Significance
In redox biology, the chemistry performed by proteins that contain the rare amino acid selenocysteine is frequently critical to the detoxification of reactive species that are harmful to cellular function. Selenocysteine and cysteine partner to form a motif featuring a sulfur–selenium covalent bond in many selenoproteins. This work demonstrates that selenium NMR, when paired with calculations, can provide critical insight concerning the local environment of these enigmatic redox motifs. It details how redox potentials, conformational preferences, and mobilities of such redox motifs change when the local environment of the selenocysteine is varied. Surprisingly, reverting selenocysteine to cysteine exerts only minor effects on redox potential. These new approaches deepen our understanding of the chemical reactivity and thermodynamic properties of selenoenzymes.
Abstract
Selenoproteins use the rare amino acid selenocysteine (Sec) to act as the first line of defense against oxidants, which are linked to aging, cancer, and neurodegenerative diseases. Many selenoproteins are oxidoreductases in which the reactive Sec is connected to a neighboring Cys and able to form a ring. These Sec-containing redox motifs govern much of the reactivity of selenoproteins. To study their fundamental properties, we have used 77Se NMR spectroscopy in concert with theoretical calculations to determine the conformational preferences and mobility of representative motifs. This use of 77Se as a probe enables the direct recording of the properties of Sec as its environment is systematically changed. We find that all motifs have several ring conformations in their oxidized state. These ring structures are most likely stabilized by weak, nonbonding interactions between the selenium and the amide carbon. To examine how the presence of selenium and ring geometric strain governs the motifs’ reactivity, we measured the redox potentials of Sec-containing motifs and their corresponding Cys-only variants. The comparisons reveal that for C-terminal motifs the redox potentials increased between 20–25 mV when the selenenylsulfide bond was changed to a disulfide bond. Changes of similar magnitude arose when we varied ring size or the motifs’ flanking residues. This suggests that the presence of Sec is not tied to unusually low redox potentials. The unique roles of selenoproteins in human health and their chemical reactivities may therefore not necessarily be explained by lower redox potentials, as has often been claimed.
Selenium in biological systems has long been studied for its role in aging, cancer and chronic diseases (1). Selenium is incorporated into proteins via the amino acid selenocysteine (Sec or U), which is structurally similar to cysteine yet more reactive (2). Selenoproteins exploit Sec to catalyze a wide range of electron-transfer reactions (3). Although selenoproteins are rare, they are essential for mammals and have important roles in antioxidant defense and redox regulation (4). The majority of selenoproteins either directly scavenge oxidants (such as the reactive oxygen and nitrogen species that lead to deleterious covalent modification of cellular components), or they may signal the cellular level of these reactive species, thereby activating defense pathways. Selenoproteins are particularly well-suited for the task of combating oxidative stress because they are both sensitive to low levels of oxidants and yet resilient to inactivation by these reactive species (3). Because of their role in longevity and human diseases, there has been sustained interest in deciphering the unique role of Sec in selenoproteins (5). However, it remains largely unclear what sets selenoproteins apart from their cysteine-containing counterparts and why, in mammals, they cannot be replaced by them. Sec commonly forms a selenenylsulfide bond with Cys, resulting in redox motifs analogous to the Cys-containing ones of nonselenoprotein oxidoreductases. Although the latter are well-studied, many of the basic properties of Sec-containing redox motifs are not known. To better understand features modulating the reactivity of selenocysteine, we set out to investigate the redox potential, conformational preferences, and dynamics of Sec-containing redox motifs using mass spectrometry, 77Se NMR spectroscopy, and density functional theory (DFT).
We have selected four representative redox motifs with different ring sizes and geometric strains. The first and second redox motifs, Ser-Cys-Sec-Ser-COOH (SCUS) and Gly-Cys-Sec-Gly-COOH (GCUG), form a highly constrained eight-membered ring in the oxidized state (6). The GCUG is naturally found in mammalian thioredoxin reductase (TrxR), whereas the more geometrically constrained SCUS motif is related to the SCCS motif in Drosophila melanogaster TrxR (7, 8). Interestingly, similar variations of these TrxR-based motifs have previously been shown to affect kinetic parameters in TrxR isoenzymes (7, 9, 10). The third and fourth motifs were selected to demonstrate effects of increased ring size; the Gly-Cys-Ala-Sec-Gly-COOH (GCAUG) and the Gly-Cys-Gly-Ala-Sec-Gly-COOH (GCGAUG) motifs form less constrained 11- and 14-membered rings, respectively. The size of the ring is directly linked to its chemical reactivity because if the strain is increased, the selenenylsulfide bond is destabilized. This in turn leads to a higher redox potential (more oxidizing), which favors a reduced protein (11, 12). The 14-membered ring is the most ubiquitous motif in the selenoproteome. The motifs were positioned at the C terminus of a redox-inactive stable protein scaffold (see below), noting that many selenoproteins use such C-terminally located active sites (13). Sec and [77Se]Sec were incorporated via heterologous expression in Escherichia coli using the native genetic selenium incorporation machinery (14).
It should be noted that the overall features of a selenoprotein are dependent upon both the intrinsic physicochemical features of the Sec residue (5, 15), and the interactions of a Sec-containing redox active motif with either substrates or other residues of the active site microenvironment. Context-dependent effects were previously demonstrated with TrxR, where the solely Cys-based motif of the Drosophila enzyme is highly active as part of that enzyme (7, 8) but inactive if placed into the mammalian enzyme (9). Such observations illustrate that the inherent features of Sec- vs. Cys-containing motifs may be difficult to judge within the context of natural selenoproteins and their active sites. Hence, to avoid the contextual difficulties associated with the use of a given specialized active site environment, we fused the four motifs within the flexible C-terminal region of a stable well-folded protein scaffold (here, a redox inactive E. coli thioredoxin mutant), so that we could probe these introduced Sec- or Cys-containing redox active motifs without confounding long-range interactions. Indeed, this approach mirrors the salient features of native selenoproteins with C-terminal redox motifs. Specifically, such native motifs are also located 1–2 aa from the C terminus and use small amino acids as bridging residues (i.e., G, A, V, S, and T). The conformational mobility and influence of the remaining protein skeleton in both model and native proteins are expected to be similar because the C terminus of proteins tends to be unstructured and hence relatively free from geometric constraints placed by secondary structure. In contrast to peptides, the N and C termini of both the native selenoproteins and the model proteins studied here are spatially sufficiently separated, so as to not influence each other (16). Thus, the trends uncovered in this work should be directly applicable to native selenoproteins with C-terminal redox motifs. Furthermore, the methodology used here can be extended to encompass studies of all selenoproteins, including those that do not contain redox motifs.
We investigated the conformational preferences and dynamics of the Sec-containing redox active motifs using 77Se NMR spectroscopy. This method directly probes the properties of Sec in a given redox motif and is highly sensitive to changes in the electronic environment. 77Se has previously been used as a spectroscopic probe of protein structure and function (17–20), but the relation between chemical shifts and the specific location or properties of Sec was not deduced in these earlier studies. Here, DFT calculations of the 77Se chemical shift were used to determine conformational preferences of the oxidized selenenylsulfide-containing rings. Such calculations have been shown to correlate well with experimental values and provide an important predictive tool (21, 22). This study is, to our knowledge, the first to combine DFT calculations with experimental selenium NMR spectroscopy to study the environment and molecular properties of selenium in biological systems and lays the foundation for comparable studies of structural and dynamic features of Sec-dependent redox motifs found in native selenoproteins (23).
In addition to reporting the NMR parameters and conformational preferences of the selenenylsulfide motifs, we also compared the redox potentials of the Sec-containing proteins with their Cys homologs. This is a critical aspect of the reactivity of many selenoproteins because reduction of the selenenylsulfide liberates the nucleophilic, and catalytically essential, selenolate (R-Se−) species. Although it was proposed that the redox potential of selenoproteins might be much lower then their Cys-containing orthologs (15), only a few Sec-containing proteins have been examined experimentally (24–26). Herein, we also provide, to our knowledge, the first quantitative comparisons of redox potentials of Sec-containing to all Cys-containing redox motifs in a range of ring sizes. Surprisingly, these systematic studies reveal a much less dramatic impact of selenium on the redox potential of these centers than was previously believed.
Results and Discussion
Preparation of 77Se-Enriched Selenoproteins.
Preparation of selenoproteins is often the limiting step in their biophysical characterization (27). For NMR experiments, the selenoproteins need to be both produced in high amounts and also enriched with the NMR active 77Se isotope, whose natural abundance is 7.6%. Here, we have used an isotopic enrichment protocol that incorporates [77Se]Sec via heterologous expression using the native genetic selenium incorporation machinery of E. coli (14). Details concerning the preparation of the selenoproteins are presented in SI Appendix, SI Materials and Methods and Figs. S1 and S2.
77Se NMR Properties of the Oxidized Selenenylsulfide Motifs.
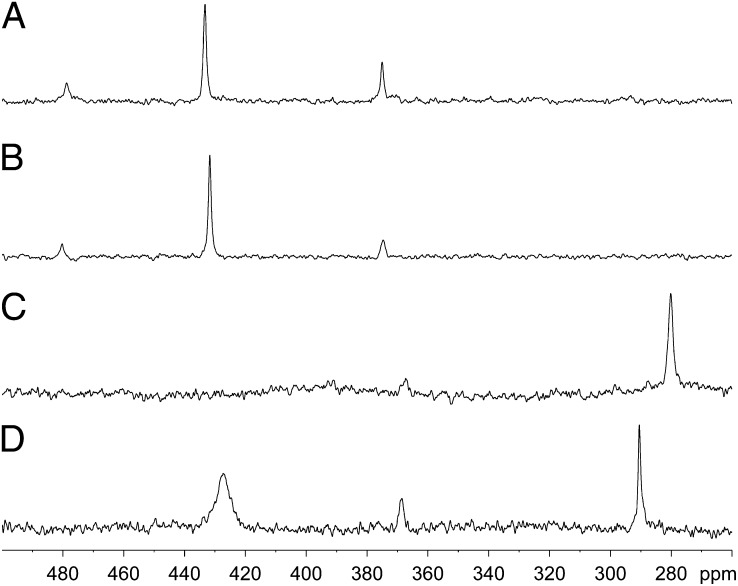
The isotopic chemical-shift values of the four redox motifs in their oxidized states were recorded using 1D proton-decoupled 77Se NMR. It should be noted that the large chemical-shift anisotropy of selenium leads to fast relaxation rates, which for selenium sites at mobile positions such as studied here leads to 1D experiments being the most sensitive. The spectra of all four motifs contained more than one resonance (Fig. 1 and Table 1). Comparing the spectra of the four rings, we found that despite their different sizes, they had commonalities: all rings had a resonance around 373 ppm, and three had a resonance around 430 ppm. The eight-membered rings had a unique configuration around 480 ppm not observed for the larger rings, which in contrast had a common resonance around 280–290 ppm. The presence of multiple resonances for each ring could not be attributed to intermolecular selenenylsulfide bonds because the protein was monomeric, as evident from size-exclusion chromatography. Mass spectrometry confirmed that no chemical modifications occurred before or following the NMR experiments. SI Appendix, Fig. S3 shows that NMR spectra of the motifs following repeated reduction and reoxidation were similar, within signal to noise, to that before reduction. These results suggested that the different resonances were inherent properties of a given redox motif and not attributable to sample heterogeneity. The multiple resonances likely derived from different ring conformations.
Fig. 1.
77Se NMR spectra of oxidized selenenylsulfide motifs. Proton-decoupled 77Se spectra were acquired at 20 °C and 14.1 T. (A) Oxidized SCUS. (B) Oxidized GCUG. (C) Oxidized GCAUG. (D) Oxidized GCGAUG.
Table 1.
NMR parameters of redox motifs
| Redox motif and form | Chemical shift, ppm | Half-height line width, Hz* | T1, s† |
| SCUS | |||
| Oxidized | 478.9 ± 0.1 | ||
| 433.7 ± 0.1 | 47 ± 11 | 0.10 | |
| 375.6 ± 0.2 | |||
| Reduced | −214.2 ± 0.1 | 410 ± 20 | |
| GCUG | |||
| Oxidized | 481.3 ± 0.1 | ||
| 432.4 ± 0.3 | 72 ± 12 | 0.13 | |
| 375.0 ± 0.2 | |||
| Reduced | −208.6 ± 0.4 | 276 ± 21 | |
| GCAUG | |||
| Oxidized | 373.0‡ | ||
| 279.7 ± 0.5 | 165 ± 20§ | 0.11 | |
| Reduced | −209.0 ± 0.2 | 255 ± 16 | |
| GCGAUG | |||
| Oxidized | 426.6 ± 0.2 | 500 ± 70 | 0.15 |
| 368.1 ± 1.3 | |||
| 290.0 ± 0.2 | 72 ± 8 | 0.09 | |
| Reduced | −212.6 ± 0.2 | 149 ± 4 |
Line width was measured without line broadening for SCUS and GCUG, 20-Hz line broadening for GCAUG, and 60-Hz line broadening for GCGAUG. Values are only reported for resonances whose signal to noise allowed reliable measurements of the peak area.
The measurement error is estimated as 5%.
Only one high signal to noise measurement is available.
Only two high signal to noise measurements are available.
Density Functional Theory Conformation Search and Gauge-Invariant 77Se Chemical Shifts.
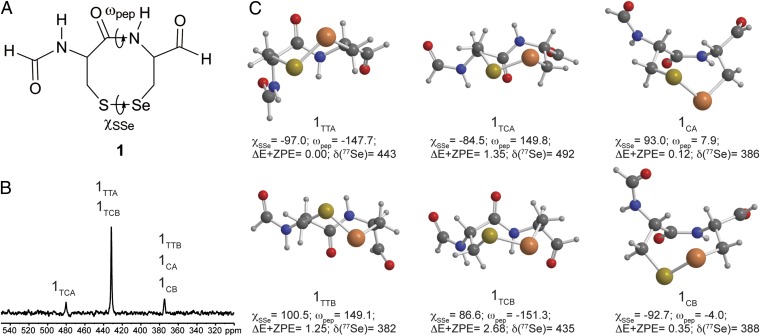
To delineate the origin of the multiple resonances, we used DFT calculations, focusing on the CU motif. A conformation search of the oxidized CU ring 1 model was performed using DFT (mPW1PW91) and an implicit solvation model (Fig. 2 and SI Appendix, Table S1). The lowest energy conformation on the energy corrected for the zero-point vibrational energy (E+ZPE) surface, twist trans conformation 1TTA, was similar to that observed for the eight-membered ring GCUG in rat TrxR, the only available selenoprotein structure with a CU motif (28). Similarly, conformations 1TCA and 1TCB were equivalent to CC rings found in the Protein Data Bank (PDB). A key feature of the trans isomers was a significantly distorted amide dihedral angle (ωpep ∼ ±150°) attributable to the high ring strain akin to trans cyclooctene. This strain distorted the selenenylsulfide dihedral angle (χSSe) relative to the idealized value from MeSeSMe (∼90°). In contrast, little ring strain was reflected in the ωpep and χSSe dihedrals of the cis isomers 1CA and 1CB. Short intraring Se···CO distances were found both computationally and experimentally for both CU- and CC-type oxidized rings and suggested a stabilizing effect by donation of electron density from the Se (or S) to the ring amide carbonyl.
Fig. 2.
(A) The structure of compound 1 and the dihedral angles that define the conformation. (B) 77Se NMR spectrum of oxidized GCUG labeled with the calculated structures that best match the experimental data. (C) DFT(mPW1PW91)–PCM conformations of the oxidized CU model 1. The dihedral angles, ΔE+ZPE (kcal/mol), and isotropic chemical shifts δ (ppm) are listed below the respective structure and in SI Appendix, Table S1.
Gauge-invariant atomic orbital DFT 77Se chemical shifts of these conformations were clustered around values similar to those observed experimentally: 380–385 ppm (1CA, 1CB, 1TTB), 435–445 ppm (1TTA, 1TCB), and 490 ppm (1TCA). The trans conformations were generally deshielded because of the effects of ring strain on the Se-S bond except for the trans twist conformer 1TTB, which was shifted up-field by 61 ppm compared with 1TTA because of shielding effects attributed to closer Se···CO interactions in the latter. From the calculated 77Se chemical shifts, the experimental 433- and 480-ppm resonances could be assigned as 1TTA and 1TCA. The experimental 375-ppm resonance was either the cis-amide conformation 1CA, 1CAB, or 1TTB. Previous conformational analysis of oxidized CC rings in peptides has suggested conformations with a cis-amide (11, 29). In contrast, analysis of CC motifs in the PDB concluded that the occurrence of the cis conformation was remarkably low (30). Hence, although the cis structures have 77Se chemical shifts in the region of interest, they may not have contributed to the observed spectra. Certainly, this conformation is not expected to be highly populated in the CU rings.
77Se NMR Properties of the Reduced Selenolthiol-Containing Motifs.
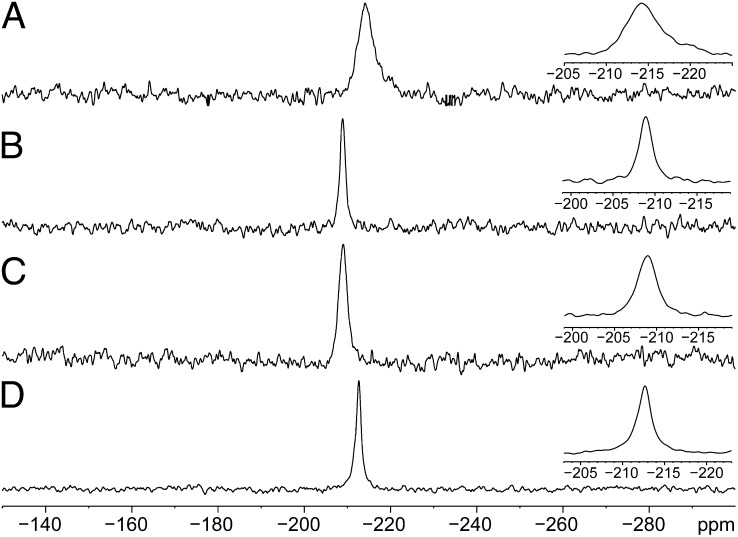
In the majority of selenoproteins, it is the reduced protein that encounters the target substrate (6). Therefore, we recorded the NMR spectra in the presence of the reducing agent dithiothreitol (DTT). The resonances of the reduced proteins appeared between −208.0 and −212.2 ppm (Table 1 and Fig. 3). The chemical shifts of SCUS and GCGAUG were up-field relative to those of GCUG and GCAUG. It should be noted that the Sec in all of these redox motifs is positioned one residue away from the charged C terminus and fully exposed to the solvent. Hence, the differences in chemical shifts and resonance widths may be attributed to variation in hydrogen bonds and/or nonbonded interactions pattern. Because the full-width at half-maximum (FWHM) of the SCUS motif was the broadest among the four motifs, we attribute the underlying phenomena to a dynamic exchange of a hydrogen bond between the charged selenolate (R-Se−) and water or with a serine hydroxyl side chain. The up-field move of the isotropic chemical shift was typical of hydrogen bond proton acceptors, although selenium hydrogen bonds are weak. It was previously proposed, and further supported here by our findings, that the hydroxyl group of Ser in SCCS or SCUS motifs may interact with the S or Se of the penultimate residue in these motifs (7). An up-field shift was also observed for GCGAUG, but in this case, the line width was the narrowest observed among the four reduced spectra. Geometrically, the Sec in this motif may be capable of hydrogen bonding with the Cys. In Cys-based redox motifs, the ability of a thiolate to form hydrogen bonds stabilizes it (31). We speculate that the 77Se spectra show similar trends, but this assumption needs to be directly corroborated by structural or distance sensitive measurements.
Fig. 3.
77Se NMR spectra of reduced selenolthiol motifs. Proton-decoupled 77Se spectra were acquired at 20 °C and 14.1 T. Samples were reduced with DTT. (A) Reduced SCUS. (B) Reduced GCUG. (C) Reduced GCAUG. (D) Reduced GCGAUG.
Conformational Mobility of the Oxidized Selenenylsulfide-Containing Motifs.
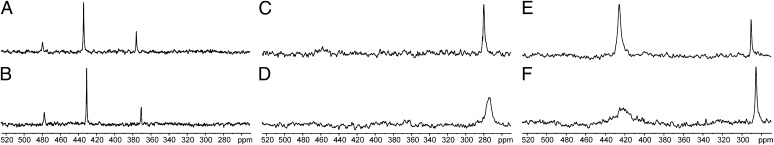
To better understand the conformational mobility of the different redox motifs, we measured the relaxation properties of Sec in the respective oxidized redox motifs (SI Appendix, Fig. S4 and Table 1). The T1 relaxation time varied between different redox motifs and within different conformations present in the same motif (e.g., GCGAUG). Differences in relaxation times were also observed for the traverse relaxation time T2, which was calculated here from the inverse of the FWHM. Here, similar to the spin-lattice relaxation time, GCGAUG exhibited a pronounced difference: about 500-Hz FWHM for the 426.6-ppm resonance vs. 70-Hz FWHM for the 290.0-ppm resonance. The variation in the calculated chemical-shift tensor (see SI Appendix, Table S1) is not sufficient to account for the differences in relaxation times. The temperature dependence of the NMR spectra points toward protein dynamics as the most likely cause. To gain more insight into these dynamics, we compared the spectra of SCUS, GCAUG, and GCGAUG at 25 and 4 °C (Fig. 4). Overall, the relative changes in isotropic chemical shifts between different temperatures were typical for the temperature-sensitive 77Se nucleus. However, for the GCGAUG ring, the downfield 426.6-ppm resonance was significantly broadened at lower temperature compared with the 290.0-ppm resonance. The magnetization loss observed for the 426.6-ppm resonance indicated a chemical-exchange process taking place at the time scale of measurements (high milliseconds to low microseconds). A likely explanation was that these were several ring conformations similar in structure that could interconvert at that time scale. The 290.0-ppm resonance could then arise from either one ring conformation or possibly a collection of several conformations that interconverted on a faster time scale than those contributing to the 426.6-ppm resonance. In the latter case, an average chemical shift would appear as that which was detected. The ring conformations corresponding to the 426.6-, 373.0-, and 290.0-ppm resonances were separated by a significant activation barrier preventing their interconversion in 20 °C. Molecular mechanics studies suggest barriers of 8–21 kcal/mol for interconversion of conformations of a CC ring (32). The differences in inherent flexibility corroborate our observed differences in T1 relaxation rate, with the 290-ppm resonance displaying the faster relaxation rate. This supported the presence of fast motion. The GCAUG motif also displayed signs of conformational mobility because the 279.7-ppm resonance was significantly broadened at lower temperature. In contrast, the dynamics of the SCUS motif was not much affected by the temperature.
Fig. 4.
77Se NMR spectra of the oxidized motifs acquired at different temperatures (the number of scans is similar). (A) Spectrum of SCUS acquired at 25 °C. (B) Spectrum of SCUS acquired at 4 °C. (C) Spectrum of GCAUG acquired at 25 °C. (D) Spectrum of GCAUG acquired at 4 °C. (E) Spectrum of GCGAUG acquired at 25 °C. (F) Spectrum of GCGAUG acquired at 4 °C.
Redox Potentials of Sec- and Cys-Containing Proteins.
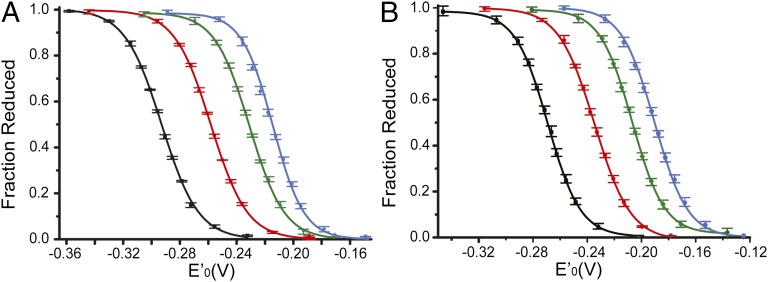
The NMR results supported the view that an increasing geometric strain in the ring of these redox motifs reduces mobility. The strain should also be expected to increase the redox potentials, but many additional factors can be involved. Among those factors are the residue pKa, solvation, weak forces, entropy, and nucleophilicity; the interplay among these factors is hard to deconvolve (5). Here, the redox potentials of Sec-containing motifs and their Cys homologs were thus measured to quantitate the effect of the geometric strain and the differences when either selenium or sulfur were involved. The proteins were equilibrated in buffers with known redox potentials, quenched, and then alkylated so that the reduced component could be quantified by mass shift (SI Appendix, Figs. S5–S7). It was feasible to measure the ratio of oxidized and reduced proteins from the mass spectrum because the alkylated forms ionized at similar efficiency as the oxidized protein forms (SI Appendix, Fig. S5). The ratios were subsequently used to determine the redox potentials of the Sec- and Cys-containing redox motifs using the Nernst equation (Fig. 5, Table 2, and SI Appendix, Figs. S8 and S9). As controls, redox potentials were also determined from gel-shift assays (SI Appendix, Figs. S10 and S11). The values derived from both methods were in good agreement but the mass spectrometry method was the more sensitive of the two and allowed us to measure the redox potentials within 1 mV (SI Appendix, Table S2).
Fig. 5.
Determination of the redox potentials of Sec- and Cys-containing motifs. (A) Sec-containing selenoproteins. GCGAUG, black; GCAUG, red; GCUG, green; SCUS, blue. (B) Sec-deficient Cys-containing variants. GCGACG, black; GCACG, red; GCCG, green; SCCS, blue. The error bars represent the range of measurements (that is, highest and lowest measurements) among three repetitions, using two independent preparations of each protein.
Table 2.
Redox potentials of representative Sec- and Cys-containing redox motifs as determined at pH 7.0
| Sec-containing redox motif | Redox potential, mV | Cys-containing redox motif | Redox potential, mV |
| SCUS | −215 ± 1 | SCCS | −190 ± 1 |
| GCUG | −231 ± 1 | GCCG | −207 ± 1 |
| GCAUG | −255 ± 1 | GCACG | −235 ± 1 |
| GCGAUG | −291 ± 1 | GCGACG | −269 ± 1 |
It is generally believed that the longer selenenylsulfide bond length would reduce the ring strain relative to that of disulfide-containing rings (15). As expected, the redox potentials of the Sec-containing proteins were lower than those of their Cys-containing homologs. The redox potentials also decreased (became more reducing) with the expansion in ring size and alleviation of geometric strain. The absolute magnitude of change was similar for the Sec- and Cys-containing rings. Notably, expansion from the 11-membered rings to the 14-membered rings resulted in the largest decreases in redox potential, suggesting that the 11-membered ring was indeed significantly strained. This is also where the values for the Sec-ring vs. the Cys-ring differed the most. Rather surprisingly, the differences in redox potentials between the Sec- and Cys-containing motifs were only between 20–25 mV. In comparison, the change of flanking residues from Gly to Ser resulted in a similar change in redox potentials–about two-thirds of that resulting from the substitution from Sec to Cys. Previous measurements of the Sec-containing forms of E. coli glutaredoxin 3 reported the differences in redox potentials between the selenium and sulfur forms to be larger (66–81 mV) (24). We believe that the smaller differences in redox potentials found here may be attributable to the positioning of the Sec at the C terminus. This location, which is frequently used by selenoproteins, is predominantly solvent-exposed, flexible, and in the immediate vicinity of the carboxylate negative charge. Similar results were recently reported for selenoprotein S, which carries a Sec at the C terminus of an intrinsically disordered domain (25). Hence, we conclude that the intrinsic redox potentials of the Sec-containing redox active ring motifs studied here are only about 20–25 mV lower than those of the corresponding Cys homologs.
Conclusions
In conclusion, 77Se isotropic chemical shifts, combined with structural information from DFT calculations, reveal the existence of several stable configurations of each Sec-containing redox motif under investigation. The results support a view in which weak, nonbonding interactions between the selenium and the amide carbon stabilize certain ring conformations. Thus, it is possible to resolve molecular features of the selenium environment that are not otherwise accessible, because other methods have been mostly unsuccessful in producing high-resolution structures of selenoproteins to date. Thus, we demonstrate herein that 77Se NMR spectroscopy is a powerful way to study the structure and function of selenium-containing proteins. The approach taken here can be extended to studies of selenium in proteins.
By comparing the Sec- and Cys-containing variants of redox motifs with different ring sizes, it is established that their redox potentials are highly dependent on the geometric strain and impact of flanking residues. In contrast, whether the motif contains Sec or Cys matters less in terms of the redox potential than generally believed. Thus, the combined information about structures, flexibilities, and redox potentials of Sec-containing redox motifs must clearly be considered for a full understanding of selenoproteins reactivities, rather than attributing the main causes of such reactivities to the sole presence of Sec. With several human diseases linked to the functions and unique reactivities of selenoproteins (1), the 77Se-based NMR methodology developed here and the results of our study will be important for future analyses of selenium-derived characteristics of selenoproteins and thereby for the understanding of their potential roles and functions in health and disease.
Materials and Methods
Expression and Purification of 77Se-Labeled Selenoproteins Carrying Different Redox Motifs.
The four Sec-containing motifs studied here were positioned as C-terminal fusion motifs on a Cys-less Trx scaffold. Trx was selected because of its stability, ease of expression, high yield, and pH tolerance (16). The two native Trx active site Cys residues were mutated to Ser, to avoid interference with our redox measurements. The desired Sec- or Cys-containing redox motifs were fused to Trx using a single Gly spacer (SI Appendix, Fig. S1). The expression vectors for the Sec-containing variants were transformed into BL21(DE3) along with the pSUABC vector that expresses the native selenium production proteins SelA, SelB, and SelC under their native promoter (14). To control the source of selenium in the growth media, we used a defined growth medium containing amino acids that was enriched with 77selenite. The full-length Sec-containing proteins were purified using ion-exchange chromatography. The final enrichment was 30–75% full-length selenium-containing protein. See SI Appendix, SI Materials and Methods for full details on the expression and purification of the proteins.
NMR Spectroscopy.
Solution-state NMR spectra were acquired on a 14.1 T Bruker AV600 spectrometer equipped with a Bruker 5-mm broadband outer coil liquid-state probe. Proton-decoupled 77Se spectra at 114.493 MHz were recorded using a spectral width of 96.1 KHz, an acquisition time of 0.085 s, with 250,000 scans, pulse delay of 0.5 s, a 90° pulse width of 15 μs, and WALTZ decoupling with a 2.78-kHz field during acquisition and 2.5-kHz field during the relaxation delay. NMR spectra were processed using backward linear prediction of the first four points of the free induction decay and 100 Hz exponential apodization. The data were processed with an exponential of about one-half of the natural line width. In Fig. 1, the line broadening applied was 50, 50, 50, and 100 Hz, respectively. In Fig. 3, the spectra were processed with 50-Hz line broadening, except for Fig. 3A, which used 150 Hz. In Fig. 4 and SI Appendix, Fig. S4, the line broadening was 100 Hz. 77Se chemical shifts are reported with respect to diphenyl diselenide used as an external secondary chemical-shift reference standard, set at 463 ppm (dimethyl selenide as primary reference at 0 ppm). The sample temperature was 20 °C, controlled to within 0.1 °C, unless otherwise specified. Protein concentrations in the NMR sample ranged from 5 to 9 mM protein (full-length and truncated forms), with an incorporation ratio of 30–75% in 20 mM Tris (pH 8.0), 50 mM NaCl, 1 mM EDTA, and 10% (vol/vol) 2H2O. When required, samples were reduced with DTT at ∼5× the molar protein concentration (i.e., 5–70 mM DTT). The chemical shifts were independent of either DTT or protein concentrations. DTT was removed by dialysis overnight using three buffer exchanges, and the spectrum was recorded again to ensure that the protein was unmodified. Measurements were repeated at least three times using samples with different incorporation, unless otherwise indicated. The chemical shifts were highly reproducible and the presented data represent the average of three to six independent protein preparations. More details about the NMR samples and measurements can be found in SI Appendix, SI Materials and Methods.
DFT Calculations.
DFT geometry optimizations were performed using Gaussian 09 (33), the mPW1PW91 exchange correlation functional, and a polarization continuum model (PCM) aqueous solvation field. The triple-zeta plus polarization functions (TZVP) basis set was used for C, N, O, and H (34). An additional set of diffuse functions was added to O and N. Sulfur and selenium were represented by the Wadt–Hay (35) and Ermler–Christiansen (36) relativistic effective core potential basis sets, respectively, augmented with polarization and diffuse functions. Each reported structure was confirmed to be a minimum on the potential energy surface by vibrational analysis. Chemical-shift calculations in aqueous solvation field were conducted from the optimized structures with the relativistic effective core potential basis set of selenium replaced by an all-electron TZVP basis set with additional polarization and diffuse functions.
Measurements of Redox Potentials.
We opted to measure the redox potential of the proteins by electrospray ionization (ESI) mass spectrometry because the different chemical species can be directly identified. We relied on alkylation of the reduced form of the protein by iodoacetamide or N-ethylmaleimide to gain a mass shift from the oxidized protein that can easily be detected. In this experiment, the protein was equilibrated in different buffers whose redox potential was poised by fixing the ratio between GSH and GSSG or reduced and oxidized DTT. Once equilibrium was reached, the protein was denatured with acid and subsequently alkylated. The oxidized protein was unmodified, whereas in the reduced protein, the exposed Cys and Sec were alkylated and acquired a mass shift. The different protein forms were easily resolved by ESI mass spectrometry (SI Appendix, Figs. S5–S7), and the ratio of oxidized to alkylated protein was subsequently determined. Using the measured ratio and the known values of the either DTTox/DTTred or GSSG/GSH ratio, the Nernst equation was used to calculate the redox potential as detailed in SI Appendix, SI Materials and Methods.
A key requisite for the validity of this approach was that the oxidized and alkylated forms of the proteins ionize at similar efficiencies. As SI Appendix, Fig. S5 demonstrates, when we mixed a 1:1 molar ratio of the oxidized and of the alkylated protein, the ionization efficiencies were the same. Hence, the detection of the different forms by ESI mass spectrometry was representative of their concentrations in the sample. Truncated proteins resulting from translational termination at the Sec-encoding UGA were readily distinguished and could be excluded from further analysis. A typical representative dataset for the SCUS protein is shown in SI Appendix, Fig. S6 as an example (and examples for all of the proteins are shown in SI Appendix, Fig. S7). The titration curves for the Sec-containing proteins are presented in SI Appendix, Fig. S8, and those for the Cys-containing proteins are presented in SI Appendix, Fig. S9. As a control, we also preformed gel-shift assays using alkylation with PEG 1200 maleimide (MM(PEG)24). Those assays relied on a 1.2-kDa mass addition per alkylation of each free Cys and Sec residue. The percentage of oxidized vs. alkylated protein could then be visualized using SDS/PAGE (SI Appendix, Figs. S10 and S11). SI Appendix, Table S2 compares the redox potentials measured by ESI mass spectrometry and by gel-shift assays.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation (NSF) Grant MCB-1054447 (to S.R.). C.A.B. acknowledges support from NSF Grant CHE-0750413. E.S.J.A. acknowledges support from the Karolinska Institutet, the Swedish Cancer Society, the Swedish Foundation for Strategic Research, and the Swedish Research Council.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319022111/-/DCSupplemental.
References
- 1.Rayman MP. Selenium and human health. Lancet. 2012;379(9822):1256–1268. doi: 10.1016/S0140-6736(11)61452-9. [DOI] [PubMed] [Google Scholar]
- 2.Hatfield DL, Tsuji PA, Carlson BA, Gladyshev VN. Selenium and selenocysteine: Roles in cancer, health, and development. Trends Biochem Sci. 2014;39(3):112–120. doi: 10.1016/j.tibs.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hondal RJ, Marino SM, Gladyshev VN. Selenocysteine in thiol/disulfide-like exchange reactions. Antioxid Redox Signal. 2013;18(13):1675–1689. doi: 10.1089/ars.2012.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu J, Holmgren A. Selenoproteins. J Biol Chem. 2009;284(2):723–727. doi: 10.1074/jbc.R800045200. [DOI] [PubMed] [Google Scholar]
- 5.Arnér ESJ. Selenoproteins-What unique properties can arise with selenocysteine in place of cysteine? Exp Cell Res. 2010;316(8):1296–1303. doi: 10.1016/j.yexcr.2010.02.032. [DOI] [PubMed] [Google Scholar]
- 6.Arnér ESJ. Focus on mammalian thioredoxin reductases—important selenoproteins with versatile functions. Biochim Biophys Acta. 2009;1790(6):495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Gromer S, et al. Active sites of thioredoxin reductases: Why selenoproteins? Proc Natl Acad Sci USA. 2003;100(22):12618–12623. doi: 10.1073/pnas.2134510100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckenroth BE, Lacey BM, Lothrop AP, Harris KM, Hondal RJ. Investigation of the C-terminal redox center of high-Mr thioredoxin reductase by protein engineering and semisynthesis. Biochemistry. 2007;46(33):9472–9483. doi: 10.1021/bi7004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson L, Arscott LD, Ballou DP, Williams CH, Jr, Arnér ESJ. Studies of an active site mutant of the selenoprotein thioredoxin reductase: The Ser-Cys-Cys-Ser motif of the insect orthologue is not sufficient to replace the Cys-Sec dyad in the mammalian enzyme. Free Radic Biol Med. 2006;41(4):649–656. doi: 10.1016/j.freeradbiomed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Hondal RJ. Using chemical approaches to study selenoproteins-focus on thioredoxin reductases. Biochim Biophys Acta. 2009;1790(11):1501–1512. doi: 10.1016/j.bbagen.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruggles EL, Deker PB, Hondal RJ. Synthesis, redox properties, and conformational analysis of vicinal disulfide ring mimics. Tetrahedron. 2009;65(7):1257–1267. doi: 10.1016/j.tet.2008.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hondal RJ, Ruggles EL. Differing views of the role of selenium in thioredoxin reductase. Amino Acids. 2011;41(1):73–89. doi: 10.1007/s00726-010-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kryukov GV, et al. Characterization of mammalian selenoproteomes. Science. 2003;300(5624):1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 14.Arnér ESJ, Sarioglu H, Lottspeich F, Holmgren A, Böck A. High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J Mol Biol. 1999;292(5):1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 15.Wessjohann LA, Schneider A, Abbas M, Brandt W. Selenium in chemistry and biochemistry in comparison to sulfur. Biol Chem. 2007;388(10):997–1006. doi: 10.1515/BC.2007.138. [DOI] [PubMed] [Google Scholar]
- 16.Lillig CH, Holmgren A. Thioredoxin and related molecules—from biology to health and disease. Antioxid Redox Signal. 2007;9(1):25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- 17.Gettins P, Crews BC. 77Se NMR characterization of 77Se-labeled ovine erythrocyte glutathione peroxidase. J Biol Chem. 1991;266(8):4804–4809. [PubMed] [Google Scholar]
- 18.House KL, Dunlap RB, Odom JD, Wu ZP, Hilvert D. Structural characterization of selenosubtilisin by Se-77 NMR-spectroscopy. J Am Chem Soc. 1992;114(22):8573–8579. [Google Scholar]
- 19.Mobli M, Morgenstern D, King GF, Alewood PF, Muttenthaler M. Site-specific pKa determination of selenocysteine residues in selenovasopressin by using 77Se NMR spectroscopy. Angew Chem. 2011;50(50):11952–11955. doi: 10.1002/anie.201104169. [DOI] [PubMed] [Google Scholar]
- 20.Schaefer SA, et al. 77Se enrichment of proteins expands the biological NMR toolbox. J Mol Biol. 2013;425(2):222–231. doi: 10.1016/j.jmb.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayse CA. The theoretical 77Se chemical shift as a probe of selenium state in selenoproteins and their mimics. Inorg Chem. 2004;43(4):1208–1210. doi: 10.1021/ic035283l. [DOI] [PubMed] [Google Scholar]
- 22.Bayse CA. Considerations for reliable calculation of Se-77 chemical shifts. J Chem Theory Comput. 2005;1(6):1119–1127. doi: 10.1021/ct050136t. [DOI] [PubMed] [Google Scholar]
- 23.Rozovsky S. 77Se NMR spectroscopy of selenoproteins. In: Bayse CA, Brumaghim JL, editors. Biochalcogen Chemistry: The Biological Chemistry of Sulfur, Selenium, and Tellurium. Washington, DC: Am Chem Soc; 2013. pp. 127–142. [Google Scholar]
- 24.Metanis N, Keinan E, Dawson PE. Synthetic seleno-glutaredoxin 3 analogues are highly reducing oxidoreductases with enhanced catalytic efficiency. J Am Chem Soc. 2006;128(51):16684–16691. doi: 10.1021/ja0661414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Li F, Rozovsky S. The intrinsically disordered membrane protein selenoprotein S is a reductase in vitro. Biochemistry. 2013;52(18):3051–3061. doi: 10.1021/bi4001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cenas N, et al. Interactions of quinones with thioredoxin reductase: A challenge to the antioxidant role of the mammalian selenoprotein. J Biol Chem. 2004;279(4):2583–2592. doi: 10.1074/jbc.M310292200. [DOI] [PubMed] [Google Scholar]
- 27.Johansson L, Gafvelin G, Arnér ESJ. Selenocysteine in proteins-properties and biotechnological use. Biochim Biophys Acta. 2005;1726(1):1–13. doi: 10.1016/j.bbagen.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Cheng Q, Sandalova T, Lindqvist Y, Arnér ESJ. Crystal structure and catalysis of the selenoprotein thioredoxin reductase 1. J Biol Chem. 2009;284(6):3998–4008. doi: 10.1074/jbc.M807068200. [DOI] [PubMed] [Google Scholar]
- 29.Creighton CJ, Reynolds CH, Lee DHS, Leo GC, Reitz AB. Conformational analysis of the eight-membered ring of the oxidized cysteinyl-cysteine unit implicated in nicotinic acetylcholine receptor ligand recognition. J Am Chem Soc. 2001;123(50):12664–12669. doi: 10.1021/ja016505m. [DOI] [PubMed] [Google Scholar]
- 30.Hudáky I, et al. Vicinal disulfide bridge conformers by experimental methods and by ab initio and DFT molecular computations. Proteins. 2004;55(1):152–168. doi: 10.1002/prot.10581. [DOI] [PubMed] [Google Scholar]
- 31.Jeng MF, Holmgren A, Dyson HJ. Proton sharing between cysteine thiols in Escherichia coli thioredoxin: Implications for the mechanism of protein disulfide reduction. Biochemistry. 1995;34(32):10101–10105. doi: 10.1021/bi00032a001. [DOI] [PubMed] [Google Scholar]
- 32.Avizonis DZ, FarrJones S, Kosen PA, Basus VJ. Conformations and dynamics of the essential cysteinyl-cysteine ring derived from the acetylcholine receptor. J Am Chem Soc. 1996;118(51):13031–13039. [Google Scholar]
- 33.Frisch MJ, et al. Gaussian 09. Wallingford, CT: Gaussian; 2009. [Google Scholar]
- 34.Dunning TH. Gaussian basis functions for use in molecular calculations. 3. Contraction of (10S6P) atomic basis sets for first-row atoms. J Chem Phys. 1971;55(2):716–723. [Google Scholar]
- 35.Wadt WR, Hay PJ. ab initio effective core potentials for molecular calculations - potentials for main group elements Na to Bi. J Chem Phys. 1985;82(1):284–298. [Google Scholar]
- 36.Hurley MM, Pacios LF, Christiansen PA, Ross RB, Ermler WC. ab initio relativistic effective potentials with spin-orbit operators. 2. K through Kr. J Chem Phys. 1986;84(12):6840–6853. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.