping there.
Actin assembly at the interface between the plasma membrane and the cytoplasm is driven by the recruitment of proteins that promote the nucleation of new actin filaments (1). A key nucleating factor driving actin assembly at the leading edge of crawling cells is the Arp2/3 complex, which creates the branched actin networks that comprise lamellipodia (2, 3). Capping protein (CP), which binds the barbed end of the actin filament with very high affinity (∼0.1 nM) to halt elongation (4), is an essential component of Arp2/3-generated branched actin networks (5). Paradoxically, although CP terminates filament elongation, it promotes Arp2/3-dependent actin network assembly and actin-based motility both in vitro and in vivo by increasing the frequency of Arp2/3-dependent nucleation (6–10). Factors that regulate the activity of CP therefore should play important roles in actin assembly and cell motility. One such factor may be the large scaffold protein CARMIL (capping protein Arp2/3 myosin I linker) (11), which contains a region of ∼75 residues that binds CP very tightly (Kd ∼1 nM) (12, 13). This region, which we named CAH3 for CARMIL Homology Domain 3, antagonizes the function of CP in vitro in two ways. First, by binding to free CP, CAH3 reduces CP’s affinity for the barbed end from ∼0.1 to ∼50 nM, thereby creating a weak barbed-end capping complex (CP:CAH3) (12, 13). Second, by binding to CP already present on the barbed end, CAH3 accelerates by ∼300-fold the rate of dissociation of CP from the end; i.e., CAH3 rapidly uncaps CP-capped filaments (12–14).
Another potentially important regulator of CP is the protein V-1, also known as myotrophin (15, 16). This 13 kDa, ankyrin-repeat protein binds CP with an affinity of ∼20 nM, creating a 1:1 complex (CP:V-1) that has no affinity for the barbed end (16–18). V-1 has the potential, therefore, to influence polymerization by reducing the extent of barbed-end capping.
Full understanding of CP regulation will require an in-depth understanding of how the activities of CARMIL and V-1 are coordinated. Relevant to this, Takeda et al. (17) showed that CAH3-like peptides readily convert the CP:V-1 complex into the CP:CAH3 complex. In terms of the mechanism of this exchange reaction, they postulated that the binding of CAH3 to the CP:V-1 complex results in the transient formation of a CP:V-1:CAH3 ternary complex. A subsequent allosteric change in CP caused by ternary complex formation then drives V-1 dissociation, yielding the CP:CAH3 complex. Importantly, the binding sites on the surface of CP for CAH3 and V-1 are nonoverlapping, consistent with the possibility that these three proteins form a ternary complex (17–21).
The first goal of this study was to define precisely how CAH3 catalyzes the exchange of CP from its sequestered state (CP:V-1) into its weak barbed-end–binding state (CP:CAH3). This effort focused on providing direct evidence for the existence of the ternary complex and on determining the relative contributions of allostery versus competitive binding to complex exchange. The second goal was to determine the effect of complex exchange on the rate of barbed-end elongation in vitro, which must be understood to predict the effect of complex exchange on actin assembly in vivo. The third goal was to measure the cellular concentrations of V-1 and CP, as these values are required to gauge the likely significance of the complex exchange reaction in vivo. Finally, the fourth goal was to further define the spatial features and temporal dynamics of CARMIL’s leading-edge localization (11, 12, 22, 23), as this should further pinpoint both the site and the timing of complex exchange. Specifically, we sought to characterize CARMIL’s leading-edge localization relative to other key leading-edge molecules and to the plasma membrane and to determine if CARMIL’s leading-edge accumulation correlates with edge dynamics, i.e., protrusion versus retraction. Overall, our results support a model in which the recruitment of CARMIL to the plasma membrane specifically at sites of active polymerization and edge protrusion drives the exchange of abundant, freely diffusing, inactive CP:V-1 complexes into membrane-bound CP:CARMIL complexes that cap the barbed end weakly. This CARMIL-driven exchange reaction should therefore serve to promote Arp2/3-dependent actin network formation at protruding edges by promoting barbed-end capping there.
Results
CAH3 Accelerates the Conversion of CP:V-1 into CP:CAH3 via Two Mechanisms.
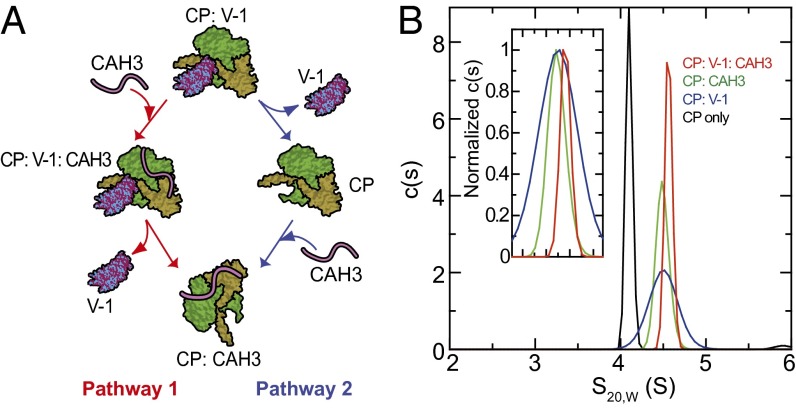
Previously, Takeda et al. (17) showed that CAH3-like peptides (see Fig. S1A for nomenclature) inhibit the binding of V-1 to CP and accelerate the rate of dissociation of V-1 from preformed CP:V-1 complexes. Consistently, when we added increasing amounts of CAH3 to bead-immobilized GST-V-1 saturated with CP, we observed the progressive appearance of CP in the bead supernatant (dissociated fraction) (Fig. S2A). Fitting yielded a value of 625 ± 103 nM for the half-maximal concentration of CAH3 required for this apparent complex exchange reaction (Fig. S2B). CAH3-driven complex exchange could occur by either or both of two pathways (Fig. 1A). In pathway 1 (allosteric pathway), the binding of CAH3 to the CP:V-1 complex creates a CP:V-1:CAH3 ternary complex that results, via an allosteric mechanism, in the weakening of V-1’s interaction with CP. This leads to V-1 dissociation, forming the CP:CAH3 complex. This pathway was previously proposed by Takeda et al. (17). In pathway 2 (competitive binding pathway), the binding of CAH3 to free CP generated by the spontaneous dissociation of V-1 from the CP:V-1 complex results in the conversion of the CP:V-1 complex into the CP:CAH3 complex.
Fig. 1.
CAH3-driven complex exchange can occur via two pathways, one of which involves the formation of a CP:V-1:CAH3 ternary complex. (A) Schematic of the two possible pathways by which CAH3 could drive the conversion of the CP:V-1 complex to the CP:CAH3 complex. (B) Data from analytical ultracentrifugation demonstrating the existence of the CP:V-1:CAH3 ternary complex. The c(s) distributions obtained for all four samples showed single dominant peaks in all cases representing over 95% of the loading concentrations, indicating that even in samples containing protein mixtures only single molecular species were detected. The c(s) distribution peak for the CP-only sample had a weight average s20,w value of 4.4 S and a calculated molar mass of 63.5 kDa, which is in good agreement with the theoretical molecular weight of CP. In the three samples containing protein mixtures, peaks representing free V-1 and CAH3 fractions were either not detectable or constituted less than 0.5% of the loading concentration. It is not possible to obtain reliable mass estimates for those complexes because the broadening of their sedimentation boundaries represent not only diffusion, but also complex exchange during sedimentation. The positions of the various complexes in the c(s) versus S-value plot are indicted by color coding (see key). (Inset) Normalized data.
Our first evidence for the existence of the ternary complex implicated in pathway 1 was the presence of small amounts of CAH3 together with GST-V-1 and CP in bound fractions (see the asterisks in Fig. S2A). More rigorous support for the existence of the ternary complex was provided by sedimentation velocity analyses using the following four samples: CP only, CP + V-1, CP + CAH3, and CP + V-1 + CAH3. For the mixed samples, equal volumes of the proteins, all at a final concentration of 10 µM, were combined and incubated for 30 min at 21 °C before initiating centrifugation. As the mass of a complex increases, so does its sedimentation speed, causing it to be shifted more to the right in a sedimentation coefficient distribution [c(S)] plot. Consistently, Fig. 1B shows that the position of the peak corresponding to the CP:V-1:CAH3 ternary complex (s20,w = 5.1 S), which has the greatest mass, is farther to the right than the peaks corresponding to the CP:CAH3 and CP:V-1 complexes (s20,w = 4.6 and 5.0 S, respectively). Together, these results confirm the existence of the ternary complex and support the idea that pathway 1 can contribute to complex exchange.
For pathway 2 to contribute significantly to complex exchange, the rate at which V-1 dissociates spontaneously from the CP:V-1 complex must be relatively fast. To measure this rate, we used fluorescence anisotropy of IAEDANS-labeled V-1 [IAEDANS-(C2)-V-1], which binds CP with normal affinity [15.4 ± 5.2 nM and 20.4 ± 4.8 nM, as estimated by steady-state fluorescence anisotropy (Fig. S3A) and isothermal titration calorimetry, respectively, versus 21 nM (17) and 14 nM (20) for wild-type V-1]. IAEDANS-(C2)-V-1 and CP (600 nM each, final) were first mixed together and incubated for 30 min at 22 °C to form the complex. Excess nonlabeled V-1 (60 µM final) was then added to prevent rebinding of labeled V-1 to CP and the rate of dissociation of IAEDANS-(C2)-V-1 from CP measured by fluorescence anisotropy (Fig. 2A). Fitting using a single exponential decay yielded a rate of spontaneous dissociation of V-1 from CP of 0.049 ± 0.015/s (black dots and line in Fig. 2A), in good agreement with a previous report using surface plasmon resonance [0.031/s (17)]. Importantly, this rate, which corresponds to a halftime of dissociation of ∼14 s, appears to be fast enough to support to some extent complex exchange via pathway 2.
Fig. 2.
The rate of spontaneous dissociation of V-1 from CP is accelerated dramatically by CAH3 via a mixture of pathways 1 and 2, with pathway 1 dominating at high concentrations of CAH3. (A) The rate of spontaneous dissociation of IAEDANS-(C2)-V-1 from a preformed complex of CP:IAEDANS-(C2)-V-1 (∼600 nM) in the presence of 60 µM unlabeled V-1, measured by fluorescence anisotropy. The estimated rate of spontaneous dissociation of IAEDANS-(C2)-V-1 from CP was k5− = 0.049 ± 0.015/s (Fig. S1B). (B) The rates of dissociation of IAEDANS-(C2)-V-1 from a preformed complex of CP:IAEDANS-(C2)-V-1 in the presence of 60 µM unlabeled V-1 and 0 (black), 25 nM (red) or 125 nM (blue) CAH3, measured by fluorescence anisotropy (data fit with single exponentials). (C) As in B, except that dissociation rates in assays containing more than 125 nM CAH3 were measured using a stopped-flow apparatus. The estimated equilibrium dissociation constant of CAH3 binding to CP:V-1 is Kd6 = 618 ± 164 nM, and the estimated rate of dissociation of V-1 from the CP:CAH3 complex is k7− = 3.57 ± 0.28/s (Fig. S1B). (D) The relative contribution of pathway 1 (ternary complex formation) to overall complex exchange (pathway 1 plus pathway 2) as a function of CAH3 concentration.
To estimate the relative contributions of pathways 1 and 2 to complex exchange, we monitored the time course of dissociation of IAEDANS-(C2)-V-1 from CP in the presence of excess nonlabeled V-1 (60 µM final) and increasing amounts of CAH3. As shown in Fig. 2B, 25 and 125 nM CAH3 accelerated in a dose-dependent manner the rate of dissociation of V-1 from CP. Importantly, this result supports an allosteric component to CAH3-driven V-1 dissociation (17), as CAH3 addition would not accelerate the rate of V-1 dissociation if the exchange reaction was driven purely by competitive binding. To perform measurements at higher concentrations of CAH3, a stop-flow apparatus was used. Fig. 2C shows that, at saturation, CAH3 accelerates the rate of dissociation of IAEDANS-(C2)-V-1 from CP by ∼73-fold from ∼0.05/s to ∼3.6/s. In other words, the halftime of V-1 dissociation from CP goes from ∼14 s in the absence of CAH3 to ∼0.2 s in the presence of saturating CAH3.
Importantly, the dissociation rates measured in Fig. 2C represent mixtures of pathways 1 and 2. To estimate the relative contributions of each pathway as a function of CAH3 concentration, we used two exponentials, one describing each pathway, and the following three rate constants: (i) the equilibrium dissociation rate constant of CAH3 binding to CP:V-1 (Kd6, 618 ± 164 nM; Fig. 2C), (ii) the dissociation rate of V-1 from CP:CAH3 (k7−, 3.57 ± 0.28/s; Fig. 2C), and (iii) the rate of spontaneous dissociation of V-1 from CP (k5−, 0.049 ± 0.015/s; Fig. 2A). Fig. 2D shows that the relative contribution of pathway 1 (ternary complex formation) to overall complex exchange increases progressively from 0% in the absence of CAH3 to ∼90% in the presence of 12 µM CAH3. Exactly which region of this curve is most reflective of the situation at the leading edge in vivo is difficult to estimate, although the local concentration of active CARMIL in the two-dimensional plane of the membrane could be on the order of several micromolars (see Discussion). In summary, we conclude that complex exchange occurs via a mixture of CAH3-triggered dissociation of V-1 from CP present in the CP:V-1:CAH3 ternary complex (pathway 1) and the spontaneous dissociation of V-1 from CP, followed by CAH3 association (pathway 2). Importantly, and in agreement with the overall concept proposed previously by Takeda et al. (17), both pathways allosterically inhibit the binding of V-1 to CP once CP is bound to CAH3.
Additional Factors Are Required for V-1 to Effectively Retrieve CP from the CP:CAH3 Complex.
The results above describe how CAH3 can effectively drive the conversion of a sequestered form of CP (CP:V-1) to a weak barbed-end–binding form of CP (CP:CAH3) (steps 1–3 in the proposed CP regulatory cycle depicted in Fig. 3A). For this regulatory cycle to be complete, the tight complex of CP: CAH3 (∼1 nM affinity) must be converted back to the complex of CP: V-1 (∼20 nM affinity) (step 4 in Fig. 3A). Based on the data above, V-1 itself would not be expected to drive the latter half of the cycle effectively. Consistently, pull-down assays in which increasing amounts of purified V-1 were added to bead-immobilized GST-CAH3 saturated with CP resulted in only a very gradual appearance of CP in the bead supernatant (dissociated fraction) (Fig. S4A). Although ∼60% of CP could eventually be dissociated from GST-CAH3:CP beads, this required a concentration of 660 µM of V-1 (Fig. S4B), an ∼100-fold molar excess over GST-CAH3. Consistently, the estimated half-maximal concentration of V-1 required for the complex exchange reaction (274 ± 65 µM) is ∼400-fold higher than for the inverse exchange reaction (Fig. S2B), further indicating that V-1 binds the complex of CP:CAH3 very weakly.
Fig. 3.
The CARMIL-1– and V-1–dependent regulatory cycle for CP and the affinity of V-1 for CP in the CP:CAH3 complex. (A) Proposed CARMIL-1– and V-1–dependent regulatory cycle for CP, based on data in this and previous studies (see Results). Step 1: V-1 inactivates a significant fraction of cellular CP by sequestering it in the CP:V-1 complex (barbed-end affinity = 0). Step 2: CARMIL drives via pathways 1 and 2 the conversion of the CP:V-1 complex into the CP:CARMIL complex, thereby partially activating CP (barbed-end affinity = ∼50 nM). Step 3: The CP:CARMIL complex caps the barbed end weakly. In contrast to free CP (barbed-end affinity = ∼0.1 nM), the CP:CARMIL complex allows barbed-end elongation, albeit at rates slower than for a free barbed end. Step 4: A factor and/or mechanism causes a large reduction in CARMIL’s affinity for CP, thereby driving the dissociation of CP from CARMIL. The free CP thus generated binds V-1 to repopulate the pool of sequestered CP. (B) The steady-state binding of a constant amount (200 nM) of IAEDANS-(MC7)-V-1 to increasing amounts (0–9 µM) of CP:CAH3 complex, estimated from the increase in the fluorescence of IAEDANS-(M7C)-V-1 that occurs upon binding, measured following 10 min of incubation at 22 °C. Of note, we could not distinguish the extent to which signal increase was from ternary complex formation or from IAEDANS-(M7C)-V-1 binding to CP following its dissociation from CAH3. That said, because V-1 dissociates very quickly from the ternary complex (3.6/s), and because CAH3 has a much higher affinity for both free CP and CP in the CP:V-1 complex than V-1 has for both free CP and CP in the CP:CAH3 complex, we assumed that the bulk of the increase in IAEDANS-(M7C)-V-1 fluorescence was due to its association in forming the ternary complex. The estimated equilibrium dissociation constant for the binding of IAEDANS-(M7C)-V-1 to the CP:CAH3 complex (Kd7) was 1.3 ± 0.3 µM (Fig. S1B).
To better quantify the affinity of V-1 for the CP:CAH3 complex, we monitored the fluorescence enhancement upon binding to this complex of IAEDANS-(M7C)-V-1, which also binds CP tightly (20). Equal concentrations of CP and CAH3 ranging from 0 to 9 µM were first mixed to form the CP:CAH3 complex. IAEDANS-(M7C)-V-1 was then added to a final concentration of 200 nM, and the steady-state fluorescent signal was measured 10 min later (Fig. 3B). The estimated equilibrium dissociation constant for the binding of IAEDANS-(M7C)-V-1 to the CP:CAH3 complex (Kd7) was 1.3 ± 0.3 µM (Fig. 3B), which is ∼65 times weaker than its affinity for free CP (assuming an equilibrium dissociation constant for Kd5 of 20 nM; Fig. S1B). In other words, V-1’s affinity for CP is attenuated tremendously when CP is in a complex with CAH3, consistent with previous work (17). Together, these results confirm that V-1 is very ineffective at retrieving CP from CAH3 and argue that V-1 requires an additional mechanism and/or cellular factor to effectively complete the regulatory cycle.
Cellular Concentrations of V-1 and CP Argue That Most CP Is Sequestered by V-1 at Steady State in Vivo.
V-1’s potency as a CP regulator should scale positively with the degree to which its cellular concentration exceeds that of CP’s, previously estimated to be ∼1 µM (1). We determined the amount of V-1 in a known number of mouse embryo fibroblasts (MEFs) by Western blotting using a monoclonal antibody to V-1 and a standard curve created using purified mouse V-1 (Fig. S5A). The average of five determinations yielded a value of 71.7 ± 20.5 ng of V-1 per 106 cells. We also estimated the amount of CP in MEFs using a monoclonal antibody to CPα1 and purified mouse CPα1β2 to create a standard curve (Fig. S5B). The average of four determinations yielded a value of 57.4 ± 13.6 ng of CPα1 per 106 cells, or ∼111 ng of CPα1β2 per 106 cells. Assuming that this amount corresponds to a cellular concentration of 1 µM for CP (1), and correcting for the difference in molecular weight between CP (64 kDa) and V-1 (13 kDa), one obtains a value of ∼3.2 µM for the cellular concentration of V-1, which is roughly three times the cellular concentration of CP. Given this result and V-1’s affinity for CP, ∼99% of cellular CP should be sequestered by V-1 at steady state, barring regulation.
Central to the model of cellular CP regulation proposed here is that active V-1, like CP (1, 4), diffuses freely in the cytoplasm, whereas active CARMIL is restricted to the leading edge. Cells stained for endogenous V-1 yield a diffuse cytoplasmic signal, consistent with the first assumption (15). Moreover, the signal for GFP-tagged V-1 is also diffuse throughout the cytoplasm (Fig. S6 A1 and A2), and it recovers from photo-bleaching with kinetics (Fig. S6 B1 and B2) similarly to that of freely diffusing GFP (24). Together, these observations argue that the sequestering activity of V-1 pervades the entire cytoplasm and that V-1 equilibrates rapidly throughout the cytoplasm by diffusion.
Complex Exchange Regulates the Rate of Actin Polymerization in Solution Assays.
The CARMIL-driven conversion of sequestered CP into weakly active CP could regulate the velocity of barbed-end elongation by creating ends that can elongate, unlike CP-capped ends, but at rates that are slower than at free ends. To obtain support for this hypothesis, we measured the rate of actin polymerization in reactions containing a set amount of CP:V-1 and increasing amounts of CAH3 (Fig. 4). First, high concentrations of CP (100 nM final) and V-1 (6 µM final) were mixed and incubated at 21 °C for 10 min to create ∼100 nM CP:V-1 complex in the presence of a vast excess of free V-1. Actin seeds (∼0.8 nM barbed ends final) were then added. Finally, actin monomer (10% pyrene-labeled) was added (2 µM final) to initiate actin polymerization (t = 0). Of note, both V-1 and CAH3 are known to have no effect on the rate of actin polymerization (12, 13, 16). Moreover, the complex of CP:V-1 has no effect on the rate of actin polymerization (Fig. S3 B and C), consistent with previous work (16, 17, 20). Importantly, the addition of 20 nM (red trace in Fig. 4A) or 200 nM (blue trace) CAH3 into already initiated assays resulted in the dose-dependent inhibition of actin polymerization relative to the control (0 nM CAH3; green trace in Fig. 4A), presumably by converting the sequestered CP into weakly active CP via complex exchange (steps 1–3 in the CP regulatory cycle in Fig. 3A).
Fig. 4.
CARMIL-driven complex exchange supports rates of barbed-end elongation in bulk solution assays that are in between those of free barbed ends and CP-capped ends. (A) Bulk actin polymerization assays performed using a solution containing ∼100 nM CP:V-1 complex, generated by incubating 100 nM CP with 6 µM V-1 for 10 min at 22 °C. Polymerization was then initiated by the addition of actin seeds (∼0.8 nM barbed ends) and 2 µM G-actin (10% pyrene-labeled). The black line shows the rate of polymerization in this control reaction. Forty seconds after initiating polymerization (arrow), we added to three parallel reactions a solution that represented 10% of the total volume of the reaction and that contained 0, 200 nM, or 2 µM CAH3, yielding final concentrations of 0 (green line), 20 nM (red line), or 200 nM (blue line) CAH3 (note: the gap following the arrow corresponds to the ∼20 s required to add CAH3 to the cuvette and return it to the fluorimeter). Note that the slight decrease in the rate for the green trace relative to the base rate (black trace) is due to the 10% dilution of the assay required for adding the solution with or without CAH3. (B) The initial rate of actin polymerization in solution assays containing 100 nM CP and 6 µM V-1 (i.e., ∼100 nM CP:V-1 complex) as a function of the amount of CAH3 present in the assay. Initial rates were estimated from a linear fitting of the increase in pyrene fluorescence during the first 40 s of the reaction. The values are expressed as a fraction of the control rate (0 nM CAH3).
To measure the effect of steady-state complex exchange on the rate of actin polymerization, CAH3 was mixed with the CP:V-1 complex in advance. Following a 10-min preincubation, actin polymerization was initiated by adding actin monomer. As in the previous experiment, CAH3 inhibited polymerization in a dose-dependent manner, with ∼100 nM CAH3 sufficient to maximally inhibit polymerization (Fig. 4B).
In terms of how CAH3 is inhibiting polymerization in these assays, we do not think formation of the ternary complex is responsible because this complex cannot associate with the barbed end (16, 17, 20). Therefore, the inhibition must be arising from some combination of (i) free CP and (ii) the weak-capping complex of CP:CAH3 formed by complex exchange. Importantly, the contribution of free CP in our assays should be minimized by the presence of the vast excess of free V-1 available to rapidly sequester it. The prediction, therefore, is that most of the progressive decrease in polymerization rate is due to the progressive increase in the fraction of barbed ends occupied by the weak-capping complex of CP:CAH3.
CP:CAH3 Complex Regulates the Rate of Barbed-End Elongation at the Level of Single Actin Filaments.
To obtain further evidence that the complex of CP:CAH3 regulates polymerization, we monitored barbed-end elongation at the single actin filament level using total internal reflection fluorescence (TIRF) microscopy. The TIRF-based assay used (14) is shown in schematic form in Fig. S7A (see legend for details). In the last step (step 4), 2 µM monomeric actin [containing 20% tetramethylrhodamine-5-maleimide (TMR)-labeled and 0.5% biotin-labeled actin], together with variable concentrations of CP (0–1 µM final) and a constant concentration of CAH3 (10 µM final), were added. Given CAH3’s strong affinity for CP (1–3 nM) (17, 19, 25), this very high concentration of CAH3 should essentially preclude inhibition of actin polymerization by free CP. Fig. 5A shows that the rate at which individual actin filament barbed ends elongated slowed progressively as the concentration of the CP:CAH3 complex in the assay increased from 0 to 200 nM (compare the slopes to the right of the arrow marked “step 4”). Fig. S7B presents still images of representative fields for assays containing 0 or 200 nM CP:CAH3 complex (Movie S1), whereas Fig. 5B shows kymographs for single, representative actin filaments at these two complex concentrations. Plotting the time course of elongation of individual actin filament barbed ends (Fig. 5C, black circles) shows clearly that the elongation rate slowed progressively as the concentration of the CP:CAH3 complex in the assay increased from 0 to 1,000 nM.
Fig. 5.

Direct observation of the regulation of barbed-end elongation by the CP:CAH3 complex using single-filament imaging. (A) Representative time plots of the rate of growth in micrometers per second of individual actin filament barbed ends before the addition of the CP:CAH3 complex (to the left of the arrow marked “step 4” and preceding the plateau in elongation corresponding to the washing step, i.e., step 3), and after the addition of 0 nM (black), 25 nM (red), 100 nM (blue), or 200 nM (green) CP:CAH3 complex (to the right of the arrow marked “step 4”). (B) Kymographs showing the barbed-end elongation of single, representative actin filaments in the presence of 0 nM (Left) or 200 nM (Right) CP:CAH3 complex. (Scale bar, 5 µm.) (C) The fractional decrease in the rate of barbed-end elongation (relative to the rate in the absence of the CP:CAH3 complex) as a function of CP:CAH3 complex concentration. The data from TIRF assays are shown with black circles (Kd3 = 37 ± 5 nM), and the data from pyrene-based bulk solution assays are shown with red circles (Kd3 = 62 ± 11 nM). For TIRF assays, average barbed-end elongation rates were estimated from the linear fitting of 20 actin filaments for each complex concentration. For pyrene assays, each data point is the average of three independent experiments for each complex concentration (error bars represent the SD). We note that the inhibition of polymerization must also be due, to some small extent, to barbed-end capping by the very small amount of free CP that must be present in the assays. Such events should, however, be very rapidly reversed in our TIRF assays by CAH3-driven uncapping, given the very high concentration of CAH3 present in the assays (within less than 10 s; see figure 3 in ref. 14).
Interestingly, we did not detect in the single filament traces any clear evidence of staircase-like changes in length, i.e., actin elongation interrupted repeatedly by pauses in elongation (Fig. 5 A and B). This observation argues that the rate at which the CP:CAH3 complex associates with, and dissociates from, the barbed end is rapid. As a result, the complex slows, in a progressive, dose-dependent manner, the overall rate of barbed-end elongation without creating obvious pauses at the single filament level in barbed-end extension rate. Consistent with this conclusion, the CP:CAH3 complex resides on the barbed end with a half-life of ∼8 s (versus ∼30 min for CP alone) (14). Given the rate of actin filament elongation (∼60 nm/s), and the temporal and spatial resolution in our TIRF assay (we imaged every 2 s with a pixel size of 80 nm), we suspected that staircase-like growth behavior might be just below the level of visual detection. Therefore, we plotted the change in length of individual actin filament barbed ends every 5 s (in bin sizes of 100 nm) at the different concentrations of the CP:CAH3 complex tested (Fig. 6, white bars). If the rate of elongation of individual filament barbed ends is being slowed because the complex is undergoing rapid association with, and dissociation from, the barbed end, then these histograms should be the sum of just two phases—elongation and pausing. Therefore, we performed global fitting of the histograms based on the assumption that they must represent two overlapping Gaussian distributions. The results of this fitting (Fig. 6) show that one Gaussian (red distribution) exhibits a median rate of 0.23 µm/5 s, which matches quite well the expected rate in 2 µM actin (∼0.26 µm/5 s), whereas the second Gaussian (blue distribution) exhibits a median rate of 0.00 µm/5 s, which corresponds to the pause phase. Importantly, the relative contribution of this pause phase increases in progressive fashion as the concentration of the CP:CAH3 complex increases. Specifically, it goes from a very minor component in the absence of the complex to the major component at 150 nM of the complex and eventually to virtually the sole component at 1,000 nM of the complex. We conclude, therefore, that the ability of the CP:CAH3 complex to rapidly associate with, and dissociate from, the barbed end is the primary mechanism responsible for its ability to slow the rate of growth of individual barbed ends.
Fig. 6.

Quantitative analysis of the inhibition in growth rate of individual actin filament barbed ends by the CP:CAH3 complex. The black histograms show for each indicated CP:CAH3 complex concentration (upper right corner in boldface type) the change in length in intervals of 100 nm of individual actin filament barbed ends every 5 s, plotted as the fraction of total 5-s intervals scored (corresponding to the n value in upper right corner). The data at each complex concentration represent the behavior of at least 20 actin filaments. The n values rise as the complex concentration rises because the progressive slowing in growth rate associated with increasing complex concentration extends the time interval for accurately scoring length changes. Global fitting to two Gaussian distributions was performed (note that the peaks of the two distributions were not predefined). The Gaussian in red, which has a median rate of 0.23 µm/5 s, corresponds to the barbed end elongation phase, whereas the Gaussian in blue, which has a median rate of 0.00 µm/5 s, corresponds to the pause phase. The vertical dashed lines in red and blue mark the peaks of these two Gaussians.
By linear fitting of the slopes of barbed-end elongation at various concentrations of CP:CAH3 obtained from the TIRF assays (Fig. 5C, black circles), we estimated an equilibrium dissociation constant (Kd3) of 37 ± 5 nM for the binding of the CP:CAH3 complex to the barbed end (Fig. S1B) (14). This estimate is in relatively good agreement with the estimate that we obtained from fitting the signal for pyrene actin in solution assays containing the same mixtures of CP and CAH3 (Kd3 = 62 ± 11 nM; Fig. 5C, red circles), as well as with previous estimates (12–14, 25). Together, therefore, the data in Figs. 4–6 argue that CAH3-driven complex exchange regulates actin polymerization by converting CP from its sequestered state (CP:V-1) to its weak barbed-end–binding state (CP:CAH3). Importantly, although the CP:CAH3 complex slows the rate of barbed-end elongation relative to free barbed ends, it allows relatively robust elongation compared with barbed ends capped with CP alone.
mCARMIL-1 Is Closely Associated with the Plasma Membrane at Leading Edges.
We next sought to define the spatial features and temporal dynamics of mCARMIL-1’s cortical localization, as this should pinpoint the site and timing of complex exchange. As shown previously (12, 22), GFP-mCARMIL-1 exhibits a bright rim of fluorescence that colocalizes with a bright rim of F-actin staining at the edges of MEFs (Fig. S8 A1–B3). Moreover, line scans of MEFs cotransfected with mGFP-mCARMIL-1 and mCherry as a volume marker (Fig. 7A1) show that mCARMIL-1, but not mCherry, exhibits a peak of fluorescence at cell edges undergoing protrusion (Fig. 7A2) [and this peak is absent at stationary edges (Fig. 7A3)]. This result confirms that mCARMIL-1 is actually concentrated at extending cell edges.
Fig. 7.
mGFP-mCARMIL-1 localizes very close to, if not on, the plasma membrane. (A1) An overlaid image of an NRK cell cotransfected with mGFP-mCARMIL-1 and mCherry as a volume marker. The results of two separate intensity line scans (marked a and b in A1) in the green (mGFP-mCARMIL-1) and red (mCherry) channels are shown using green and red traces, respectively, in A2 (line scan a) and A3 (line scan b) (vertical black lines mark the cell boundary). (A2) Line scan a was at an edge undergoing active protrusion, and (A3) line scan b was at a stationary edge (based on the video sequences). (B1–B7) Overlaid images of representative NRKs cotransfected with mCherry-mCARMIL-1 and GFP-actin (B1), GFP-Cortactin (B2), GFP-Cofilin (B3), GFP-Arp3 (B4), GFP-CPβ (B5), GFP-VASP (B6), or GFP-mCARMIL-1 (B7). A magnification of the boxed region in B6 is shown as separate red, green, and overlaid images at the bottom of that panel. (Scale bars, 5 µm.)
To examine in more detail mCARMIL-1’s relationship with the plasma membrane, we stained different MEFs individually for endogenous mCARMIL-1, vasodilator-stimulated phosphoprotein (VASP), F-actin, CP, or cortactin (Fig. 8 A–E). Line scans performed on these cells (Fig. 8F) show that the averaged fluorescence signals for mCARMIL-1 and VASP are the first to appear as the scan moves into the cell. In addition, the peaks of fluorescence for these two proteins occur closer to the leading-edge plasma membrane than the peaks for actin, CP, and cortactin. Given that VASP is known to associate with the plasma membrane (26–28), whereas CP and (especially) cortactin are known to permeate most of the branched actin network (1, 10, 29), these results argue that mCARMIL-1, like VASP, is associated with the leading-edge plasma membrane.
Fig. 8.
Distribution of endogenous mCARMIL-1 relative to other lamellipodia-resident proteins and the cell boundary. Representative immunofluorescence images of different primary MEFs stained for endogenous mCARMIL-1 (A), VASP (B), CP (D), or cortactin (E) or for F-actin (C) using phalloidin. (F) The distributions of fluorescence intensities for these four proteins and F-actin (color coded as indicated), obtained from intensity line scans beginning outside the cell and crossing into the cell (the cell boundary is indicated by the vertical black line). Each distribution represents the mean of 12–18 intensity line scans obtained from at least three cells. Colored arrowheads mark the peak of each intensity plot. (Scale bars, 5 µm.)
To extend this result, we cotransfected normal rat kidney cells (NRKs) with mCherry-mCARMIL-1 and GFP-tagged versions of actin, cortactin, cofilin, Arp2/3, CP, or VASP (Fig. 7 B1–B6, respectively). The overlaid images show in every case, except VASP, a thin rim of red mCARMIL-1 signal at the very outermost edge of the cell. Importantly, this red rim is not due to an offset in the red and green channels because NRKs cotransfected with mCherry-mCARMIL-1 and mGFP-mCARMIL-1 do not exhibit a red rim (Fig. 7B7). Together, these observations argue that mCARMIL-1, like VASP (26–28), is intimately associated with the plasma membrane at extending lamellipodia.
mCARMIL-1 Localizes to the Plasma Membrane Only at Edges Undergoing Protrusion.
Typically, only portions of the cell edge exhibit enhanced fluorescence for both endogenous and exogenous mCARMIL-1. This heterogeneity could mean that mCARMIL-1’s edge accumulation is tied to the polymerization state at the edge, such that its edge recruitment is prominent only in advancing edges where actin assembly is robust. To begin to address this possibility, we sought ways to promote robust leading-edge advance before imaging mCARMIL-1. Fig. S9 A1–B3 shows that the extent to which endogenous mCARMIL-1 exhibits edge accumulation in NRKs increases dramatically when the cells are allowed to recover from ATP depletion, a method known to promote robust, synchronous polymerization at the cell edge (30). Fig. S9C shows that this recruitment is rapid. Similar results were obtained for endogenous mCARMIL-1 in MTLn3 cells allowed to recover from serum starvation (Fig. S9 D1–E4), another treatment known to promote widespread polymerization at the cell edge (31).
To examine in more dynamic fashion the temporal and spatial correlation between mCARMIL-1’s edge recruitment and the polymerization state at the edge, we used PtK1 cells. Importantly, these cells exhibit rhythmic protrusions and retractions of their leading edge (32) that are evident in kymographs of cells expressing GFP-actin [Fig. 9A1 and the corresponding kymograph in Fig. 9A2; protrusions (P; green boxes), retractions (R; red boxes)] and that correlate with periods of actin polymerization and depolymerization, respectively (Movie S2). In movies of PtK1 cells expressing mGFP-mCARMIL-1 (Fig. 9B1 and Movie S3), mCARMIL-1 is seen to appear and then to disappear from the cell edge over time. Importantly, the three kymographs from this cell shown in Fig. 9 B2–B4 (which correspond to arrows 2–4 in Fig. 9B1) show that the accumulation of mGFP-mCARMIL-1 at the cell edge is tied to the activity state of the edge. Specifically, it is there during periods of edge protrusion (green boxes) and largely disappears during periods of edge retraction (red boxes) (Fig. 9 B2–B4). Moreover, dynamic two-color imaging of PtK1 cells coexpressing mCherry-mCARMIL-1 and GFP-actin show that mCARMIL-1’s leading-edge accumulation is evident only during periods of actin polymerization, as it largely disappears from the edge as soon as the edge stops advancing and actin retrograde flow ceases (Fig. 9 C1–C3 and Movie S4).
Fig. 9.
mGFP-mCARMIL-1 is recruited to the plasma membrane only at cell edges undergoing active protrusion and actin polymerization. (A1) A representative image of a PtK1 cell expressing GFP-actin (Movie S2). The arrow marks the position used to obtain the kymograph in A2. (A2) A kymograph for GFP-actin. The green boxes marked with a “P” in this and subsequent panels denote periods of edge protrusion (seen as positive slopes in the kymograph), and the red boxes marked with an “R” denote periods of edge retraction (seen as negative slopes in the kymograph). (B1) A representative image of a PtK1 cell expressing mGFP-mCARMIL-1 (Movie S3). Arrows 2–4 mark the positions used to obtain the kymographs in B2–B4, respectively. (C1–C3) A representative kymograph from a PtK1 cell coexpressing GFP-actin (C1) and mCherry-mCARMIL-1 (C2) (Movie S4). The overlaid image is shown in C3. (B5–B8) Representative kymographs from PtK1 cells expressing GFP-VASP (B5) (Movie S5), GFP-CPβ (B6 and B7) (Movies S6 and S7), or GFP-cortactin (B8) (Movie S8). The time and distance scales for the kymographs in A2, B2–B8, and C1–C3 are indicated to the right of B8. (Scale bars: A1, 2 µm; B1, 5 µm.)
Finally, comparison of kymographs from cells expressing mGFP-mCARMIL-1 with different cells expressing GFP-VASP, GFP-CPβ, or GFP-cortactin reveals two distinct classes of leading-edge accumulation. The first, exemplified by mGFP-mCARMIL-1 (Fig. 9 B2–B4 and Movie S3) and GFP-VASP (Fig. 9B5 and Movie S5), has the proteins accumulated at the cell edge during protrusion and then rapidly diminished or lost from the edge during subsequent retraction. The second, exemplified by GFP-CPβ (Fig. 9 B6 and B7 and Movies S6 and S7) and GFP-cortactin (Fig. 9B8 and Movie S8), has the proteins accumulated at the cell edge during both the protrusion and the retraction phases, with only minimal loss during the later phase. These results argue that the leading-edge accumulation of both mCARMIL-1 and VASP is driven primarily by their association with the plasma membrane rather than with F-actin, as this latter interaction would cause them to be retained at retracting edges, as seen for stable components of the lamellipodial actin network like CP and (especially) cortactin (1, 10, 29). Moreover, these results suggest that the recruitment of mCARMIL-1 to the plasma membrane specifically at sites of edge protrusion is driven by the same signaling molecules (e.g., active Rho GTPases and phosphoinositides) that recruit de novo nucleators of actin assembly there (33). Similarly, VASP may be recruited to extending edges (at least in part) via its interaction with lamellipodin, whose PH domain binds phosphoinositides in the plasma membrane (28).
Discussion
Complex Exchange.
We presented three biochemical findings regarding the CAH3-driven exchange of CP from its sequestered state (the CP:V-1 complex) to its weak barbed-end–binding state (the CP:CAH3 complex). First, we confirmed and extended the elegant work of Takeda et al. (17) by providing physical evidence for the existence of the ternary complex formed by CP, CAH3, and V-1. Second, we determined the relative contributions to complex exchange of ternary complex formation/allostery versus competitive binding at different concentrations of CAH3. Finally, using both bulk solution assays and single-filament imaging, we showed that complex exchange regulates actin filament assembly in vitro. Specifically, we demonstrated that the CP:CAH3 complex created by complex exchange slows in a dose-dependent manner the rate of barbed-end elongation. Kinetic analyses, together with previous work (12–14), all argue that this effect is due to the rapid association with, and dissociation from, the barbed end of the CP:CAH3 complex, whose affinity for the barbed end is ∼50 nM, compared with ∼0.1 nM for CP alone.
To complete the CP regulatory cycle in vivo, V-1 must retrieve CP from CARMIL to replenish the pool of sequestered CP. Consistent with the fact that CAH3 binds CP much more tightly than does V-1, and that CAH3 inhibits in allosteric fashion the binding of V-1 to the CP:CAH3 complex, V-1 is unable on its own to effectively retrieve CP from CAH3 in vitro. Clearly, V-1 requires an additional mechanism and/or cellular factor to effectively retrieve CP from CARMIL in vivo. One distinct possibility is that full-length CARMIL undergoes a conformational change following its dissociation from the plasma membrane in vivo that results in a large reduction in its affinity for CP.
Interestingly, the CP regulatory cycle described here has many parallels with a regulatory cycle described previously for G-actin (34–36). In this latter cycle, profilin drives the conversion of the ADP-G-actin:thymosin β-4 complex, where the actin is sequestered in a nonpolymerizable state into the ADP-G-actin:profilin complex. Once the actin monomer is on profilin, the ADP is rapidly exchanged for ATP, and the ATP-G-actin:profilin complex is efficiently funneled onto the growing barbed end. This profilin-driven conversion of sequestered G-actin (ADP-G-actin:thymosin β-4) into active G-actin (ATP-G-actin:profilin) is similar, therefore, to the CARMIL-driven conversion of sequestered CP (CP:V-1) into weakly active CP (CP:CARMIL). Moreover, both of these exchange reactions are driven by a combination of competitive binding and an allosteric effect following ternary complex formation (34–36).
CARMIL Localization.
Knowing where and when CARMIL is active in the cell is crucial for gauging the possible physiological significance of the CP regulatory cycle. Our imaging of mCARMIL-1 yielded two insights regarding the spatial and temporal aspects of mCARMIL-1’s leading-edge localization. First, within the limitations of light microscopy, our data argue that mCARMIL-1 associates with the plasma membrane at the leading edge. Most telling, the peak of fluorescence for mCARMIL-1 mirrored that of VASP, which is known to be recruited to the plasma membrane (26–28), and was closer to the membrane than the peak for cortactin, which permeates most of the lamellipodial actin network (10, 29). Second, we showed that mCARMIL-1 localizes only to the edges of lamellipodia that are protruding, i.e., that are undergoing active polymerization. Once robust nucleation and assembly cease and the edge begins to retreat, mCARMIL-1 largely disappears from the edge. Importantly, VASP exhibited identical behavior, consistent with a previous study (26), whereas stable components of the lamellipodial actin network like cortactin did not disappear during edge retraction. These observations argue strongly that CARMIL and VASP are recruited to the plasma membrane and not simply to cortical F-actin and that their membrane recruitment is restricted to regions of the plasma membrane undergoing active actin assembly. This spatial and temporal pattern of plasma membrane recruitment should serve to focus the functions of CARMIL and VASP within a narrow zone at advancing edges. Of particular note, a recent report describing an unconventional PH domain in mCARMIL-1 that promotes mCARMIL-1’s association with the plasma membrane (37) further corroborates our conclusions.
In terms of CARMIL regulation, we think it is very likely that cytoplasmic CARMIL is inactive as a consequence of being auto-inhibited through intramolecular folding and that the protein is simultaneously recruited to the plasma membrane, unfolded, and activated by the same key signaling molecules (e.g., phosphoinositides and active Rho GTPases) that recruit, unfold, and activate key players in actin assembly, such as formins and the Arp2/3 complex activator Wasp (2, 3, 33, 38). Consistent with this hypothesis, mCARMIL-1 contains an N-terminal polybasic sequence that might bind phosphoinositides in the plasma membrane (Fig. S1A). Moreover, mCARMIL-1 contains two CRIB-like sequences that in other proteins mediate interactions with active, membrane-bound Rho GTPases (Fig. S1A). Importantly, the CP regulatory activities of full-length Acanthamoeba CARMIL are greatly attenuated relative to its isolated CAH3 domain (13). This fact, together with proteolysis data in the same study, provide significant support for the idea that full-length CARMIL is negatively regulated by intramolecular folding (although see ref. 12).
Model for CP Regulation.
Our working model for the regulation of CP by CARMIL and V-1 in vivo is based on four key observations and one assumption. First, the cellular concentrations of CP and V-1, together with their affinity for each other, argue that vast majority of cellular CP is probably sequestered by V-1 at steady state. Second, this large pool of sequestered CP is freely diffusing in the cell. Third, although CP can dissociate from V-1 spontaneously, CARMIL accelerates this rate by >70-fold at saturation, in the process creating the weak-capping complex of CP: CARMIL. Fourth, CARMIL is recruited to the plasma membrane, and only at edges undergoing protrusion. Assuming CARMIL is active only when recruited to the plasma membrane, we can make a series of linked predictions that culminate in the overall model. First, CARMIL-driven complex exchange should occur exclusively at protruding edges. Second, because complex exchange converts inactive CP into active CP, complex exchange should increase the rate of barbed-end capping at protruding edges. Finally, because higher rates of barbed-end capping promote Arp2/3-dependent actin network assembly and actin-based motility in vitro and in vivo (6–10), CARMIL-driven complex exchange should promote Arp2/3-dependent actin network assembly at protruding edges. Consistently, actin network assembly and network-based cell motility are inhibited in cells where CARMIL function has been attenuated (11, 12, 22, 23). Finally, we note that our model incorporates two apparent paradoxes. The first—that CP halts assembly at the barbed end in vitro but is required for Arp2/3-dependent actin network assembly in vitro and in vivo—was explained previously (8). The second—that CARMIL reduces CP’s ability to halt assembly at the barbed end in vitro but, in collaboration with V-1, should promote Arp2/3-dependent actin network formation in vivo—is explained by this study.
In the absence of CP regulation, barbed ends at protruding edges would be capped by CP alone, which has a half-life on the end of ∼30 min (39). CARMIL-driven complex exchange, on the other hand, will populate barbed ends at protruding edges with a cap that has a half-life of ∼8 s (14). Therefore, a second functional prediction from our work is that CARMIL-driven complex exchange should promote actin assembly at protruding edges relative to unregulated CP by creating a cap that dissociates fairly rapidly, allowing relatively rapid resumption of assembly. Although complex exchange in principle is not required for weak-cap formation because CARMIL can bind directly to free CP, it should be very important in vivo because most of the cell’s CP is probably sequestered by V-1. Moreover, the V-1–dependent sequestration of CP and subsequent CARMIL-driven complex exchange reaction that liberates this sequestered CP will greatly magnify the net effect of the weak-capping complex at the plasma membrane:cytoplasm interface by reducing the competition from free CP for available barbed ends. Stated another way, CARMIL-driven complex exchange at protruding edges should increase the rate of barbed-end capping, but decrease the potency of the cap created (relative to unregulated CP). This latter point is consistent with measurements of CP’s half-life on barbed ends near the plasma membrane in vivo. Specifically, the values obtained [∼2 s (40) and ∼15 s (10)] are much closer to the half-life of the CP:CAH3 complex on the barbed end in vitro (∼8 s) than to the half-life of CP in vitro (∼30 min).
Importantly, CARMIL-driven complex exchange should act synergistically with other components to promote actin network assembly at protruding edges. Specifically, branched actin network assembly at protruding edges is initiated by the recruitment and activation of nucleation-promoting factors to these edges, which then strongly activate Arp2/3-dependent branched nucleation at the plasma membrane:cytoplasm interface (1, 2, 38). Actin assembly at protruding edges should be further promoted by the dramatic recruitment of VASP to these edges because VASP acts as a processive polymerase and protects barbed ends from CP (41). Finally, actin network assembly at protruding edges should also be promoted by CARMIL-driven complex exchange via the two mechanisms discussed above.
Finally, we note that important pieces of the puzzle are still missing. For example, how is the connection between CARMIL and CP broken to complete the regulatory cycle? Do membrane binding and release affect the biochemical properties of full-length CARMIL? To what degree does the spontaneous dissociation of CP from V-1 contribute to barbed-end capping? To what degree does CARMIL’s ability to uncap CP-capped ends contribute to actin dynamics in vivo? These are some of the important questions to address in future work.
Materials and Methods
Proteins, Antibodies, and Plasmids.
Rabbit skeletal muscle actin was purified and labeled as described previously (14). GST-tagged CAH3 and untagged mouse V-1 were prepared as in refs. 14 and 20, respectively. To generate IAEDANS-(C2)-V-1, cysteines C45 and C83 were changed to serines, and the remaining cysteine (C2) was labeled with IAEDANS as described in ref. 20. Recombinant, untagged mouse CP α1/β2 and GST-V-1 were expressed in Escherichia coli and purified as in refs. 14 and 20, respectively. The following primary antibodies were used: rabbit anti-mCARMIL1 (1:300; raised against the C-terminal 14 residues of mCARMIL-1), rabbit anti-CPβ (1:1,000; same peptide antigen as for antibody R26 in ref. 42), rabbit anti-p34 (1:200; Millipore, 07–227), rabbit anti-VASP (1:500; gift of F. Gertler, Massachusetts Institute of Technology, Cambridge, MA), mouse anti-cortactin (1:1,000; Millipore, 05–180), and mouse anti-mV-1 (1:50; BD Biosciences, 611830). Secondary antibodies were species-specific and conjugated with Alexa Fluor 595 (1:1,000, Invitrogen, A11037). F-actin was stained using Alexa Fluor 488-labeled phalloidin (1:400; Invitrogen, A12379). GFP-VASP, GFP-Arp3, GFP-cofilin, GFP-cortactin, GFP-CPβ, and GFP-actin were kind gifts of F. Gertler, M. Welch (Department of Molecular and Cell Biology, University of California, Berkeley, CA), J. Condeelis (Albert Einstein College of Medicine, Yeshiva University, Bronx, NY), T. Uruno (Department of Immunobiology and Neuroscience, Kyushu University, Fukuoka, Japan), D. Schaefer (Department of Biology, University of Virginia, Charlottesville, VA), and C. Ballestrem (Faculty of Life Sciences, University of Manchester, Manchester, UK), respectively. The mGFP- and mCherry-tagged versions of full-length mCARMIL-1 were generated by restriction enzyme-assisted fusion of two mouse CARMIL-1 EST clones (AI852804 and 4014128).
Analytical Methods.
Pull-down assays were calibrated and performed as described in ref. 13. Fluorescence anisotropy experiments using IAEDANS-(C2)-V-1 were performed as described (20) except that a Fluorolog-3 spectrophotofluorometer (SPEX) connected to a stopped-flow minimixer (KinTek) was used for experiments containing >125 nM CAH3. The fraction of CP that remained bound to IAEDANS-(C2)-V-1 over time was estimated by dividing the observed value by the maximum anisotropy value at time 0. The binding of V-1 to the complex of CP:mCAH3ab was estimated from the change in fluorescence intensity of IAEDANS-(M7C)-V-1, as described (20). Sedimentation velocity experiments were conducted using a Beckman Optima XL-I analytical ultracentrifuge (Beckman) equipped with a four-hole An60Ti rotor and cells with 12-mm double-sector Epon centerpieces and sapphire windows. Proteins were dialyzed overnight in the KMEI buffer. For all experiments, 0.4 mL of the protein sample and dialyzate buffer were loaded into sample and reference centerpiece channels, respectively. The rotor was accelerated to 50,000 × g after thermal equilibrium (20 °C) was reached. Absorbance scans (280 nm) were started immediately after the rotor reached the set speed and collected until no further sedimentation boundary movement was observed. Density and viscosity of the buffer were determined with the DMA-58 densitometer and AMVn viscometer (Anton-PAAR KG). Data were analyzed in terms of continuous c(s) distributions using the SEDFIT program.
Actin Polymerization Assays.
Solution-based actin polymerization assays using pyrene-labeled actin were performed as described previously (13, 14), except that actin seeds were used at a final concentration of 0.8 µM (∼0.8 nM barbed ends). TIRF-based actin polymerization assays were performed as described (14), except that filaments were attached by coating the glass with streptavidin and using filaments assembled from a mixture of G-actin, TMR-labeled actin, and biotinylated actin at molar ratios of 79.5:20:0.5%, respectively.
Cell Culture, Transfection, Immunofluorescence, and Imaging.
MEFs, NRKs (ATCC, CRL-6509), and PtK1s (ATCC, CRL-6493) were cultured in DMEM (Invitrogen, 11965118) supplemented with 10% (vol/vol) FBS (Invitrogen, 10082147) at 37 °C in 5% CO2. MTLn3 cells stably transfected with GFP-actin (gift of J. Condeelis, Albert Einstein College of Medicine, Yeshiva University, Bronx, NY) were cultured as in ref. 31. NRKs were transfected using Lipofectamine 2000 (Invitrogen, 11668027), and MEFs and PtK1s were transfected by Amaxa nucleofection (Lonza, AAB-1001). For live-cell imaging, cells were plated on chambered coverglass (Lab-TEK, 155382) coated with fibronectin (10 μg/mL; Invitrogen, 33016015). MEFs and NRKs were imaged using a 63× (1.4 N.A.) objective on a Zeiss LSM 510 confocal microscope equipped with a 37 °C/5% CO2 environmental chamber. PtK1s were imaged using a 150× (1.45 N.A.) objective on an Olympus IX81 microscope fitted with a Yokogawa CSU-X1 spinning disk confocal unit and a Photometrics QuantEM 512SC camera. For dynamic imaging, temperature control was achieved using a Nevtek stage heater. For kymograph analyses, a line (3-pixels width) was drawn from the cell center across the cell edge using MetaMorph software. ATP depletion/recovery experiments and EGF treatments were performed as described in refs. 30 and 31, respectively. Immunofluorescence staining was performed as described in ref. 43.
Supplementary Material
Acknowledgments
We thank Edward D. Korn for his comments on the manuscript; members of the J.A.H. laboratory (especially Melanie Barzik) for their support; and Xufeng Wu and the National Heart, Lung, and Blood Institute Light Microcopy Imaging Core for assistance. We acknowledge that the original idea for the CP regulatory cycle described in this work came from a former member of the J.A.H. laboratory, Takehito Uruno. Finally, we thank one anonymous reviewer whose comments greatly improved our manuscript.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313738111/-/DCSupplemental.
References
- 1.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 2.Chhabra ES, Higgs HN. The many faces of actin: Matching assembly factors with cellular structures. Nat Cell Biol. 2007;9(10):1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- 3.Campellone KG, Welch MD. A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11(4):237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper JA, Sept D. New insights into mechanism and regulation of actin capping protein. Int Rev Cell Mol Biol. 2008;267:183–206. doi: 10.1016/S1937-6448(08)00604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401(6753):613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 6.Hug C, et al. Capping protein levels influence actin assembly and cell motility in dictyostelium. Cell. 1995;81(4):591–600. doi: 10.1016/0092-8674(95)90080-2. [DOI] [PubMed] [Google Scholar]
- 7.Serio AW, Jeng RL, Haglund CM, Reed SC, Welch MD. Defining a core set of actin cytoskeletal proteins critical for actin-based motility of Rickettsia. Cell Host Microbe. 2010;7(5):388–398. doi: 10.1016/j.chom.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133(5):841–851. doi: 10.1016/j.cell.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mejillano MR, et al. Lamellipodial versus filopodial mode of the actin nanomachinery: Pivotal role of the filament barbed end. Cell. 2004;118(3):363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol. 2007;17(5):395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung G, Remmert K, Wu X, Volosky JM, Hammer JA., III The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol. 2001;153(7):1479–1497. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang C, et al. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev Cell. 2005;9(2):209–221. doi: 10.1016/j.devcel.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uruno T, Remmert K, Hammer JA., III CARMIL is a potent capping protein antagonist: Identification of a conserved CARMIL domain that inhibits the activity of capping protein and uncaps capped actin filaments. J Biol Chem. 2006;281(15):10635–10650. doi: 10.1074/jbc.M513186200. [DOI] [PubMed] [Google Scholar]
- 14.Fujiwara I, Remmert K, Hammer JA., III Direct observation of the uncapping of capping protein-capped actin filaments by CARMIL homology domain 3. J Biol Chem. 2010;285(4):2707–2720. doi: 10.1074/jbc.M109.031203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taoka M, et al. V-1, a protein expressed transiently during murine cerebellar development, regulates actin polymerization via interaction with capping protein. J Biol Chem. 2003;278(8):5864–5870. doi: 10.1074/jbc.M211509200. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharya N, Ghosh S, Sept D, Cooper JA. Binding of myotrophin/V-1 to actin-capping protein: Implications for how capping protein binds to the filament barbed end. J Biol Chem. 2006;281(41):31021–31030. doi: 10.1074/jbc.M606278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeda S, et al. Two distinct mechanisms for actin capping protein regulation: Steric and allosteric inhibition. PLoS Biol. 2010;8(7):e1000416. doi: 10.1371/journal.pbio.1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukman S, Robinson RC, Wales D, Verma CS. Conformational dynamics of capping protein and interaction partners: simulation studies. Proteins. 2012;80(4):1066–1077. doi: 10.1002/prot.24008. [DOI] [PubMed] [Google Scholar]
- 19.Zwolak A, Uruno T, Piszczek G, Hammer JA, III, Tjandra N. Molecular basis for barbed end uncapping by CARMIL homology domain 3 of mouse CARMIL-1. J Biol Chem. 2010;285(37):29014–29026. doi: 10.1074/jbc.M110.134221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zwolak A, Fujiwara I, Hammer JA, III, Tjandra N. Structural basis for capping protein sequestration by myotrophin (V-1) J Biol Chem. 2010;285(33):25767–25781. doi: 10.1074/jbc.M110.135848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez-Valladares M, et al. Structural characterization of a capping protein interaction motif defines a family of actin filament regulators. Nat Struct Mol Biol. 2010;17(4):497–503. doi: 10.1038/nsmb.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Y, Niederstrasser H, Edwards M, Jackson CE, Cooper JA. Distinct roles for CARMIL isoforms in cell migration. Mol Biol Cell. 2009;20(24):5290–5305. doi: 10.1091/mbc.E08-10-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards M, Liang Y, Kim T, Cooper JA. Physiological role of the interaction between CARMIL1 and capping protein. Mol Biol Cell. 2013;24(19):3047–3055. doi: 10.1091/mbc.E13-05-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, et al. Clathrin exchange during clathrin-mediated endocytosis. J Cell Biol. 2001;155(2):291–300. doi: 10.1083/jcb.200104085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim T, Ravilious GE, Sept D, Cooper JA. Mechanism for CARMIL protein inhibition of heterodimeric actin-capping protein. J Biol Chem. 2012;287(19):15251–15262. doi: 10.1074/jbc.M112.345447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rottner K, Behrendt B, Small JV, Wehland J. VASP dynamics during lamellipodia protrusion. Nat Cell Biol. 1999;1(5):321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]
- 27.Bear JE, Gertler FB. Ena/VASP: Towards resolving a pointed controversy at the barbed end. J Cell Sci. 2009;122(Pt 12):1947–1953. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krause M, et al. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell. 2004;7(4):571–583. doi: 10.1016/j.devcel.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 29.Weaver AM, et al. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol. 2001;11(5):370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- 30.Svitkina TM, Neyfakh AA, Jr, Bershadsky AD. Actin cytoskeleton of spread fibroblasts appears to assemble at the cell edges. J Cell Sci. 1986;82:235–248. doi: 10.1242/jcs.82.1.235. [DOI] [PubMed] [Google Scholar]
- 31.Chan AY, Bailly M, Zebda N, Segall JE, Condeelis JS. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J Cell Biol. 2000;148(3):531–542. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burnette DT, et al. A role for actin arcs in the leading-edge advance of migrating cells. Nat Cell Biol. 2011;13(4):371–381. doi: 10.1038/ncb2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev. 2010;90(1):259–289. doi: 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- 34.Goldschmidt-Clermont PJ, et al. The control of actin nucleotide exchange by thymosin beta 4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Mol Biol Cell. 1992;3(9):1015–1024. doi: 10.1091/mbc.3.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pantaloni D, Carlier MF. How profilin promotes actin filament assembly in the presence of thymosin beta 4. Cell. 1993;75(5):1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- 36.Yarmola EG, Parikh S, Bubb MR. Formation and implications of a ternary complex of profilin, thymosin beta 4, and actin. J Biol Chem. 2001;276(49):45555–45563. doi: 10.1074/jbc.M105723200. [DOI] [PubMed] [Google Scholar]
- 37.Zwolak A, et al. CARMIL leading edge localization depends on a non-canonical PH domain and dimerization. Nat Commun. 2013;4:2523. doi: 10.1038/ncomms3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chesarone MA, Goode BL. Actin nucleation and elongation factors: Mechanisms and interplay. Curr Opin Cell Biol. 2009;21(1):28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhn JR, Pollard TD. Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J Biol Chem. 2007;282(38):28014–28024. doi: 10.1074/jbc.M705287200. [DOI] [PubMed] [Google Scholar]
- 40.Miyoshi T, et al. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: Evidence of frequent filament severing. J Cell Biol. 2006;175(6):947–955. doi: 10.1083/jcb.200604176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol. 2010;191(3):571–584. doi: 10.1083/jcb.201003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schafer DA, Korshunova YO, Schroer TA, Cooper JA. Differential localization and sequence analysis of capping protein beta-subunit isoforms of vertebrates. J Cell Biol. 1994;127(2):453–465. doi: 10.1083/jcb.127.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu XS, et al. Identification of an organelle receptor for myosin-Va. Nat Cell Biol. 2002;4(4):271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]