Significance
Staphylococcus aureus is a ubiquitous pathogen that causes disease in a variety of tissues. Our studies provide insight into how this pathogen uses the SaeS sensor kinase to recognize innate immunity signals and induce a transcriptional response tailored for its environment. Our results demonstrate that the specificity of these responses is determined by individual amino acids predicted to be in an extracellular domain. These amino acids include aromatic anchors and a methionine residue essential for activation of target gene transcription and virulence. Our findings provide a putative mechanism for the ability of bacterial sensory systems to integrate and diversify the responses to host stimuli. They underscore the exquisite nature of bacterial signaling and the complexity of host-pathogen interactions.
Keywords: host-pathogen interactions, membrane proteins
Abstract
Two-component systems (TCSs) are highly conserved across bacteria and are used to rapidly sense and respond to changing environmental conditions. The human pathogen Staphylococcus aureus uses the S. aureus exoprotein expression (sae) TCS to sense host signals and activate transcription of virulence factors essential to pathogenesis. Despite its importance, the mechanism by which the histidine kinase SaeS recognizes specific host stimuli is unknown. After mutagenizing the predicted extracellular loop of SaeS, we discovered one methionine residue (M31) was essential for the ability of S. aureus to transcribe sae target genes, including hla, lukAB/lukGH, and hlgA. This single M31A mutation also significantly reduced cytotoxicity in human neutrophils to levels observed in cells following interaction with ΔsaeS. Another important discovery was that mutation of two aromatic anchor residues (W32A and F33A) disrupted the normal basal signaling of SaeS in the absence of inducing signals, yet both mutant kinases had appropriate activation of effector genes following exposure to neutrophils. Although the transcriptional profile of aromatic mutation W32A was consistent with that of WT in response to human α-defensin 1, mutant kinase F33A did not properly transcribe the γ-toxin genes in response to this stimulus. Taken together, our results provide molecular evidence for how SaeS recognizes host signals and triggers activation of select virulence factors to facilitate evasion of innate immunity. These findings have important implications for signal transduction in prokaryotes and eukaryotes due to conservation of aromatic anchor residues across both of these domains and the important role they play in sensor protein structure and function.
The Gram-positive bacterial pathogen, Staphylococcus aureus is capable of causing a remarkable range of acute and chronic infections in almost every tissue of the human body (1). The highly adaptable nature of S. aureus, including the rapid development of antibiotic resistance and large tissue tropism, has resulted in it being one of the leading causes of infection in the United States (2). This versatility is made possible by the large array of virulence factors encoded in the S. aureus genome and on mobile genetic elements that are regulated by a complex, interactive network of regulatory systems including two-component signal transduction systems (TCSs) (3–5).
Following its discovery almost 20 y ago, the sae TCS was recognized as a major regulator of secreted toxins and exoenzymes and was named the S. aureus exoprotein expression (sae) system (6–9). More recently, the system has been shown to be essential for virulence in multiple animal models of infection and to play a critical role in evasion of the innate immune response (10–12). SaeR, the response regulator, and SaeS, the sensor kinase, comprise the traditional machinery of the TCS (13). SaeR is a member of the OmpR family of response regulators and binds a direct DNA repeat sequence upon phosphorylation by SaeS to activate transcription of genes in the sae regulon (11, 14). SaeS, a member of the HisKA family of histidine protein kinases, is predicted to contain a sensing domain composed of only two transmembrane segments connected by a short extracellular loop (15). Work on the antimicrobial peptide sensing TCS ApsXRS in Staphylococcus epidermidis suggests the nine amino acid extracellular (EC) loop of the sensor kinase ApsS may be sufficient to detect the presence of specific host antimicrobial peptides. Antisera raised against the extracellular loop rendered the bacteria unable to transcribe ApsR target genes in response to human β-defensin 3, demonstrating the importance of the sensor kinase residues exposed to the external environment regardless of the size of the extracellular domain (16). The topology of ApsS and SaeS is similar, resulting in both being classified as intramembrane-sensing kinases. This class of histidine kinases has traditionally been thought only capable of sensing perturbations to the plasma membrane or receiving signals through auxiliary proteins (17). Indeed, the sae system encodes two additional genes, saeP and saeQ, immediately upstream of saeRS and autoinduces their transcription on activation of the sae system (13). SaeP and SaeQ, a lipoprotein and integral membrane protein, respectively, have recently been shown to form a protein complex with SaeS, which would seem to support the classification of SaeS as an intramembrane-sensing kinase requiring the presence of the sae auxiliary proteins to sense extracellular signals (18). However, formation of the protein complex results in repression of the sae system and not activation. In addition, saeP is not required for the sae system to activate (15). Therefore, how SaeS senses host signals known to activate the sae system is unclear.
In this study, we investigate the mechanism by which the histidine kinase SaeS is able to specifically detect α-defensin 1 (HNP-1) and human polymorphonuclear leukocytes (PMNs). We mutagenized every residue in the predicted extracellular loop of SaeS and evaluated activation of the mutant kinases in response to each signal by analyzing expression of sae target genes. In addition, we evaluated PMN survival when exposed to S. aureus USA300 strain LAC expressing each mutant kinase. Our results demonstrate specific residues in the predicted extracellular loop of SaeS are essential to downstream activation of SaeR-regulated genes. Moreover, this study identifies the importance of individual residues for sensing specific host stimuli by demonstrating differential activation of sae targets by the mutant kinases in response to the different stimuli.
Results
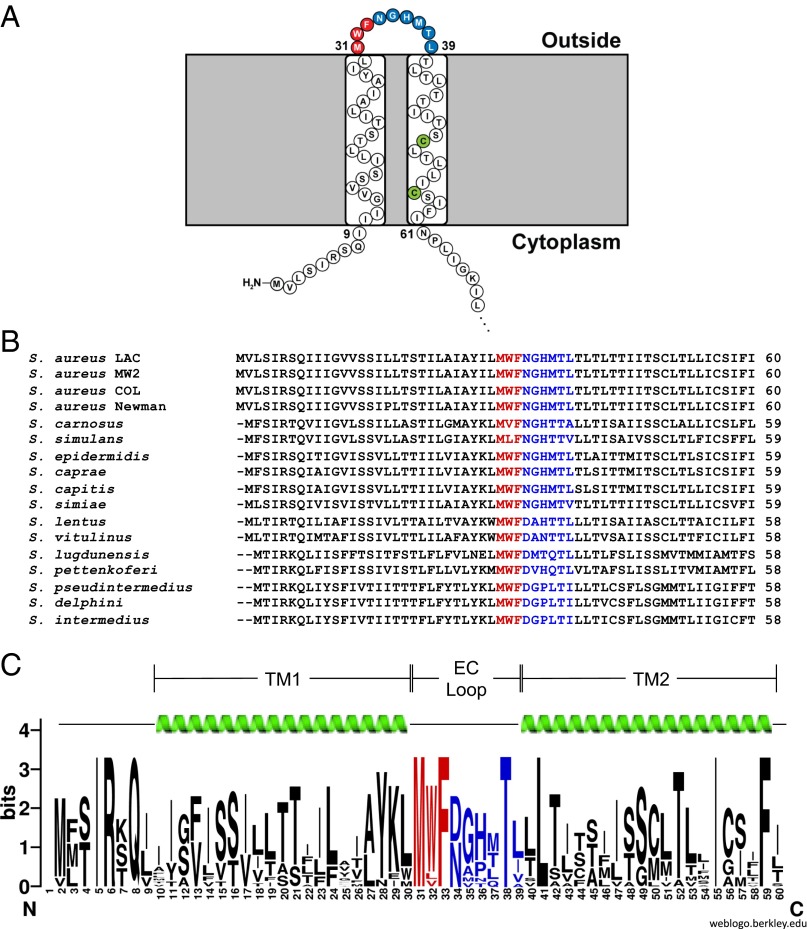
Determining the Topology of SaeS.
The exact topology of SaeS in the cytoplasmic membrane has not been reported. To address this, we used the substituted cysteine accessibility method (SCAM) (19), which is a technique that allows membrane protein topology to be determined with single amino acid resolution and has recently been adapted for use in S. aureus (20). SCAM requires protein constructs that contain only one cysteine residue, which is labeled with a maleimide ring conjugated to biotin (MPB). By engineering individual cysteines throughout a protein, the topology can be determined based on the accessibility of each cysteine residue to labeling. SaeS contains three native cysteines, which needed to be removed to perform SCAM topology analysis. Residues C50 and C56 are near the N terminus of SaeS (depicted green in Fig. 1A), whereas C226 is near the C terminus of the 351 amino acid protein (Fig. S1B). The WT saeS gene from USA300 strain LAC (21) was cloned into pEPSA5 (22) with a C-terminal T7-tag for detection by Western blot. Site-directed mutagenesis was used to mutate each cysteine residue to a serine to cause minimal structural perturbation, eventually producing constructs with each native cysteine present as the only cysteine in the protein (Table S1). These constructs were used for preliminary SCAM studies in a LAC ΔsaePQRS strain to ensure SaeP, SaeQ, and chromosomally encoded SaeS did not interfere with labeling (SI Materials and Methods). All mutant SaeS constructs expressed at levels similar to WT (Fig. S1A, anti-T7). The inability of C50 and C56 to react with the MPB labeling reagent indicates these residues are likely embedded in the cytoplasmic membrane (Fig. S1A, Strep-HRP), whereas the labeling pattern of residue C226 is consistent with a cytoplasm location (Fig. S1). Interestingly, when either transmembrane cysteine was mutated to a serine either alone or together, the resulting protein ran at a slightly higher molecular weight than WT, as determined by immunoblotting (Fig. S1A). Denaturing and reducing agents did not alter the mobility of SaeS in the gel. Because the altered migration rate indicates a structural change we are unable to fully explain, these constructs were not used for further SCAM analysis.
Fig. 1.
Residues in the EC loop of SaeS are conserved across staphylococcal strains and species. (A) Cartoon representation of the predicted topology of LAC’s SaeS sensing domain from TOPCONS. (B) A sequence alignment of the sensing domain of SaeS from all staphylococcal species and strains of S. aureus identified as having sae systems shows a high level of conservation across the entire domain. SaeS sequences from species other than S. aureus were identified by comparison with the S. aureus LAC sequence using BLAST. The first three predicted extracellular residues (depicted in red) are completely conserved with the exception of W32 in S. carnosus and S. simulans. Strain sequences used for each species are as follows: S. carnosus, TM300; S. simulans, ACS-120-V-Sch1; S. epidermidis, M23864:W1; S. caprae, C87; S. capitis, SK14; S. simiae, CCM 7213; S. lugdunensis, M23590; S. pettenkoferi, VCU012; S. pseudintermedius, HKU10-03. Other species not listed did not have strains designated. (C) An SaeS Weblogo constructed from the sequences listed in A. For S. aureus, only the LAC sequence was used. TM1, transmembrane domain 1; TM2, transmembrane domain 2.
As an alternative method to obtain the topology for SaeS and validate our SCAM assignments, we used the SaeS amino acid sequence from LAC for TOPCONS analysis, a consensus topology prediction program (23). A small sensing domain composed of two transmembrane passes connected by a nine amino acid extracellular loop was predicted (depicted in red and blue in Fig. 1A). Next, alignment and Weblogo (24) analysis of the SaeS sensing domain from all 14 staphylococcal species currently identified as having the saeS gene demonstrated that residues across the entire SaeS sequence are conserved, particularly in the predicted EC loop (Fig. 1 B and C).
Mutagenesis of the Predicted Extracellular Loop of SaeS Identified Residues Important for Kinase Activation.
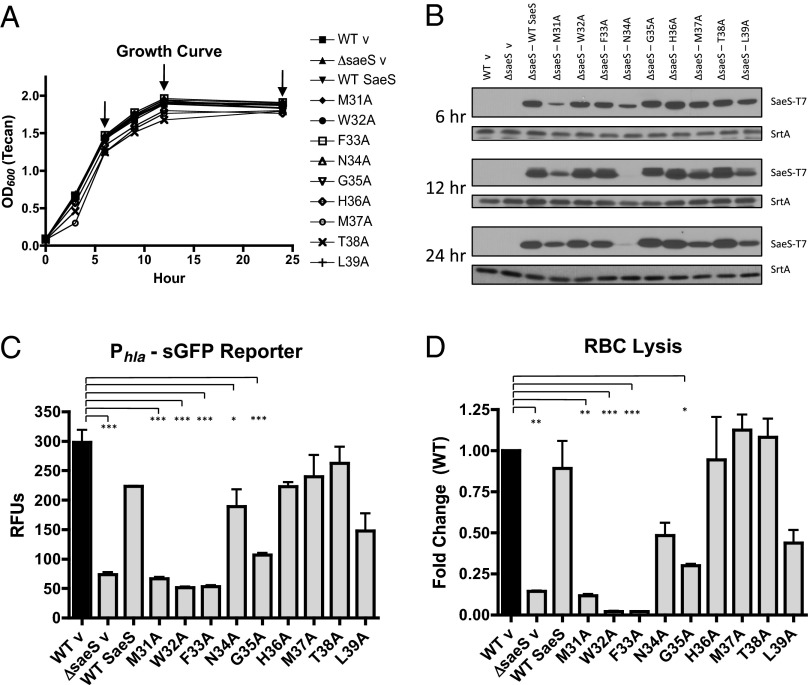
To determine the biological relevance of conserved residues in the sensing domain, we performed an alanine scan of the extracellular loop of SaeS and evaluated activation of the sae system by the mutant kinases. To this end, we first constructed a markerless ΔsaeS mutant. This strain grew as well as WT LAC but was unable to produce any Phla-dependent fluorescence or red blood cell (RBC) lysis after 24 h of growth (Fig. 2 and Fig. S2). This observation is consistent with previous work showing sae regulates transcription of hla (11, 25). Both WT LAC and ΔsaeS containing the hla reporter were transformed with the empty complementation vector pEPSA5 (WT v and ΔsaeS v, respectively), and complementation of the ΔsaeS mutant with WT SaeS restored both Phla fluorescence and RBC lysis (Fig. 2 C and D). Using the SaeS complementing plasmid as a template, each of the nine extracellular residues were mutated to alanines and transformed into the ΔsaeS strain containing the integrated Phla-sGFP reporter (SI Materials and Methods and Table S1). These strains were evaluated for growth, expression of the mutant kinases, fluorescence from the hla reporter, and RBC lysis activity (Fig. 2). None of the mutations in the EC loop caused any changes in SaeS SDS/PAGE migration like those observed for the transmembrane cysteine mutants (Fig. 2B and Fig. S1A). All mutant protein constructs were expressed; however, N34A demonstrated reduced protein expression at later time points (Fig. 2B). This result may explain the slight decrease observed in RBC lysis for this kinase at 24 h (Fig. 2D).
Fig. 2.
In vitro analys is of SaeS EC loop mutations. (A) Growth of LAC and ΔsaeS expressing WT and EC loop mutant copies of SaeS. (B) T7 immunoblot of cell pellets from strains depicted in A to evaluate kinase expression levels during in vitro growth at 6, 12, and 24 h (indicated by arrows). Sortase A was used as a loading control. (C) Fluorescence from a Phla-sGFP reporter at 24 h, expressed as relative fluorescence units (RFUs, fluorescence divided by OD600). (D) Lysis of RBCs following incubation with bacterial supernatants harvested after 24 h. Error bars are SEM of two biological replicates examined in triplicate. Asterisks denote significance as evaluated by a one-way ANOVA with Tukey’s posttest based on P values as follows: *0.01–0.05, **0.001–0.01, and ***<0.001.
The most striking observation was the complete inability of single mutations M31A, W32A, and F33A (depicted red in Fig. 1) to complement the saeS deletion (Fig. 2 C and D). These constructs did not cause any growth defects (Fig. 2A), expression of saeR was not disrupted, and saeS was transcribed in a manner similar to the other plasmid based sensor kinases (Fig. S3). Even with its reduced protein expression, the N34A mutation was able to complement ΔsaeS (Fig. S3), suggesting concentration is likely not the issue with the inability of M31A, W32A, and F33A mutant kinases to signal. Although M31 and F33 are completely conserved among staphylococcal species, two substitutions can be found for W32. Staphylococcus simulans and Staphylococcus canosus encode a leucine and a valine, respectively, maintaining the hydrophobicity of the position without the aromatic residue (Fig. 1B). Although the effect that these specific mutations have on S. aureus gene expression is unknown and will require further study, we identified the importance of the alanine substitutions in the SaeS EC loop in vitro and next evaluated their significance in an ex vivo model.
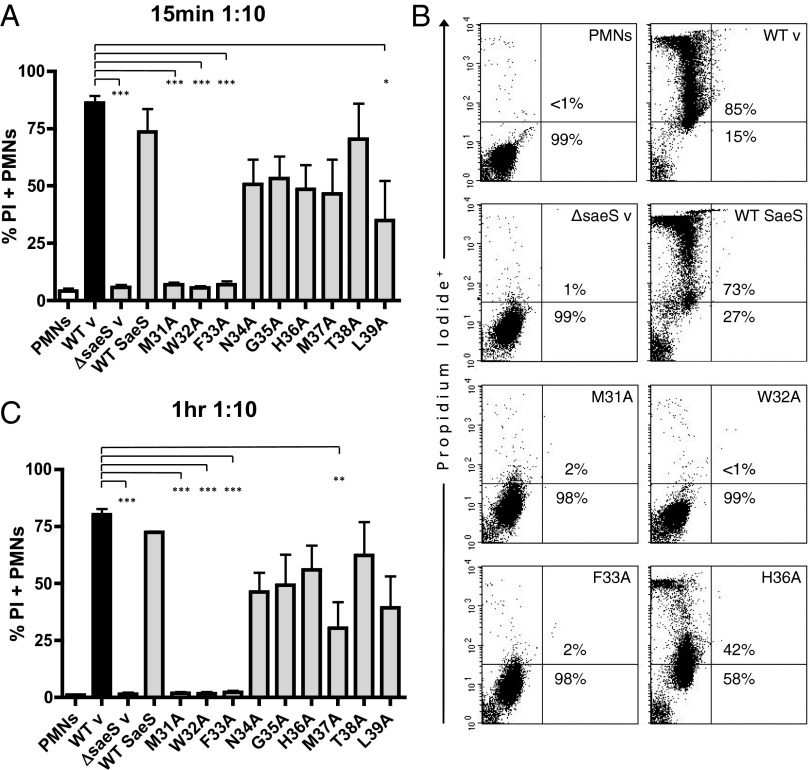
Supernatants from M31A, W32A, and F33A Mutant Strains Are Unable to Cause PMN Plasma Membrane Permeability.
SaeRS regulates transcription of multiple toxins, including γ-toxin (hlgA, hlgB, hlgC), LukSF-PVL, and LukAB/LukGH, which have been shown to contribute to PMN lysis (26–31). Additionally, transcription of the saePQRS operon and SaeR target genes are activated in response to PMN phagocytosis and PMN components (32–34). Collectively, these data suggest sensing and responding to PMNs is a central function of the sae system. To evaluate the effect of SaeS EC mutations on the cytolytic capacity of bacterial supernatants, we exposed PMNs to spent media from strains containing the SaeS EC point mutations for varied periods of time. PMN membrane permeability was assessed using propidium iodide (PI) (Fig. 3). Consistent with previous observations, the WT LAC spent media caused ∼85% of PMNs to be PI positive within 15 min, with similar results after 1 h (Fig. 3) (35). Almost no cytotoxicity was observed for ΔsaeS, and complementation with WT SaeS was able to return cytotoxicity to WT levels. Although all other EC loop mutations retained cytotoxicity, the single point mutations M31A, W32A, and F33A were unable to cause any membrane damage, similar to the RBC lysis assay (Fig. 2D), suggesting they are important for SaeS-mediated expression of cytolytic toxins.
Fig. 3.
Supernatants from M31A, W32A, and F33A mutant strains are unable to cause PMN plasma membrane permeability. (A) PMN plasma membrane permeability at 15 min after incubation with bacterial supernatants. (B) Representative dot plots of PMNs stained with PI following 1-h incubation with spent supernatants. (C) PMN plasma membrane permeability at 1 h after incubation with bacterial supernatants. Results are the mean ± SEM of three separate PMN donors. Asterisks denote statistical significance evaluated by a one-way ANOVA with Tukey’s posttest based on the following P values: *0.01–0.05, **0.001–0.01, and ***<0.001.
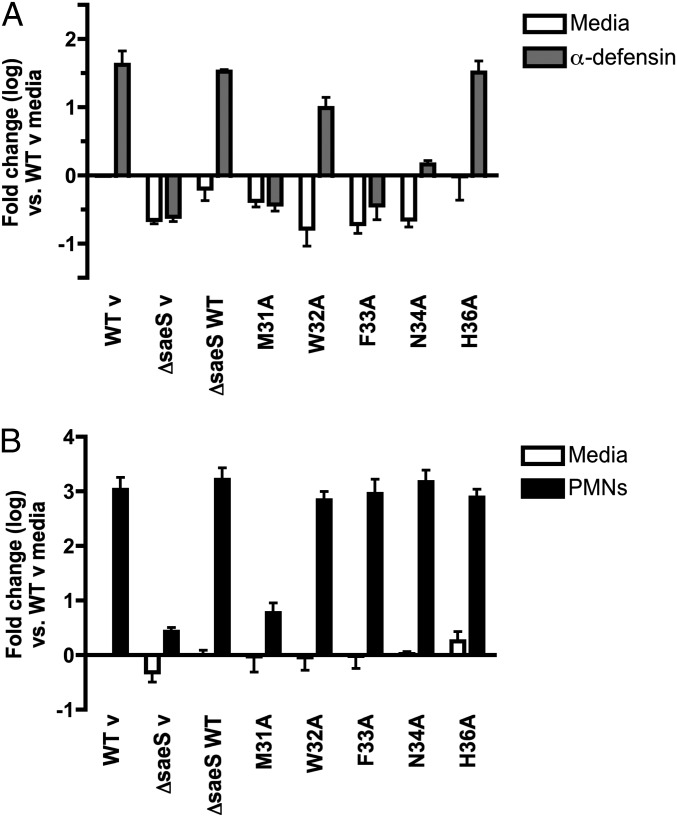
Activation Profile of Specific SaeS EC Mutants Is Signal Dependent.
Differential transcription of sae regulated genes has been shown to be dependent on specific host stimuli including HNP-1 and PMNs, implying another level of target gene regulation via specific stimuli (34). To investigate the role of the SaeS EC residues in the transcriptional response previously identified (34), we exposed mutant strains that were unable to activate the hla reporter or cause PMN cytotoxicity (M31A, W32A, and F33A) to a subinhibitory concentration of HNP-1 or human PMNs and measured transcript levels of sae target gene hlgA by real-time quantitative PCR (Fig. 4). To ensure the reduced protein expression of M31A (Fig. 2B) did not influence target gene transcription, we included N34A as a copy level control that also demonstrated reduced SaeS protein expression (Fig. 2B). Additionally, we included point mutation H36A that did not appear to alter activation of the hla reporter or PMN cytotoxicity. As expected, α-defensin induced an increase in hlgA transcription compared with media controls in a saeS-dependent manner (Fig. 4A), and mutation of residue H36 had no impact on hlgA expression (Fig. 4A). Consistent with previous experiments (Figs. 2D and 3), mutants M31A and F33A were unable to activate; however, W32A was able to induce a 1.2 log increase in transcription of hlgA in response to α-defensin. The N34A mutant elicited a weak response to α-defensin, suggesting it may play a role in sensing α-defensin or alternatively weak expression resulted in an inability to fully activate sae target gene expression.
Fig. 4.
Expression of sae target gene hlgA in mutant kinase strains following exposure to human α-defensin 1 and human PMNs. (A) Relative gene expression of hlgA in SaeS EC mutants was analyzed following a 30-min exposure to a subinhibitory concentration of human α-defensin 1 (0.48 μM). All transcript levels are normalized to gyrB and calibrated to transcript abundance in WT strains in media only. Data are shown as mean fold change of two biological replicates analyzed in triplicate. (B) Transcription of hlgA was measured 30 min after PMN phagocytosis (10:1 MOI). Data are reported as mean fold change of two to four biological replicates analyzed in duplicate.
Transcription of hlgA was also analyzed following a 30-min exposure to human PMNs. PMNs induced a robust increase in hlgA expression in the WT strain, whereas M31A transcription mirrored ΔsaeS (Fig. 4B). Unlike α-defensin, N34A and both aromatic mutations (W32A and F33A) were able to activate transcription of hlgA to WT levels in response to human PMNs. F33A and N34A had similar activation profiles following exposure to α-defensin and PMNs, confirming that reduced SaeS protein expression was not the reason the M31A mutant was unable to activate sae target gene transcription following either stimulus. Taken together, these data suggest that the M31 residue is essential for activation of the sae system. In addition, it appears that single residues in the EC domain control the ability of SaeS to integrate and diversify responses to host stimuli.
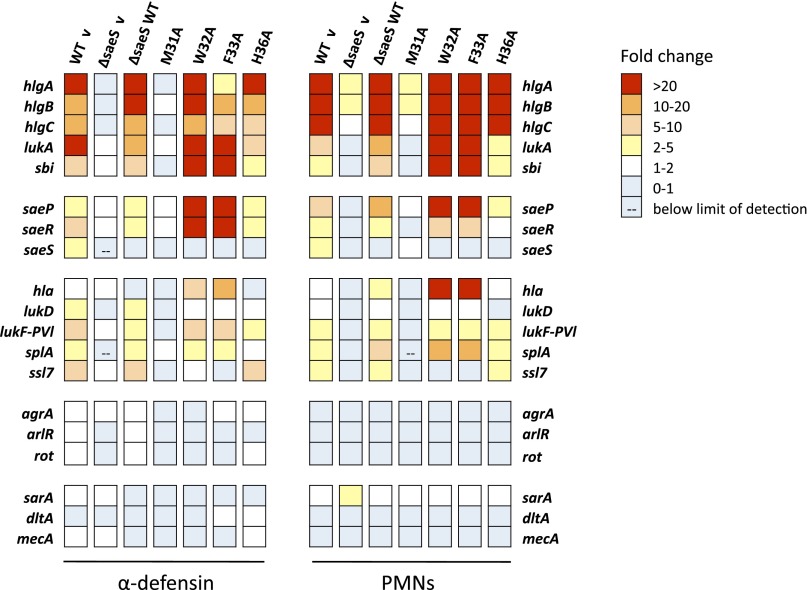
To further examine this hypothesis, we analyzed the expression profile of a large number of sae target genes for point mutations M31A, W32A, F33A, and H36A in response to both α-defensin and PMNs using the QuantiGene 2.0 assay (34). Changes in gene expression in response to different host stimuli were determined by comparing levels in S. aureus SaeS EC point mutants treated with the described stimulus to those in the same strain exposed to media only (Fig. 5 and Tables S2 and S3). Due to autoregulation of the sae system from the P1 promoter, transcript levels of saeR and saeS increase moderately on activation of the system (13, 15). An increase in expression of saeR was observed in all strains capable of activating sae-dependent gene expression (WT v, ΔsaeS-WT SaeS, W32A, F33A, and H36A; Tables S2 and S3). Because complementation was plasmid based with a nonnative promoter, up-regulation of saeS in response to stimuli was only observed in the WT strain. Consistent with previous reports, both stimuli caused an increase in expression of several genes containing the SaeR binding domain upstream of their coding sequence in the WT, and ΔsaeS complemented with WT SaeS and H36A strains but not in ΔsaeS (Fig. 5) (11, 34). The M31A mutation was unable to activate expression of genes in the sae regulon and had profiles similar to ΔsaeS in the presence of either stimulus. W32A and F33A had a larger fold increase in expression of sbi, saeP, saeR, and hla than WT in response to both stimuli. Intriguingly, F33A was only able to weakly transcribe hlgA and hlgC in response to α-defensin, yet both genes had high levels of induction following PMN exposure, similar to the quantitative RT-PCR (qRT-PCR) results (Fig. 4). Genes that do not contain a SaeR binding domain upstream of their coding sequence (agrA, sarA, dltA, and mecA) were not induced after either stimulus in any of the strains, demonstrating specificity of the SaeS-mediated responses. These data demonstrate the essential nature of residue M31 for SaeS sensing and signaling and suggest other residues in the extracellular loop may refine the pathogen’s response to external stimuli including α-defensin.
Fig. 5.
Residue M31 is essential to differential regulation of sae target genes following exposure to α-defensin 1 and PMN phagocytosis. Relative gene expression in SaeS EC mutants was analyzed following a 30-min exposure to a subinhibitory concentration of human α-defensin 1 (0.48 μM) or human PMNs (10:1 MOI) using the QuantiGene 2.0 assay. Transcript levels were normalized to gyrB, and each strain was calibrated to its matched sample grown in the absence of stimulus (media only). Data shown represent mean fold changes of two independent experiments (for PMN assays, two donors were used) analyzed in duplicate or triplicate.
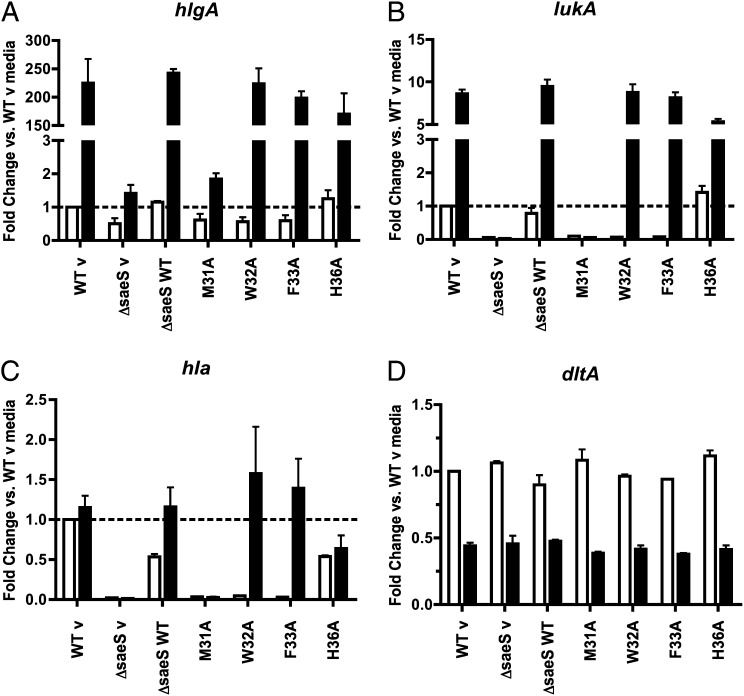
To evaluate the effect the SaeS EC mutants have on overall gene expression, the QuantiGene 2.0 data for each point mutation in the presence or absence of either host stimulus were normalized to the WT v media control. The expression profile of hlgA was consistent with qRT-PCR analysis showing all strains had a large increase in transcription, with the exception of ΔsaeS and M31A, following exposure to PMNs (Fig. 6A). Expression of lukA in M31A, W32A, and F33A was slightly reduced compared with that of WT in the absence of any signal, but contact with PMNs restored expression in W32A and F33A (Fig. 6B). Hla is expressed at a very high level in USA300 strains (36), making it difficult to measure induced expression. Consistent with this, little change was observed in hla transcript levels in WT or H36A following incubation with PMNs (Fig. 6C). Expression of hla dropped substantially in ΔsaeS, M31A, W32A, and F33A mutants in the absence of any stimulus (supported by Fig. 2), yet PMNs were able to induce overall transcript levels similar to WT in W32A and F33A. As a control, the expression profile of dltA was assessed to ensure the SaeS point mutations were not causing any changes in gene expression unrelated to the sae system (Fig. 6D) (37). Results for α-defensin–induced gene expression followed the same trends as the PMN results with the exception of hlgA in F33A, which had a robust response to PMNs and a diminished response to α-defensin (Fig. S4). These data demonstrate that, although the aromatic point mutations W32A and F33A are able to activate sae-dependent gene expression in a signal-specific manner, basal expression of several sae-regulated toxins is reduced in these mutants.
Fig. 6.
SaeS mutant kinase strains have lower basal gene expression of sae-regulated toxins. QuantiGene 2.0 assay results from PMN induced gene expression in S. aureus strains were normalized to gyrB, calibrated to WT v without stimulus (media control), and displayed as mean fold changes. Error bars are SEM of two biological replicates analyzed in duplicate. White bars, media control; black bars, PMNs; dashed lines, calibrator (WT v without stimulus). (A) hlgA, (B) lukA, (C) hla, and (D) dltA (control, gene without SaeR binding domain).
SaeS Mutant M31A Is Unable to Induce Cytotoxicity in Human PMNs Following Phagocytosis.
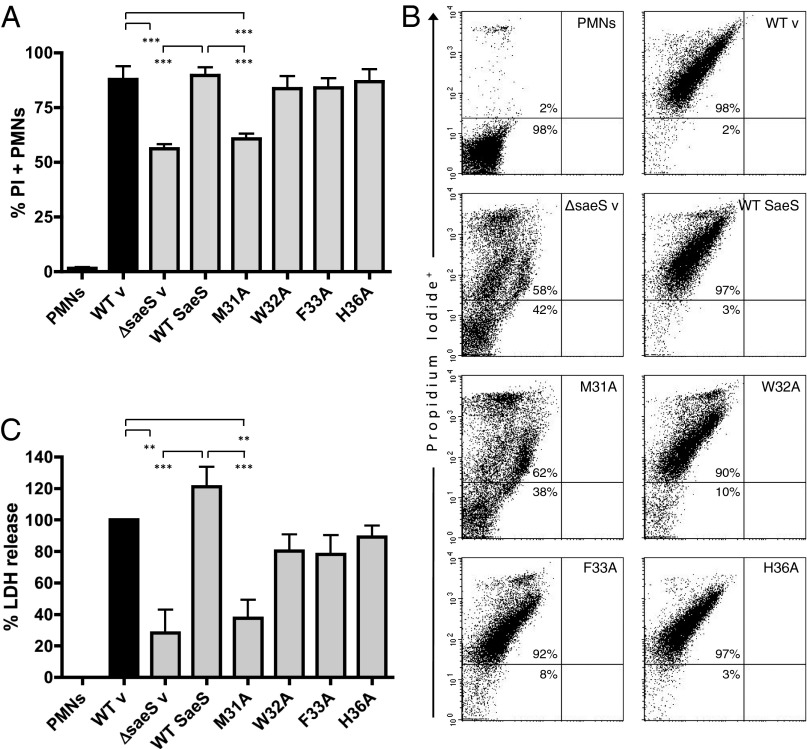
To investigate whether our transcriptional findings translate to a pathogenesis phenotype, we infected human neutrophils with the SaeS EC mutant strains and assessed PMN cytotoxicity by evaluating membrane permeability and lactate dehydrogenase (LDH) release (Fig. 7). In contrast to the >90% cytotoxicity caused by WT, the M31A mutant was only able to induce membrane permeability in ∼50% of PMNs and was indistinguishable from the attenuated ΔsaeS strain. Consistent with transcriptional data following human neutrophil interaction, mutants W32A, F33A, and H36A showed no significant differences compared with WT in their ability to cause membrane damage in neutrophils. Congruent with these results, LDH assays showed significant decreases in PMN lysis following incubation with ΔsaeS and M31A strains compared with WT. These data demonstrate a single methionine residue in the SaeS sensing domain is essential for detecting human PMNs and activating the sae regulon for bacterial defense and pathogenesis. In addition to this finding, we observed a distinct difference between basal and signal-dependent expression of virulence factors in the aromatic mutants.
Fig. 7.
Residue M31 in the SaeS sensing domain is essential for SaeS-induced PMN plasma membrane permeability and lysis. (A) Complied results representing three separate experiments investigating PMN plasma membrane permeability 3 h after infection with SaeS EC mutants (10:1 MOI). (B) Representative dot plots of PMNs stained with PI 3 h after infection. (C) LDH assays of matching supernatants from membrane permeability studies in A showing PMN cytotoxicity normalized to WT v. Results are the mean ± SEM of three PMN donors. Significance was evaluated by a one-way ANOVA with Tukey’s posttest. P values are represented as **0.001–0.01 and ***<0.001.
Discussion
Unraveling the remarkably sensitive and specific nature of bacterial two-component systems is important not only for our understanding of signal transduction but also for the development of new treatments against human pathogens. The sae system is a central regulator of many S. aureus toxins, exoenzymes, and immunomodulatory proteins known to be important for the pathogenesis of S. aureus. However, the exact mechanism of how SaeS senses its environment is largely unknown. To address this, we used a combination of in silico, in vitro, and ex vivo techniques to define the sensing domain of SaeS, assess the importance of individual amino acids for sensing host signals, and evaluate the importance of these residues to S. aureus pathogenesis.
Transcript analysis of sae target genes in SaeS mutant strains M31A, W32A, and F33A following exposure to α-defensin or human PMNs, both of which are known inducers of the sae system (34, 38), demonstrated that these residues are fundamental to the appropriate activation of sae target gene transcription. A major finding of this study was the observation that M31 was essential for activation of the sae regulon. Notably, mutation of either aromatic residue did not completely abolish the ability of the kinases to activate following induction by neutrophil-derived stimuli; however, basal expression of sae targets was dramatically reduced in W32A and F33A in the absence of stimuli. These data suggest that mutation of either aromatic anchor residue disrupts the normal function of the kinases in the absence of a specific signal. The molecular basis for this disruption is unknown, but the presence of an inducing agent, for example, highly charged antimicrobial peptides, may provoke a conformational change, allowing these mutant kinases to recover most of their functionality and respond to other unidentified signals. Another interesting finding was the altered transcriptional profile in F33A. Gene expression elicited by PMNs in the F33A mutant was similar to WT, yet a subinhibitory concentration of α-defensin induced a considerably weaker response from the γ-toxin genes, suggesting another activating signal is present in human PMNs. In contrast, mutation of the neighboring aromatic W32 did not result in a deficiency in gene expression in response to either host stimulus. These results, along with the finding that residues M31 and F33 are completely conserved in all staphylococcal species known to encode saeS, support the hypothesis that they play a critical role in specific signaling events.
The curious observation that expression of several toxins was reduced in W32A and F33A in the absence of host signals suggests the unstimulated state of SaeS is altered in some way. Recent biochemical studies of synthetic model peptides have shown the importance of aromatic residues at the lipid interface of membrane proteins and have been suggested to stabilize or anchor the structure and orientation of transmembrane helices (39). Furthermore, mutation of the tandem aromatic anchors in the Escherichia coli aspartate/maltose chemoreceptor Tar resulted in an altered baseline signaling state of the chemoreceptor and a divergent chemotactic response to each stimulus (40, 41). Interestingly, it appears the anchoring and signaling functions of aromatics are not restricted to prokaryotes. A screen of the transient receptor potential (TRP) homolog TRPY1 in the budding yeast Saccharomyces cerevisiae identified the necessity of aromatic anchors in the sixth transmembrane helix, which are widely conserved in other TRP homologs in both fungal and vertebrate species (42). Additionally, an alanine scan of the second extracellular loop of the human vasopressin receptor V1a found four aromatic residues to be important for ligand binding and receptor activation (43). Together these findings demonstrate the global importance of aromatic amino acids in the transmembrane and extracellular domains of sensing proteins in a wide range of organisms and hint at potential mechanisms for the altered transcriptional profile of the sae regulon in S. aureus SaeS mutants. It is also conceivable that aromatic residues W32 and F33 may directly interact with inducing agents, including α-defensin or other unidentified signals in PMNs. Another possibility is that mutation of W32 or F33 may alter protein-protein interactions within the SaeS homodimer or the SaePQS complex, resulting in the altered gene expression we observed. The recent finding that auxiliary proteins SaeP and SaeQ stimulate the phosphatase activity of the bifunctional kinase to modulate expression of the sae regulon supports this hypothesis (18). More studies are needed to investigate these possibilities and to examine the effect of other point mutations on the refinement of the pathogen response to stimuli.
Inasmuch as we observed different abilities of the mutant kinases to activate downstream effectors of the sae system, we evaluated the virulence of these mutant strains during interaction with human PMNs. Reflective of transcript induction of target genes following exposure to human PMNs, we observed that W32A and F33A kinase mutants had cytolytic capabilities equivalent to the WT strain, whereas following PMN phagocytosis, the M31A mutant strain produced significantly less cytotoxicity. These data combined with the lack of cytolytic activity of supernatants from all three mutant kinases harvested during in vitro growth illustrate the importance of not only specific residues in the predicted extracellular domain but the essential nature of the host stimulus to trigger virulence in S. aureus. Thus, it would be interesting to examine the SaeS EC point mutations in other infections models, notably the mouse skin, as the transcriptional profile for the sae regulon has already been shown to be modified in this environment compared with PMNs (34).
Although our findings have exciting implications for the importance of aromatic residues in signal transduction and pathogenesis, the significance of the M31A mutation is intriguing. Future studies will elucidate exactly why this methionine is essential to the SaeS sensing domain. Because this kinase appears to be completely unable to activate transcription of sae target genes, suggesting it may be in in a “locked off” conformation, it would be informative to combine the M31A mutation with the constitutively active mutation L18P found in strain Newman (15, 44, 45). It should be noted that, although we predict M31 to be EC regardless of its exact localization, we identified a residue that is essential for signaling in a major virulence regulatory system in S. aureus.
This study advances our understanding of how S. aureus can sense and respond to changing environments within the host and provides an explanation for the coordination of multiple signals via specific residues within a single sensing domain. Therefore, our findings support the hypothesis that individual residues in the sensing domain of SaeS are able to dictate the outcome of infections based on the presence of specific host stimuli. These discoveries will be valuable for the development of novel antivirulence therapeutics, because we identified residues within SaeS that are essential for proper activation of the sae system and for S. aureus pathogenesis.
Materials and Methods
Bacterial Strains and Growth Conditions, Plasmid and Strain Construction, Substituted Cysteine Accessibility Method (SCAM), hla Reporter Construction, SaeS Extracellular Point Mutations, Immunoblotting, and Statistics.
Detailed protocols are described in SI Materials and Methods and Table S4.
Fluorescent hla Reporter Assays.
Overnight cultures were diluted 1:100 in tryptic soy broth (TSB) containing appropriate antibiotics and incubated in a 37 °C shaker. At each time point, 200 μL was removed and placed in a black 96-well plate in triplicate. The OD600 and fluorescence (480-nm excitation/515-nm emission) was measured in a Tecan Infinite M200 plate reader (Tecan). The 24-h fluorescence readings were divided by the corresponding OD600 and plotted as relative fluorescence units (RFUs). At 6, 12, and 24 h, 1-mL samples were pelleted and stored at −20 °C for immunoblot analysis (SI Materials and Methods) to evaluate expression of mutant SaeS constructs. At 24 h, supernatants were filter sterilized with 0.22-μm Spin-X columns (Costar) and stored at −20 °C for future evaluation by the RBC lysis assay, as described below.
RBC Lysis Assay.
RBC lysis activity in bacterial supernatants was determined using the previously described RBC lysis titration assay with slight modifications (46). Culture supernatants from the fluorescent hla reporter assay (described above) were twofold serially diluted across a 96-well plate. Rabbit erythrocytes from HemoStat Laboratories were washed in PBS until the supernatant was clear. Washed rabbit RBCs were resuspended in PBS to create a 3% (vol/vol) solution. Seventy microliters of the freshly diluted 3% RBC solution was aliquoted into 96-well plates containing 30 μL of serially diluted culture supernatant. Plates were incubated statically at room temperature for 1 h. Hemolytic activity was evaluated by measuring the loss of turbidity at 630 nm using the Tecan Infinite M200 plate reader. The resulting curves were fit with a four-parameter logistic fit using Prism (version 4.0c for Mac; GraphPad Software) to determine the midpoint. Midpoints of each fit were normalized to WT, and all measurements were performed in triplicate.
PMN Cytotoxicity Assays.
PMNs (or neutrophils) were isolated from healthy human donors following procedures described elsewhere (33). All procedures were performed in accordance with a protocol approved by the Institutional Review Board for Human Subjects at Montana State University. S. aureus-induced cytotoxicity assays were performed as previously described with minor adjustments (47). PMNs (1 × 106) were resuspended in RPMI + 5 mM Hepes and plated in 96-well plates precoated with 20% (vol/vol) human serum diluted in Dulbecco's phosphate buffered saline (DPBS). For assays using spent supernatants, S. aureus overnight cultures were diluted 1:100 and grown for 5 h. Spent supernatants were filter sterilized and diluted 1:10 in RPMI. Diluted supernatants were added to plated PMNs, synchronized (400 × g, 8 min, 4 °C), and then incubated statically at 37 °C with 5% CO2. Membrane permeability and cytotoxicity caused by whole bacteria were evaluated as described using flow cytometry (11, 47) or the CytoTox 96 Non-Radioactive Cytotoxicity assay (Promega) according to the manufacturer’s instructions (31, 33).
Analysis of Gene Expression Following Exposure to Host Stimuli.
Overnight cultures were diluted 1:100 and grown to an OD600 = 1.5. Bacteria were washed in DPBS and then plated in 96-well round bottom plates at 5 × 107 CFUs per well. A subinhibitory concentration of human α-defensin 1 [0.48 μM, as previously determined (34); Millipore] was added to bacteria, and plates were incubated at 37 °C while shaking (250 rpm) for 30 min. RNA was harvested using RNeasy Mini Kits (Qiagen) as described elsewhere (48). For PMN-induced gene expression, PMNs were plated in a 96-well plate at 2 × 106 cells per well, and bacteria were added at a 10:1 bacteria:PMN ratio. Infections were synchronized (400 × g, 8 min, 4 °C), and plates were incubated statically at 37 °C. After 30 min, RNA was purified from bacteria as previously described (48).
QuantiGene 2.0 Assays.
QuantiGene 2.0 assays (Affymetrix) were performed on RNA harvested from α-defensin and PMN experiments as previously described (33, 34). Briefly, magnetic microsphere beads containing capture probes unique to 15 genes or operons with the SaeR binding sequence (hlgA, hlgB, hlgC, lukA, sbi, saeP, saeR, saeS, hla, lukD, lukF-PVl, splA, ssl7, arlR, and rot) and five genes without the SaeR binding site (gyrB, sarA, agrA, mecA, and dltA) were incubated with purified RNA samples (50 ng) and target-specific DNA probes. DNA probes contained biotinylation sites that allow amplification of the fluorescent signal. Following signal amplification, the fluorescence intensity of each capture bead was detected using a Luminex flow cytometer (BioRad) and reported as the mean fluorescent signal for each gene-specific bead. Two biological replicates of each experiment were examined in duplicate, and gene expression levels were normalized to gyrB. Changes in gene expression in response to different host stimuli were determined by comparing fluorescence levels in S. aureus SaeS EC point mutants treated with the described stimulus to those of the same strain in media only. To evaluate the effect the SaeS point mutations have on overall gene expression, transcript levels for each point mutation in the presence and absence of stimuli were compared with the WT media control.
Stimulus-dependent gene expression was verified by TaqMan real-time RT-PCR on bacterial RNA harvested from the aforementioned α-defensin and PMN phagocytosis experiments as previously described (11, 31). Relative quantification of hlgA was expressed as the log10 change relative to the WT v media control for each stimulus. Two to four biological replicates of each experiment were analyzed in duplicate (PMN) or triplicate (α-defensin).
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health (NIH) Grants NIH-RR020185 and NIH-R01 Award A1090046-01, Molecular Biosciences Fellowship P20RR16455-07 (to O.W.Z.), Montana State University Agriculture Experiment Station funds, and an equipment grant from the Murdoch Charitable Trust. Additional support for C.E.F., C.L.M., and A.R.H. was from project 3 of NIH Grant AI083211.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322125111/-/DCSupplemental.
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.DeLeo FR, Chambers HF. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest. 2009;119(9):2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong Y-Q. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol. 2004;40(1):1–9. doi: 10.1016/S0928-8244(03)00309-2. [DOI] [PubMed] [Google Scholar]
- 4.Bronner S, Monteil H, Prévost G. Regulation of virulence determinants in Staphylococcus aureus: Complexity and applications. FEMS Microbiol Rev. 2004;28(2):183–200. doi: 10.1016/j.femsre.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Beier D, Gross R. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol. 2006;9(2):143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Giraudo AT, Raspanti CG, Calzolari A, Nagel R. Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can J Microbiol. 1994;40(8):677–681. doi: 10.1139/m94-107. [DOI] [PubMed] [Google Scholar]
- 7.Giraudo AT, Rampone H, Calzolari A, Nagel R. Phenotypic characterization and virulence of a sae- agr- mutant of Staphylococcus aureus. Can J Microbiol. 1996;42(2):120–123. doi: 10.1139/m96-019. [DOI] [PubMed] [Google Scholar]
- 8.Rampone H, Martínez GL, Giraudo AT, Calzolari A, Nagel R. In vivo expression of exoprotein synthesis with a Sae mutant of Staphylococcus aureus. Can J Vet Res. 1996;60(3):237–240. [PMC free article] [PubMed] [Google Scholar]
- 9.Giraudo AT, Calzolari A, Cataldi AA, Bogni C, Nagel R. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol Lett. 1999;177(1):15–22. doi: 10.1111/j.1574-6968.1999.tb13707.x. [DOI] [PubMed] [Google Scholar]
- 10.Liang X, et al. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect Immun. 2006;74(8):4655–4665. doi: 10.1128/IAI.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nygaard TK, et al. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J Infect Dis. 2010;201(2):241–254. doi: 10.1086/649570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watkins RL, Pallister KB, Voyich JM. The SaeR/S gene regulatory system induces a pro-inflammatory cytokine response during Staphylococcus aureus infection. PLoS ONE. 2011;6(5):e19939. doi: 10.1371/journal.pone.0019939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novick RP, Jiang D. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology. 2003;149(Pt 10):2709–2717. doi: 10.1099/mic.0.26575-0. [DOI] [PubMed] [Google Scholar]
- 14.Sun F, et al. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol. 2010;192(8):2111–2127. doi: 10.1128/JB.01524-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adhikari RP, Novick RP. Regulatory organization of the staphylococcal sae locus. Microbiology. 2008;154(Pt 3):949–959. doi: 10.1099/mic.0.2007/012245-0. [DOI] [PubMed] [Google Scholar]
- 16.Li M, et al. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci USA. 2007;104(22):9469–9474. doi: 10.1073/pnas.0702159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mascher T. Intramembrane-sensing histidine kinases: A new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol Lett. 2006;264(2):133–144. doi: 10.1111/j.1574-6968.2006.00444.x. [DOI] [PubMed] [Google Scholar]
- 18.Jeong DW, et al. The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol Microbiol. 2012;86(2):331–348. doi: 10.1111/j.1365-2958.2012.08198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogdanov M, Zhang W, Xie J, Dowhan W. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM(TM)): Application to lipid-specific membrane protein topogenesis. Methods. 2005;36(2):148–171. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thoendel M, Horswill AR. Random mutagenesis and topology analysis of the autoinducing peptide biosynthesis proteins in Staphylococcus aureus. Mol Microbiol. 2013;87(2):318–337. doi: 10.1111/mmi.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS ONE. 2010;5(4):e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forsyth RA, et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol. 2002;43(6):1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 23.Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: Consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37(Web Server issue) suppl 2:W465–W468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho H, Jeong DW, Li C, Bae T. Organizational requirements of the SaeR binding sites for a functional P1 promoter of the sae operon in Staphylococcus aureus. J Bacteriol. 2012;194(11):2865–2876. doi: 10.1128/JB.06771-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menestrina G, et al. Ion channels and bacterial infection: The case of β-barrel pore-forming protein toxins of Staphylococcus aureus. FEBS Lett. 2003;552(1):54–60. doi: 10.1016/s0014-5793(03)00850-0. [DOI] [PubMed] [Google Scholar]
- 27.Prévost G, et al. Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect Immun. 1995;63(10):4121–4129. doi: 10.1128/iai.63.10.4121-4129.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dumont AL, et al. Characterization of a new cytotoxin that contributes to Staphylococcus aureus pathogenesis. Mol Microbiol. 2011;79(3):814–825. doi: 10.1111/j.1365-2958.2010.07490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ventura CL, et al. Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS ONE. 2010;5(7):e11634. doi: 10.1371/journal.pone.0011634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rigby KM, DeLeo FR. Neutrophils in innate host defense against Staphylococcus aureus infections. Semin Immunopathol. 2012;34(2):237–259. doi: 10.1007/s00281-011-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voyich JM, et al. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J Infect Dis. 2009;199(11):1698–1706. doi: 10.1086/598967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palazzolo-Ballance AM, et al. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J Immunol. 2008;180(1):500–509. doi: 10.4049/jimmunol.180.1.500. [DOI] [PubMed] [Google Scholar]
- 33.Voyich JM, et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175(6):3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 34.Zurek OW, et al. 2014. The role of innate immunity in promoting SaeR/S-mediated virulence in Staphylococcus aureus. J Innate Immun 6(1):21–30.
- 35.Nygaard TK, DeLeo FR, Voyich JM. Community-associated methicillin-resistant Staphylococcus aureus skin infections: advances toward identifying the key virulence factors. Curr Opin Infect Dis. 2008;21(2):147–152. doi: 10.1097/QCO.0b013e3282f64819. [DOI] [PubMed] [Google Scholar]
- 36.Kennedy AD, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: Recent clonal expansion and diversification. Proc Natl Acad Sci USA. 2008;105(4):1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herbert S, et al. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 2007;3(7):e102. doi: 10.1371/journal.ppat.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geiger T, Goerke C, Mainiero M, Kraus D, Wolz C. The virulence regulator Sae of Staphylococcus aureus: Promoter activities and response to phagocytosis-related signals. J Bacteriol. 2008;190(10):3419–3428. doi: 10.1128/JB.01927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gleason NJ, et al. Tyrosine replacing tryptophan as an anchor in GWALP peptides. Biochemistry. 2012;51(10):2044–2053. doi: 10.1021/bi201732e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Draheim RR, Bormans AF, Lai RZ, Manson MD. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry. 2006;45(49):14655–14664. doi: 10.1021/bi061259i. [DOI] [PubMed] [Google Scholar]
- 41.Adase CA, Draheim RR, Manson MD. The residue composition of the aromatic anchor of the second transmembrane helix determines the signaling properties of the aspartate/maltose chemoreceptor Tar of Escherichia coli. Biochemistry. 2012;51(9):1925–1932. doi: 10.1021/bi201555x. [DOI] [PubMed] [Google Scholar]
- 42.Zhou X, et al. Yeast screens show aromatic residues at the end of the sixth helix anchor transient receptor potential channel gate. Proc Natl Acad Sci USA. 2007;104(39):15555–15559. doi: 10.1073/pnas.0704039104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conner M, et al. Systematic analysis of the entire second extracellular loop of the V(1a) vasopressin receptor: Key residues, conserved throughout a G-protein-coupled receptor family, identified. J Biol Chem. 2007;282(24):17405–17412. doi: 10.1074/jbc.M702151200. [DOI] [PubMed] [Google Scholar]
- 44.Schäfer D, et al. A point mutation in the sensor histidine kinase SaeS of Staphylococcus aureus strain Newman alters the response to biocide exposure. J Bacteriol. 2009;191(23):7306–7314. doi: 10.1128/JB.00630-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mainiero M, et al. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J Bacteriol. 2010;192(3):613–623. doi: 10.1128/JB.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pang YY, et al. agr-Dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J Innate Immun. 2010;2(6):546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nygaard TK, et al. Alpha-toxin induces programmed cell death of human T cells, B cells, and monocytes during USA300 infection. PLoS ONE. 2012;7(5):e36532. doi: 10.1371/journal.pone.0036532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voyich JM, Sturdevant DE, DeLeo FR. Analysis of Staphylococcus aureus gene expression during PMN phagocytosis. Methods Mol Biol. 2008;431:109–122. doi: 10.1007/978-1-60327-032-8_9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.