Significance
Hantavirus pulmonary syndrome (HPS) is a rare but often fatal disease caused by infection with New World hantaviruses. A limitation to understanding the pathogenesis of HPS and developing medical countermeasures against this disease is a lack of experimental disease models. In this study we describe the characterization of a novel nonhuman primate model of HPS. After infection with deer mouse-only–passaged Sin Nombre virus, macaques developed severe respiratory disease indicative of HPS. Viremia and hematological abnormalities were the earliest markers of ensuing disease, and the hyperpermeability associated with the onset of respiratory distress coincided with dysregulation of host responses exclusively in the pulmonary endothelium. This model will help advance our understanding of HPS and preclinical development of therapeutic strategies.
Keywords: New World hantaviruses, emerging pathogen, disease modeling
Abstract
The pathophysiology of hantavirus pulmonary syndrome (HPS) remains unclear because of a lack of surrogate disease models with which to perform pathogenesis studies. Nonhuman primates (NHP) are considered the gold standard model for studying the underlying immune activation/suppression associated with immunopathogenic viruses such as hantaviruses; however, to date an NHP model for HPS has not been described. Here we show that rhesus macaques infected with Sin Nombre virus (SNV), the primary etiological agent of HPS in North America, propagated in deer mice develop HPS, which is characterized by thrombocytopenia, leukocytosis, and rapid onset of respiratory distress caused by severe interstitial pneumonia. Despite establishing a systemic infection, SNV differentially activated host responses exclusively in the pulmonary endothelium, potentially the mechanism leading to acute severe respiratory distress. This study presents a unique chronological characterization of SNV infection and provides mechanistic data into the pathophysiology of HPS in a closely related surrogate animal model. We anticipate this model will advance our understanding of HPS pathogenesis and will greatly facilitate research toward the development of effective therapeutics and vaccines against hantaviral diseases.
Hantavirus pulmonary syndrome (HPS), otherwise referred to as “hantavirus cardiopulmonary syndrome,” is an acute respiratory disorder that was described initially in 1993 during an outbreak in the Four Corners region of the United States (1). The etiological agent was identified quickly as a novel hantavirus and subsequently was named “Sin Nombre virus” (SNV) (2). Over the last two decades several species of hantaviruses have been documented throughout the Americas, many of which are etiological agents of HPS (3). More than 2,000 cases of HPS have been reported with mortality rates ranging from 30 to 50% (4). Although HPS often occurs as isolated and sporadic incidents, outbreaks have been documented, most recently in Yosemite National Park, which resulted in nine cases of HPS with three deaths and an international public health response (5–8). The Yosemite outbreak highlights the importance of developing effective medical counter measures against HPS; none are currently available.
The genus Hantavirus (family Bunyaviridae) comprises a unique group of viruses that are maintained in nature and transmitted to humans from specific small-mammal reservoirs (4). In addition to HPS, pathogenic hantaviruses also cause hemorrhagic fever with renal syndrome (HFRS), which can result from infection with Old World hantaviruses. Hantavirus infections are associated with vascular leakage that is believed to be primarily immune mediated, although the target organs for each syndrome differ; HPS primarily affects the lung, whereas HFRS targets the kidneys. Our understanding of the pathogenesis of these viruses is limited, in large part because of a lack of animal models (4, 9, 10). Currently the only model of lethal hantavirus disease is the Syrian hamster, which, after infection with Andes virus, develops severe disease that faithfully recapitulates the cardiopulmonary phase of HPS in humans (11–13). Nonhuman primates (NHPs) often are viewed as the gold-standard model for the study of emerging viral pathogens, especially those that are immunopathogenic, because these animals are believed to recapitulate most accurately the underlying deleterious host immune responses associated with disease progression in humans. To date, attempts at developing an NHP model for HPS or HFRS have largely been unsuccessful (14, 15). The lone NHP model for hantavirus infection is a mild HFRS-like disease that follows infection of cynomolgus macaques with Puumala virus propagated in bank voles (16). Cynomolgus monkeys develop mild signs of disease including lethargy, proteinuria, and microhematuria, indicative of acute nephropathy (17, 18). Here we have developed an NHP model for HPS based on infection of rhesus macaques with SNV passaged in deer mice. This model provided the first, to our knowledge, in-depth pathogenesis study in the nearest surrogate host and identified virus replication, systemic hematologic abnormalities, and lung-specific proinflammatory responses as hallmarks of HPS pathogenesis. This model will advance the development of intervention strategies combating infections with HPS-causing hantaviruses.
Results
Disease Progression.
Seventeen rhesus macaques (Macaca mulatta), all negative for anti-hantavirus antibodies, were inoculated with clarified lung homogenates prepared from deer mice infected with a deer mouse-only–passaged SNV (19) (DM-SNV, n = 10), naive (uninfected) deer mouse lung homogenates (n = 3), or purified SNV derived from propagation in Vero cells (VA-SNV, n = 4) via multiple routes using a protocol optimized for infection with respiratory viruses (20). Signs of infection were not apparent until 6 d postinoculation (dpi), at which point one animal infected with the VA-SNV (NHP no. VA-SNV 1) demonstrated mild, transient signs of illness (slightly elevated respiration rate) which resolved within 48 h. None of the remaining three animals in the VA-SNV group (VA-SNV 2, VA-SNV 3, VA-SNV 4) and none of the mock-infected animals (Mock 1, Mock 2, Mock 3) demonstrated any signs of infection throughout the entire study. In contrast, seven NHPs (DM-SNV 1, DM-SNV 3, DM-SNV 5, DM-SNV 6, DM-SNV 7, DM-SNV 8, and DM-SNV 10) infected with DM-SNV developed severe respiratory disease indicative of HPS (Table S1). Pulmonary manifestations were noted first around 14–16 dpi and initially presented as coughing and abnormal breathing patterns (rapid and shallow abdominal breathing) with occasional chest crackles apparent upon physical examination. Within 24–72 h of onset, respiratory disease progressed rapidly to acute severe respiratory distress. At the time of euthanasia, animals were hypoxic, as suggested by pale pink or more commonly bluish mucus membranes and had elevated temperature (average, 39.8 °C; range, 37.3–40.5 °C). The average time to severe HPS in NHPs was 18 d (range, 15–22 d), which is strikingly similar to the incubation period in humans (21, 22). Six animals that developed HPS had detectable anti-hantavirus IgG antibodies in terminal sera samples with ELISA titers ranging from 400 to ≥12,800 (Table S1). A single macaque (DM-SNV 3, euthanized at 18 dpi) remained seronegative, and two other animals (DM-SNV 1 and 10, euthanized at 15 and 16 dpi, respectively) were serologically equivocal with titers of 100. Of the three animals inoculated with DM-SNV that did not develop HPS, one did not seroconvert (DM-SNV 4), and the other two had anti-hantavirus IgG titers of 400 (DM-SNV 2) and ≥12,800 (DM-SNV 9). All four animals infected with VA-SNV were seropositive with titers of 3,200 (VA-SNV 2) or ≥12,800 (VA-SNV 1, VA-SNV 3, VA-SNV 4). The development of HPS in NHPs after inoculation with DM-SNV was statistically significant compared with animals inoculated with VA-SNV (70% versus 0%, P = 0.0350 by Fisher’s exact test).
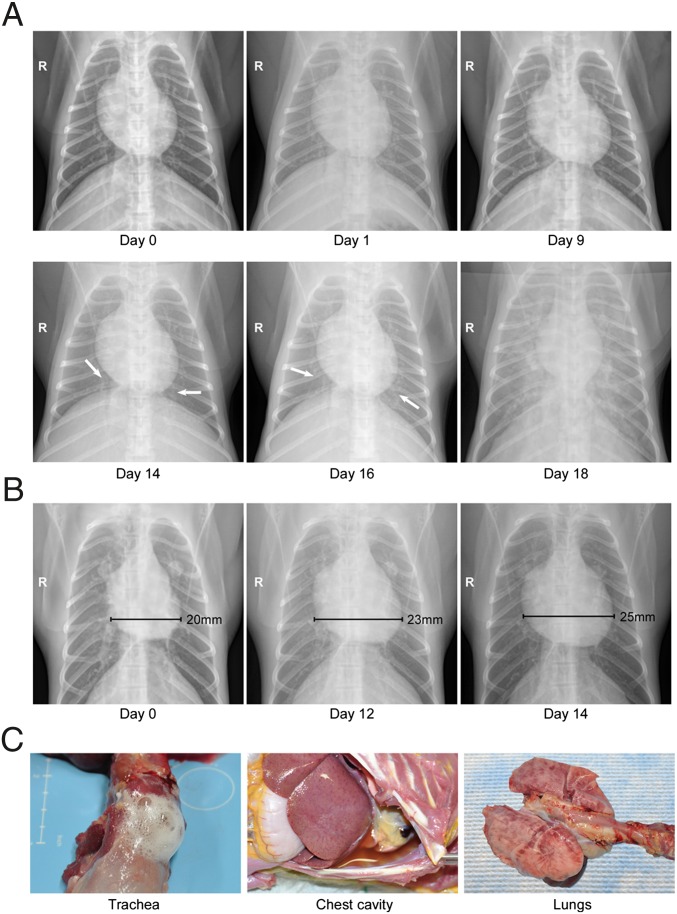
The development of respiratory disease was monitored by digital radiographic imaging. Beginning at 6 or 9 dpi (approximately 10 d before respiratory distress), small areas of increased density indicating interstitial infiltrates were noted in the right lower lobe of the majority of infected NHPs (Fig. 1A). The predilection for the early involvement of the right lung likely reflects the anatomy of the respiratory tract favoring infection in the right side through intratracheal installation and has been observed previously in other NHP models of respiratory disease (23). Over the next 6 d, the affected areas spread to include right and left lower and middle lobes. At the time when respiratory signs were apparent (∼24–72 h before the animals were euthanized), severe interstitial infiltrates were observed throughout the lungs, as is consistent with severe pulmonary edema and pneumonia. In addition, pleural fissure lines suggestive of pleural effusion as well as cardiomegaly, particularly right ventricle enlargement, were noted in radiographs taken within 48 h of euthanasia (Fig. 1B). Combined, the radiographs suggest edema from cardiac failure, although echocardiography would be necessary to characterize cardiac function and differentiate between pericardial effusion and dilated cardiomyopathy. In accordance with the radiographical observations, infected animals developed tachypnea over the course of the study with an average rate of 72 breaths per minute recorded at the time of euthanasia compared with 36 breaths per minute prechallenge. Nasal, oral, and rectal swabs collected from all animals were uniformly negative for viral RNA, suggesting that even at the peak of disease SNV is unlikely to transmit via close contact.
Fig. 1.
Radiographical examinations and gross pathology of macaques that developed HPS. Rhesus macaques infected with DM-SNV developed severe respiratory distress indicative of HPS. (A) Radiographic imaging. Chest X-rays were taken at regular intervals during the course of infection. Shown are serial images from a single representative animal demonstrating a rapid progression from small areas of increased density suggestive of light interstitial infiltrates to diffuse bilateral consolidation indicating severe pulmonary edema or pneumonia. (B) Right ventricle enlargement. Several animals that developed HPS demonstrated an expanded right ventricle suggestive of acute cardiac failure. (C) Gross pathology. At the time of euthanasia the majority of macaques that developed HPS presented with frothy tracheal exudates and red-tinged pleural effusions. The lungs were edematous, firm, and failed to collapse. There was multifocal consolidation and dark red discoloration that was most prevalent on the dorsal surfaces of the lung. These findings are consistent with interstitial pneumonia.
Histopathological Analysis.
Gross pathological abnormalities were most prominent in the thoracic cavity of animals that developed HPS. Confirming the radiographical observations, severe pleural and pericardial effusion was noted in most animals, with abundant straw-colored fluid present (Fig. 1C). Moderate edema also was present in the mediastinum of several NHPs, and frothy fluid was observed in the trachea and bronchi. The mediastinal and bronchial lymph nodes were generally swollen and edematous. The most striking gross pathological changes were in the lungs, which were diffusely dense and wet with multifocal to coalescing areas of hyperemia. Mild hepatomegaly was noted in a minority (n = 2) of NHPs necropsied. No other irregularities were noted.
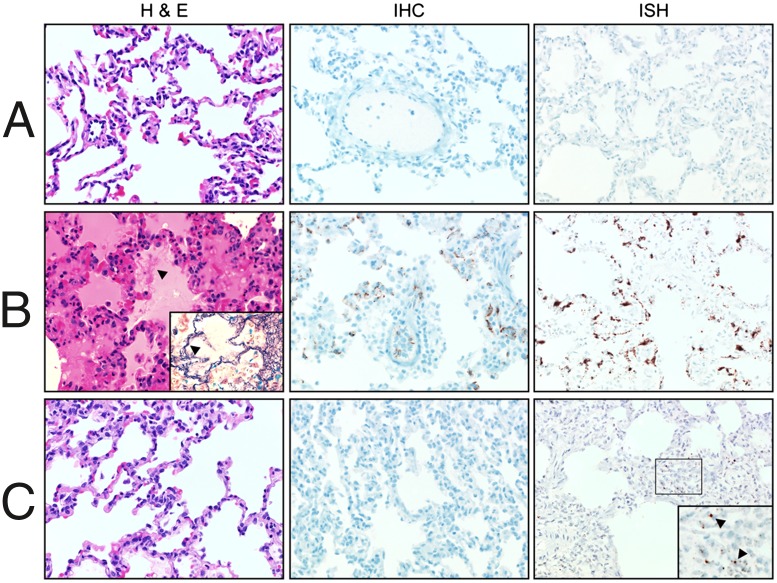
Histologically, the most prominent changes were noted in the lungs of NHPs that developed HPS. Consistent with the human condition (24), HPS in NHPs was characterized by moderate to severe interstitial pneumonia (Fig. 2). In most instances the changes were multifocal to coalescing and were characterized by thickening of the alveolar septae with edema, fibrin, macrophages, and fewer neutrophils. Multifocal type II pneumocyte hyperplasia also was noted in these animals. The remaining tissues analyzed demonstrated no discernable pathological changes, with the exception of liver samples from two animals that, in addition to the histological changes noted in lung samples, developed multifocal hepatic coagulative necrosis with acute inflammation (Fig. S1B). This lesion is consistent with acute viral hepatitis and is most likely a result of SNV infection. Although liver pathology is not commonly associated with SNV infection of humans, similar abnormalities have been noted in the hamster model of HPS (11, 25). No histological abnormalities were noted in any tissue samples collected from the mock-infected animals or in tissue samples collected from NHPs inoculated with VA-SNV.
Fig. 2.
Histopathology of lungs from control and infected macaques. Rhesus macaques were inoculated with clarified lung homogenates from naive deer mice (A) or with DM-SNV (B) or VA-SNV (C). Shown are lung samples collected from representative animals in each group necropsied between 18 and 21 dpi and stained with H&E or tested for the presence of virus by IHC (anti-N antigen) or ISH (anti-N RNA). Consistent with the clinical presentation, lung samples from macaques infected with DM-SNV demonstrated multifocal to coalescing interstitial pneumonia characterized by thickening of the alveolar septae with edema, fibrin (arrowhead; Inset is stained for fibrin with phosphotungstic acid-hematoxylin), macrophages, and fewer neutrophils. Diffuse, abundant viral antigen and RNA were detected in the pulmonary endothelium in these animals. In contrast, matched samples from animals inoculated with VA-SNV demonstrated no histological abnormalities, with drastically reduced detection of viral RNA by ISH and no detectable viral antigen by IHC.
Despite the localized histological changes, SNV RNA was readily detectable by real-time RT-PCR in all tissue samples collected from animals that developed HPS, confirming that DM-SNV achieved widespread and systemic infection (Fig. S1A). Consistent with the quantitative RT-PCR (qRT-PCR) analysis, NHPs infected with DM-SNV demonstrated diffuse, abundant viral RNA by in situ hybridization (ISH) in endothelial cells in all organs analyzed, including lung, liver, spleen, kidney, and heart (Fig. 2B and Fig. S1B). Similar results were obtained for viral antigen by immunohistochemistry (IHC) with hantaviral nucleocapsid protein (N) present in the endothelial cells of these organs. Confirming the tropism for pulmonary endothelial cells, N antigen was observed predominantly in CD31+ cells (Fig. S2). In contrast, VA-SNV infection either was focal, primarily in bronchial lymph nodes, lung, and spleen samples, or cleared rapidly from animals, with low levels of viral RNA detected by qRT-PCR. ISH demonstrated drastically reduced numbers of SNV RNA-positive cells in lung samples (Fig. 2C), and no viral RNA was detected in other organs analyzed. Likewise, viral antigen was not detected in any organs from these animals analyzed by IHC. SNV was isolated from bronchial lymph nodes, lung, and spleen samples from animals infected with DM-SNV but not from animals receiving VA-SNV.
Viremia, Hematology, Serum Biochemistry, and Coagulation Parameters.
Viremia, as measured by the detection of viral RNA, was the earliest indicator of the ensuing disease and was noted in all animals that demonstrated signs of illness, including the lone NHP (VA-SNV 1) that experienced mild, transient signs of infection following inoculation with VA-SNV. In animals that developed HPS, SNV RNA was detected first by RT-PCR in whole-blood samples and sera between 12 and 16 dpi and preceded respiratory distress and death by 4–10 d (average of 5.2 d). Peak titers were between 104 and 105 S-segment copies/mL of whole blood and were observed generally in samples collected 2 or 3 d before euthanasia. Hematology revealed thrombocytopenia in NHPs that developed HPS (Table S2). In addition, these animals demonstrated leukocytosis, which was caused predominantly by neutrophilia, eosinophilia, and monocytosis. The remaining hematological parameters evaluated did not demonstrate any discernible changes (Table S2). Throughout the course of the experiment, clinically relevant changes were not observed in the majority of biochemical mediators or coagulation parameters monitored. Increased activated partial thromboplastin time and glucose and aspartate aminotransferase concentrations and decreased alkaline phosphatase concentration were noted in several NHPs; however, these changes were observed only perimortem and therefore are not likely to have contributed to the development of HPS. d-dimers were not detected in any of the plasma samples collected.
Immunological Responses to Infection.
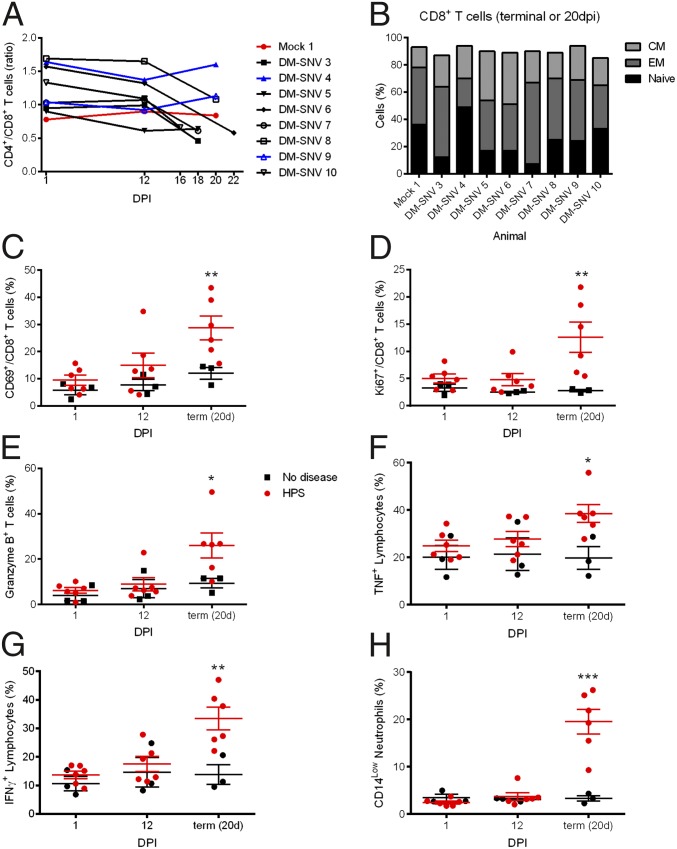
To examine the activation of the immune response, polychromatic flow cytometry was performed on peripheral blood mononuclear cells (PBMCs) isolated at 1, 12, and 20 dpi or at the time when animals that developed HPS were euthanized, from a single mock-inoculated animal (Mock 1), and from eight NPHs inoculated with DM-SNV (DM-SNV 3–DM-SNV 10). Although the proportion of CD4+/CD8+ T cells remained constant in the healthy group, this ratio decreased markedly in animals that developed HPS, especially between 12 dpi and the terminal time point, indicating a proportionally high abundance of cytotoxic T lymphocytes late in infection (Fig. 3A). To characterize the phenotype of the CD8+ responses, we examined their expression of effector and memory markers (CD28 and CD95). The two animals that did not seroconvert (Mock 1 and DM-SNV 4) had the highest percentage of naive CD8+ T cells, whereas infected NHPs generally had higher percentages of memory cells (often effector memory cells, but in some cases there was an increase in central memory cells) (Fig. 3B). CD69 is one of the most sensitive and earliest markers of activation, and its presence indicates recently activated antigen-specific lymphocytes (26). The proportion of CD8+ T cells expressing CD69 was elevated slightly at 12 dpi and was elevated significantly at the terminal time point in diseased NHPs as compared with healthy animals (Fig. 3C). Similarly, the percent of cells expressing Ki67, a reliable measure of cell proliferation (27), was significantly elevated uniquely in the DM-SNV–inoculated NHPs that developed HPS, but only at the terminal time point (Fig. 3D). To assess the effector potential of total T cells (CD3+), cells were stained for granzyme B expression. Only T cells in the diseased group at the terminal stage showed an increase in the proportion of cells expressing this cytolytic factor (Fig. 3E).
Fig. 3.
Animals that develop HPS mount a robust immune response at the time of disease. PBMCs isolated from macaques at 1, 12, and 20 dpi (for survivors) or at the time of euthanasia were stained with panels of antibodies to distinguish cell populations. (A) The ratio of CD4+ and CD8+ T cells (CD3+ cells) decreased in animals that developed HPS. (B) Memory cells were determined by their CD28 and CD95 expression on CD8+ T cells (CD3+) at the terminal stages of disease (or at 20 dpi for survivors). Animals that developed HPS had an increase in memory (effector and/or memory) cell percentages. (C–H) Cells were stained with the indicated antibodies (y axis), and percentages of cells expressing markers are shown. (E–G) PBMCs were stimulated with phorbol12-myristate13-acetate and ionomycin for 6 h, with the addition of brefeldin A for the last 4.5 h, and were stained with the indicated antibodies. Cytokine expression was increased in PBMCs from animals that developed HPS. *P < 0.05, **P < 0.01, ***P < 0.0.001.
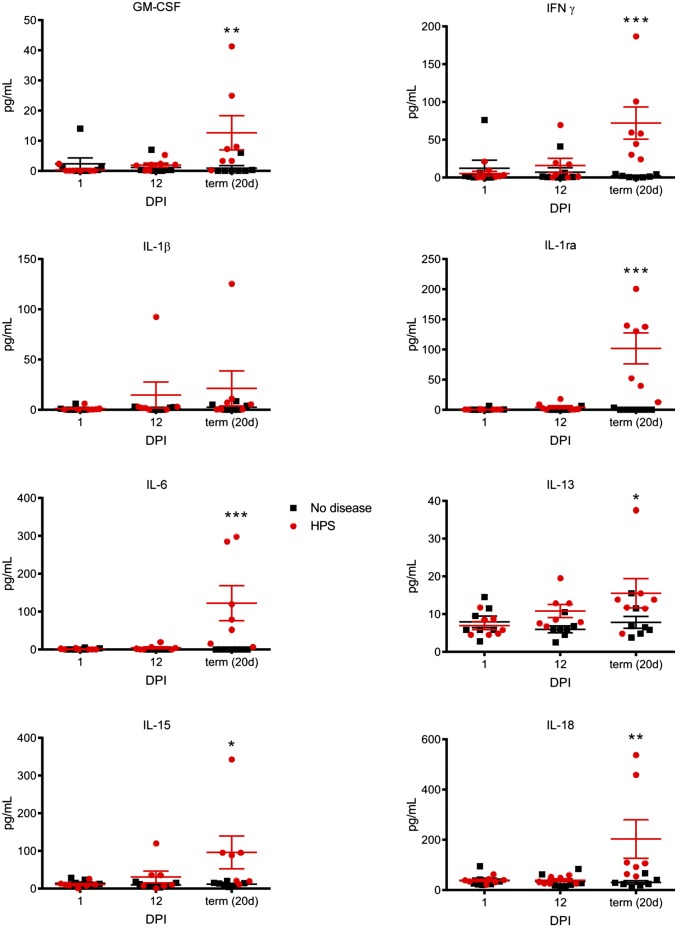
Increased cytokine expression is thought to contribute to the capillary leakage observed in HPS. Analysis of serum concentrations of immunological mediators revealed increases in proinflammatory Th1 and Th2 cytokines including IFN-γ, IL-1β, IL-6, IL-18, and IL-13, the anti-inflammatory IL-1 receptor antagonist IL-15, and GM-CSF in NHPs that developed HPS, although only at the time when respiratory signs of disease were apparent (Fig. 4). Serum concentrations of VEGF remained static throughout the course of the experiment. To determine whether circulating lymphocytes secreted proinflammatory cytokines, the proportion of cells producing TNF or IFN-γ in response to nonspecific stimulation was assessed. Both TNF and IFN-γ were increased slightly at 12 dpi but increased significantly at the terminal stage of disease as compared with NHPs that did not develop disease (Fig. 3 F and G). Neutrophilia, with a left-shift, often is associated with HPS and is used as a supportive diagnostic tool in human diagnoses (28). PBMCs were stained for CD14, and the relative abundance of neutrophils in these samples was determined using previously reported gating strategies (29). Only NHPs with terminal HPS displayed neutrophilia, and these cells were increased drastically (Fig. 3H). These neutrophils did not express significant levels of proliferation (Ki67) and activation (CD69) markers, suggesting an immature phenotype, likely in transit from the bone marrow to infected tissues. In all cases, the activation of the immune response was limited primarily to the terminal stage of disease and was activated only slightly at 12 dpi, a time only days before the onset of disease in these animals.
Fig. 4.
Animals that develop HPS have increased concentrations of immune mediators in serum. Concentrations of cytokines and chemokines were determined in serum samples collected from macaques at 1, 12, and 20 dpi (for survivors) or at the time of euthanasia using a multiplex NHP cytokine detection kit. At the time of euthanasia, animals that developed HPS had significantly increased levels of several immune mediators, including proinflammatory Th1 and Th2 cytokines (IFN-γ, IL-1β, IL-6, IL-18, and IL-13); the anti-inflammatory IL1 receptor antagonist (IL-1ra); and IL-15 and GM-CSF. *P < 0.05, **P < 0.01, ***P < 0.0.001.
Despite a systemic infection of endothelial cells in all organs analyzed, host responses were elevated primarily in lung samples (Fig. 5), coinciding with the pathological abnormalities noted in these animals and the clinical presentation of HPS. CD3 antigen was increased significantly in lung specimens collected from NHPs that developed HPS and was detected primarily in lymphocytes found within bronchiolar-associated lymphoid tissue (BALT) and in pulmonary circulation (Fig. 5 A and B). IL-4 and IFN-γ also were detected in lymphocytes of BALT and pulmonary circulation, and medium-sized lymphocytes were found within the alveolar spaces. Both antigens were increased dramatically (2.5- and sixfold, respectively) in diseased animals (Fig. 5C). TNF was detected in alveolar macrophages and fewer lymphocytes and was increased nearly threefold in animals that developed HPS. Heart, kidney, spleen, and liver samples from a subset of NHPs were evaluated similarly, and no distinguishable differences were observed between animals that developed HPS and control animals.
Fig. 5.
Animals that develop HPS have an increase in immune markers in the lung. Lung sections were stained with the indicated antibody. (A) CD3 was detected primarily in lymphocytes found within the BALT (boxed area) and in circulation. IL-4 and IFN-γ were detected predominantly in lymphocytes within BALT, in circulation, and—for IFN-γ—within alveolar spaces. TNF was observed largely in cells resembling macrophages (arrows) and in a few lymphocytes (arrowhead). (B) IHC slides were scanned with an Aperio ScanScope XT and analyzed using the ImageScope Positive Pixel Count algorithm to measure the intensity of staining. Shown are representative data postprocessing from animals Mock 1 (Upper) and DM-SNV 5 (Lower). (C) Data obtained from ImageScope software from three sections (upper, middle, and lower lung lobes) per animal were averaged, and the percent of positive pixels was determined for animals that did not develop disease (black squares) and for animals that developed HPS (red circles). **P < 0.01, ***P < 0.0.001.
Genetic Analysis of SNV.
To determine the genetic differences between the seemingly attenuated VA-SNV and the pathogenic DM-SNV, viruses were sequenced from cell culture supernatant or directly out of sera from an NHP that developed HPS using next-generation sequencing (NGS) technologies. Samples from multiple NHPs that developed HPS were processed, but only sera from a single animal yielded RNA of sufficient quality/quantity for NGS. Interestingly, the viruses that were sequenced differed by only two nucleotides, both of which were in the polymerase-coding region. Specifically, Vero propagation resulted in nucleotide changes at positions 166 (A to G), which resulted in an amino acid change (Lys to Glu), and 5364 (G to A), which was silent (GenBank accession IDs KF537001–KF537006).
Discussion
A common view of HPS and HFRS pathogenesis is that both conditions are believed to be primarily immunopathogenic with disease progression associated with intense and deleterious host immune responses (4, 10). This hypothesis is based largely on analysis of a limited number of clinical samples from confirmed symptomatic cases and cell culture models (9, 30–33). A limitation in addressing this hypothesis process experimentally has been a lack of appropriate animal models for hantaviral disease. Using the hamster model of HPS, we previously demonstrated that disease progression is associated with increased transcriptional activity of specific host responses (13); however, because of a deficiency of hamster-specific reagents, further characterization of these responses is not possible at present. In addition, the hamster model is of limited value for studying host responses to infection in relation to human disease, which could be accomplished better in NHPs as the closest surrogate model.
The current study describes the only NHP model of HPS and provides, to our knowledge, the first in-depth temporal analysis of serial samples from individual animals collected throughout the course of SNV infection and ensuing HPS. Our results identified a unique combination of virus replication, hematological abnormalities (thrombocytopenia, neutrophilia, eosinophilia, and monocytosis), and the development of uncontrolled, tissue-specific proinflammatory host responses as the mechanisms of HPS pathogenesis. The timing of these key parameters overlaps with the rapid progression from minor respiratory disease to severe respiratory distress, which is the hallmark of the cardiopulmonary phase of HPS, and supports their role in the hyperpermeability associated with late-stage disease. Viremia was the earliest indicator of disease and was first detected 4–10 d before the onset of respiratory distress and immediately before the observed hematological abnormalities and activated host immune responses. Viremia also has been implicated in the development of HPS in humans and hamsters (13, 25, 34, 35). In humans, the degree of viremia is correlated with the severity of disease and with increased hemoconcentration and low platelet counts (34). In hamsters, viremia precedes HPS manifestations by 5–7 d, and disease onset correlates with increased host immune responses in target organs (13). Interestingly, to date all attempts to protect hamsters with neutralizing antibodies or antiviral agents after the onset of viremia have failed (36, 37), as is consistent with the immunological observations and the presumed importance of the host immune responses in disease development. The NHP model will help in the future to establish more reliable markers for early treatment against hantavirus infections.
Previous attempts to develop an NHP model for HPS have used viruses propagated in vitro in Vero cells; for Puumala virus this method has been shown to result in genetic adaptation of the virus characterized by minimal changes in noncoding regions of the S segment and point mutations in the polymerase (L segment) (38, 39). Unfortunately, a lack of structural models and a reverse genetics system precludes the further characterization of the polymerase mutations. Nevertheless, Vero propagation appears to alter the biological properties of Puumala virus in that it no longer is reproducibly able to infect bank voles, the natural reservoir (38). Although preliminary, our results suggest that Vero propagation of SNV similarly results in genetic changes that are localized in the polymerase and potentially attenuates the virus. The development of NHP models for other etiological agents of HPS and HFRS will rely on propagating these pathogens in a manner that retains virulence. Currently, the only proven method for doing so is passaging these agents exclusively in the natural rodent reservoirs.
In summary, identification of the pathophysiological mechanisms using the only NHP model for HPS will open new avenues for the development of therapeutic and prophylactic approaches against this rare but frequently fatal disease with public health implications in the Americas. The strategy for model development outlined here also may translate readily into the development of disease models for other highly pathogenic zoonotic viruses, such as the Crimean-Congo hemorrhagic fever virus, for which NHP models of disease have not been described.
Materials and Methods
Ethics Statement.
Animal experiments were approved by the Animal Care and Use Committee of the Rocky Mountain Laboratories and were performed following the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care, International (AAALAC) and the National Institutes of Health Guide for the Care and Use of Laboratory Animals (40) by certified staff in an AAALAC approved facility.
Animal Infection.
Seventeen rhesus macaques (Macaca mulatta, 3.3–6.7 kg) were exposed to lung homogenates (10% wt/vol) from naive (uninfected) deer mice (n = 3) or from deer mice infected with DM-SNV (n = 10), or VA-SNV (n = 4) using an established protocol of simultaneous installation (20). The challenge dose was 6 × 106 focus-forming units (FFU) for VA-SNV or the equivalent to 6 × 106 FFU for DM-SNV. Briefly, the DM-SNV inoculum was standardized to the VA-SNV based on real-time qRT-PCR analysis of S genomic segment copies. Mock-infected animals received a similar amount of naive lung homogenates. For detailed information on inoculum preparation, clinical scoring, and sample collection, see SI Materials and Methods.
Hematology, Serum Biochemistry, and Coagulation Parameters.
Hematology was performed on EDTA blood with the HemaVet 950FS+ hematology analyzer (Drew Scientific). Serum biochemistries were measured using a Piccolo Blood Analyzer (Abaxis). Coagulation parameters were measured in plasma on a STart4 instrument (Diagnostica Stago). For detailed information, see SI Materials and Methods.
Virus Detection and Isolation.
Clinical specimens were screened for the presence of SNV RNA as previously described (19). A subset of RT-PCR–positive tissues were homogenized and used to infect triplicate monolayers of Vero cells. Cells were incubated for 14 d and scored for cytopathic effect.
Histopathology, IHC, and in Situ ISH.
Tissues were stained with H&E or phosphotungstic acid-hematoxylin using standard methods. Viral antigen and RNA was detected by IHC or ISH as previously described (41–43). CD3, IL-4, TNF, and INF-γ were stained in selected tissue specimens using standard IHC methodologies. Sections were scanned with an Aperio ScanScope XT (Aperio Technologies, Inc.) and quantified using the ImageScope Positive Pixel Count algorithm (version 9.1). For detailed information, see SI Materials and Methods.
Serum Cytokines and Chemokines.
Cytokines and chemokines were measured in serum samples using a NHP Cytokine kit (Millipore). Data were analyzed using a two-way ANOVA with a Bonferroni posttest.
Polychromatic Flow Cytometric Analysis of PBMCs.
PBMCs were stained for CD3, CD8α, Ki67, CD69, TNF, IFN-γ, Granzyme B, CD28, CD95, CD14, or CD4. Polychromatic analysis was performed using a LSR II flow cytometer equipped with FACS DiVA software (BD), and data were analyzed using FlowJo software (TreeStar). For detailed information, see SI Materials and Methods.
Serology.
Sera collected from NHPs prechallenge and at the time of necropsy were tested for the presence of hantavirus-specific IgG antibodies by standard ELISA methodologies and using a bacterially expressed, recombinant SNV N protein as antigen, essentially as previously described (44).
NGS.
Genomic sequences of VA-SNV and DM-SNV were determined using NGS technology. For detailed information, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Edward Schreckendgust, Rocky Rivera, Sandy Skorupa, Kathleen Meuchel, Jayne Faris, Amanda Weidow, Rachael LaCasse, Kimberly Meade-White, Tina Thomas, Dan Long, Shelly Robertson, Aaron Carmody, Sarah L. Anzick, Stacy M. Ricklefs, and Dan Bruno for scientific discussions and technical assistance and Anita Mora and Heather Murphy for preparing the graphics. This study was financially supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KF537001–KF537006).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401998111/-/DCSupplemental.
References
- 1.Duchin JS, et al. The Hantavirus Study Group Hantavirus pulmonary syndrome: A clinical description of 17 patients with a newly recognized disease. N Engl J Med. 1994;330(14):949–955. doi: 10.1056/NEJM199404073301401. [DOI] [PubMed] [Google Scholar]
- 2.Nichol ST, et al. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262(5135):914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 3.Macneil A, Nichol ST, Spiropoulou CF. Hantavirus pulmonary syndrome. Virus Res. 2011;162(1-2):138–147. doi: 10.1016/j.virusres.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 4.Jonsson CB, Figueiredo LT, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev. 2010;23(2):412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention (CDC) Hantavirus pulmonary syndrome in visitors to a national park—Yosemite Valley, California, 2012. MMWR Morb Mortal Wkly Rep. 2012;61(46):952. [PubMed] [Google Scholar]
- 6.Webster D, et al. Cluster of cases of hantavirus pulmonary syndrome in Alberta, Canada. Am J Trop Med Hyg. 2007;77(5):914–918. [PubMed] [Google Scholar]
- 7.MacNeil A, Ksiazek TG, Rollin PE. Hantavirus pulmonary syndrome, United States, 1993-2009. Emerg Infect Dis. 2011;17(7):1195–1201. doi: 10.3201/eid1707.101306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roehr B. US officials warn 39 countries about risk of hantavirus among travellers to Yosemite. BMJ. 2012;345:e6054. doi: 10.1136/bmj.e6054. [DOI] [PubMed] [Google Scholar]
- 9.Borges AA, et al. Hantavirus cardiopulmonary syndrome: Immune response and pathogenesis. Microbes Infect. 2006;8(8):2324–2330. doi: 10.1016/j.micinf.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 10.Mackow ER, Gavrilovskaya IN. Hantavirus regulation of endothelial cell functions. Thromb Haemost. 2009;102(6):1030–1041. doi: 10.1160/TH09-09-0640. [DOI] [PubMed] [Google Scholar]
- 11.Hooper JW, Larsen T, Custer DM, Schmaljohn CS. A lethal disease model for hantavirus pulmonary syndrome. Virology. 2001;289(1):6–14. doi: 10.1006/viro.2001.1133. [DOI] [PubMed] [Google Scholar]
- 12.Safronetz D, Ebihara H, Feldmann H, Hooper JW. The Syrian hamster model of hantavirus pulmonary syndrome. Antiviral Res. 2012;95(3):282–292. doi: 10.1016/j.antiviral.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Safronetz D, et al. Pathogenesis and host response in Syrian hamsters following intranasal infection with Andes virus. PLoS Pathog. 2011;7(12):e1002426. doi: 10.1371/journal.ppat.1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanagihara R, et al. Experimental hantavirus infection in nonhuman primates. Arch Virol. 1988;101(1-2):125–130. doi: 10.1007/BF01314657. [DOI] [PubMed] [Google Scholar]
- 15.McElroy AK, Bray M, Reed DS, Schmaljohn CS. Andes virus infection of cynomolgus macaques. J Infect Dis. 2002;186(12):1706–1712. doi: 10.1086/345768. [DOI] [PubMed] [Google Scholar]
- 16.Groen J, et al. A macaque model for hantavirus infection. J Infect Dis. 1995;172(1):38–44. doi: 10.1093/infdis/172.1.38. [DOI] [PubMed] [Google Scholar]
- 17.Klingström J, Plyusnin A, Vaheri A, Lundkvist A. Wild-type Puumala hantavirus infection induces cytokines, C-reactive protein, creatinine, and nitric oxide in cynomolgus macaques. J Virol. 2002;76(1):444–449. doi: 10.1128/JVI.76.1.444-449.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sironen T, et al. Pathology of Puumala hantavirus infection in macaques. PLoS ONE. 2008;3(8):e3035. doi: 10.1371/journal.pone.0003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Botten J, et al. Experimental infection model for Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus) Proc Natl Acad Sci USA. 2000;97(19):10578–10583. doi: 10.1073/pnas.180197197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobasa D, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445(7125):319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 21.Young JC, et al. The incubation period of hantavirus pulmonary syndrome. Am J Trop Med Hyg. 2000;62(6):714–717. doi: 10.4269/ajtmh.2000.62.714. [DOI] [PubMed] [Google Scholar]
- 22.Vial PA, et al. Incubation period of hantavirus cardiopulmonary syndrome. Emerg Infect Dis. 2006;12(8):1271–1273. doi: 10.3201/eid1208.051127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Safronetz D, et al. Pandemic swine-origin H1N1 influenza A virus isolates show heterogeneous virulence in macaques. J Virol. 2011;85(3):1214–1223. doi: 10.1128/JVI.01848-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaki SR, et al. Hantavirus pulmonary syndrome. Pathogenesis of an emerging infectious disease. Am J Pathol. 1995;146(3):552–579. [PMC free article] [PubMed] [Google Scholar]
- 25.Wahl-Jensen V, et al. Temporal analysis of Andes virus and Sin Nombre virus infections of Syrian hamsters. J Virol. 2007;81(14):7449–7462. doi: 10.1128/JVI.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Testi R, D’Ambrosio D, De Maria R, Santoni A. The CD69 receptor: A multipurpose cell-surface trigger for hematopoietic cells. Immunol Today. 1994;15(10):479–483. doi: 10.1016/0167-5699(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 27.Shedlock DJ, et al. Ki-67 staining for determination of rhesus macaque T cell proliferative responses ex vivo. Cytometry A. 2010;77(3):275–284. doi: 10.1002/cyto.a.20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mertz GJ, et al. Diagnosis and treatment of new world hantavirus infections. Curr Opin Infect Dis. 2006;19(5):437–442. doi: 10.1097/01.qco.0000244048.38758.1f. [DOI] [PubMed] [Google Scholar]
- 29.Lafont BA, Gloeckler L, D’Hautcourt JL, Gut JP, Aubertin AM. One-round determination of seven leukocyte subsets in rhesus macaque blood by flow cytometry. Cytometry. 2000;41(3):193–202. doi: 10.1002/1097-0320(20001101)41:3<193::aid-cyto6>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 30.Borges AA, et al. Role of mixed Th1 and Th2 serum cytokines on pathogenesis and prognosis of hantavirus pulmonary syndrome. Microbes Infect. 2008;10(10-11):1150–1157. doi: 10.1016/j.micinf.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Geimonen E, et al. Pathogenic and nonpathogenic hantaviruses differentially regulate endothelial cell responses. Proc Natl Acad Sci USA. 2002;99(21):13837–13842. doi: 10.1073/pnas.192298899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mori M, et al. High levels of cytokine-producing cells in the lung tissues of patients with fatal hantavirus pulmonary syndrome. J Infect Dis. 1999;179(2):295–302. doi: 10.1086/314597. [DOI] [PubMed] [Google Scholar]
- 33.Spiropoulou CF, Albariño CG, Ksiazek TG, Rollin PE. Andes and Prospect Hill hantaviruses differ in early induction of interferon although both can downregulate interferon signaling. J Virol. 2007;81(6):2769–2776. doi: 10.1128/JVI.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terajima M, et al. High levels of viremia in patients with the Hantavirus pulmonary syndrome. J Infect Dis. 1999;180(6):2030–2034. doi: 10.1086/315153. [DOI] [PubMed] [Google Scholar]
- 35.Xiao R, et al. Sin Nombre viral RNA load in patients with hantavirus cardiopulmonary syndrome. J Infect Dis. 2006;194(10):1403–1409. doi: 10.1086/508494. [DOI] [PubMed] [Google Scholar]
- 36.Safronetz D, Haddock E, Feldmann F, Ebihara H, Feldmann H. In vitro and in vivo activity of ribavirin against Andes virus infection. PLoS ONE. 2011;6(8):e23560. doi: 10.1371/journal.pone.0023560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Custer DM, Thompson E, Schmaljohn CS, Ksiazek TG, Hooper JW. Active and passive vaccination against hantavirus pulmonary syndrome with Andes virus M genome segment-based DNA vaccine. J Virol. 2003;77(18):9894–9905. doi: 10.1128/JVI.77.18.9894-9905.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundkvist A, et al. Cell culture adaptation of Puumala hantavirus changes the infectivity for its natural reservoir, Clethrionomys glareolus, and leads to accumulation of mutants with altered genomic RNA S segment. J Virol. 1997;71(12):9515–9523. doi: 10.1128/jvi.71.12.9515-9523.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nemirov K, Lundkvist A, Vaheri A, Plyusnin A. Adaptation of Puumala hantavirus to cell culture is associated with point mutations in the coding region of the L segment and in the noncoding regions of the S segment. J Virol. 2003;77(16):8793–8800. doi: 10.1128/JVI.77.16.8793-8800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Committee on Care and Use of Laboratory Animals (1985) Guide for the Care and Use of Laboratory Animals (Natl Inst Health, Bethesda), DHHS Publ No (NIH) 85-23.
- 41.Medina RA, Mirowsky-Garcia K, Hutt J, Hjelle B. Ribavirin, human convalescent plasma and anti-beta3 integrin antibody inhibit infection by Sin Nombre virus in the deer mouse model. J Gen Virol. 2007;88(Pt 2):493–505. doi: 10.1099/vir.0.82459-0. [DOI] [PubMed] [Google Scholar]
- 42.Safronetz D, et al. Hamster-adapted Sin Nombre virus causes disseminated infection and efficiently replicates in pulmonary endothelial cells without signs of disease. J Virol. 2013;87(8):4778–4782. doi: 10.1128/JVI.03291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang F, et al. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14(1):22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feldmann H, et al. Utilization of autopsy RNA for the synthesis of the nucleocapsid antigen of a newly recognized virus associated with hantavirus pulmonary syndrome. Virus Res. 1993;30(3):351–367. doi: 10.1016/0168-1702(93)90101-r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.