Significance
Environmental microorganisms are a source of diverse antibiotic resistance determinants. With the appropriate selection pressure, these resistance genes can be mobilized to clinically relevant pathogens. Identifying and characterizing elements of the environmental antibiotic resistome provides an early warning of what we may expect to encounter in the clinic. We uncover a conserved genetic element associated with various rifamycin antibiotic-inactivating mechanisms. This element led to the identification of a new resistance gene and associated enzyme responsible for inactivating rifamycin antibiotics by phosphorylation. Cryptic orthologous genes are also found in pathogenic bacteria but remain susceptible to the drug. This study reveals a new antibiotic resistance protein family and the unexpected prevalence of a silent rifamycin resistome among pathogenic bacteria.
Keywords: drug inactivation, phosphorylation, gene regulation, silent resistome
Abstract
Many environmental bacteria are multidrug-resistant and represent a reservoir of ancient antibiotic resistance determinants, which have been linked to genes found in pathogens. Exploring the environmental antibiotic resistome, therefore, reveals the diversity and evolution of antibiotic resistance and also provides insight into the vulnerability of clinically used antibiotics. In this study, we describe the identification of a highly conserved regulatory motif, the rifampin (RIF) -associated element (RAE), which is found upstream of genes encoding RIF-inactivating enzymes from a diverse collection of actinomycetes. Using gene expression assays, we confirmed that the RAE is involved in RIF-responsive regulation. By using the RAE as a probe for new RIF-associated genes in several actinomycete genomes, we identified a heretofore unknown RIF resistance gene, RIF phosphotransferase (rph). The RPH enzyme is a RIF-inactivating phosphotransferase and represents a new protein family in antibiotic resistance. RPH orthologs are widespread and found in RIF-sensitive bacteria, including Bacillus cereus and the pathogen Listeria monocytogenes. Heterologous expression and in vitro enzyme assays with purified RPHs from diverse bacterial genera show that these enzymes are capable of conferring high-level resistance to a variety of clinically used rifamycin antibiotics. This work identifies a new antibiotic resistance protein family and reinforces the fact that the study of resistance in environmental organisms can serve to identify resistance elements with relevance to pathogens.
Our control of infectious disease is increasingly challenged because of the emergence and dissemination of antibiotic resistance (1). This problem is exacerbated because of a dwindling number of new antimicrobials entering the clinic (2). The majority of clinically used antibiotics originates from microorganisms that reside in the environment (3). Recent studies have established that antibiotic resistance is ancient, and contemporary environmental bacteria are multidrug-resistant (4–6). There is also growing evidence that environmental bacteria, including antibiotic producers, are the progenitors of antibiotic-resistant determinants from pathogenic bacteria (7–9). Exploring and characterizing the antibiotic resistome, the collection of all antibiotic resistance genes and their precursors from the global microbiota, will facilitate our ability to anticipate and counter antibiotic resistance before its emergence into the clinic (10). With this knowledge, we may begin to focus efforts on circumventing these determinants by strategic drug development, resistance inhibitor combinations, and prudent administration programs.
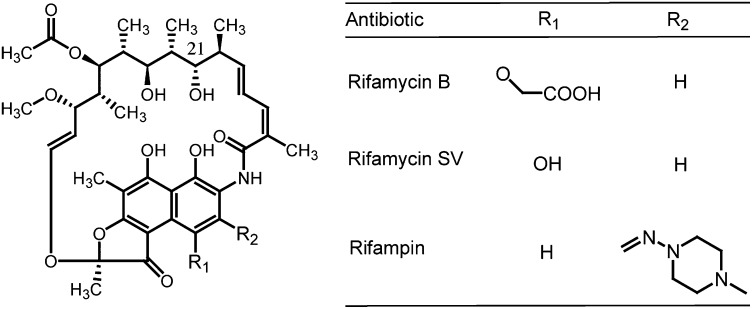
Rifamycin B, the initial member of the rifamycin family of antibiotics, was first described in the late 1950s and produced from the soil actinomycete Amycolatopsis mediterranei (11). Although the parent natural product has modest antibiotic activity, semisynthetic derivatives of this antibiotic family have experienced great success in the clinic. The most well-known is rifampin (RIF), introduced into the clinic almost 50 y ago (Fig. 1). RIF continues to be a frontline treatment of tuberculosis and other mycobacterial infections and increasingly, is as a fallback therapy for drug-resistant Staphylococcal infections (12). The rifamycins are broad-spectrum antibiotics that inhibit prokaryotic RNA polymerase (RNAP) by binding to a highly conserved region within the β-subunit (encoded by the gene rpoB), blocking the exit tunnel from the growing RNA nucleotide chain, and thus, sterically hindering the extension of nascent mRNA transcripts (13). Recently, synthetic exploration of the rifamycin scaffold has generated a number of new analogs designed to treat a much broader repertoire of bacterial infections. These drugs include rifaximin, which is administered for the treatment of travelers’ diarrhea and irritable bowel syndrome (14, 15), rifalazil, used for chlamydial infections (16), and a number of additional compounds (reviewed in ref. 12). This recent extension of the rifamycin family of antibiotics will likely apply pressure for the selection of an expanded repertoire of rifamycin resistance determinants (17).
Fig. 1.
Rifamycin antibiotics.
In mycobacteria, point mutations of the drug target are the most prevalent RIF resistance mechanism (18). Rifamycin resistance develops quite rapidly (frequency of 10−8–10−9 per bacterium per cell division), where single amino acid changes in the β-subunit significantly decrease binding of the antibiotic to its target (19). In other microbes, diverse RIF-inactivating mechanisms have been described from both pathogenic and nonpathogenic bacteria. Three RIF group transfer mechanisms of resistance have been described: glycosylation, ADP ribosylation, and phosphorylation (17). These resistance enzymes chemically modify the ansa polyketide bridge of RIF, attenuating its affinity for the β-subunit of RNAP. In addition, the decomposition of RIF initiated by an RIF monooxygenase has also been described (20, 21). Despite the fact that RIF phosphorylation has been known for 20 y, the gene encoding the RIF phosphotransferase (rph) has yet to be identified (22).
Here, we describe the discovery of a conserved upstream nucleotide element associated with various genes encoding RIF-inactivating enzymes from actinomycetes. By exploiting this conservation, we successfully identified the heretofore unrecognized rph gene. Through a combination of phylogenetics, heterologous protein expression, and in vitro enzyme assays, we establish that RPH orthologs are prevalent in both environmental and pathogenic bacteria, conferring high-level and broad resistance against clinically used rifamycin antibiotics. This effort highlights the power of screening genome data for noncoding elements in the discovery of new resistance elements.
Results
A Conserved Upstream Nucleotide Element Is Associated with RIF-Inactivating Genes from Actinomycetes.
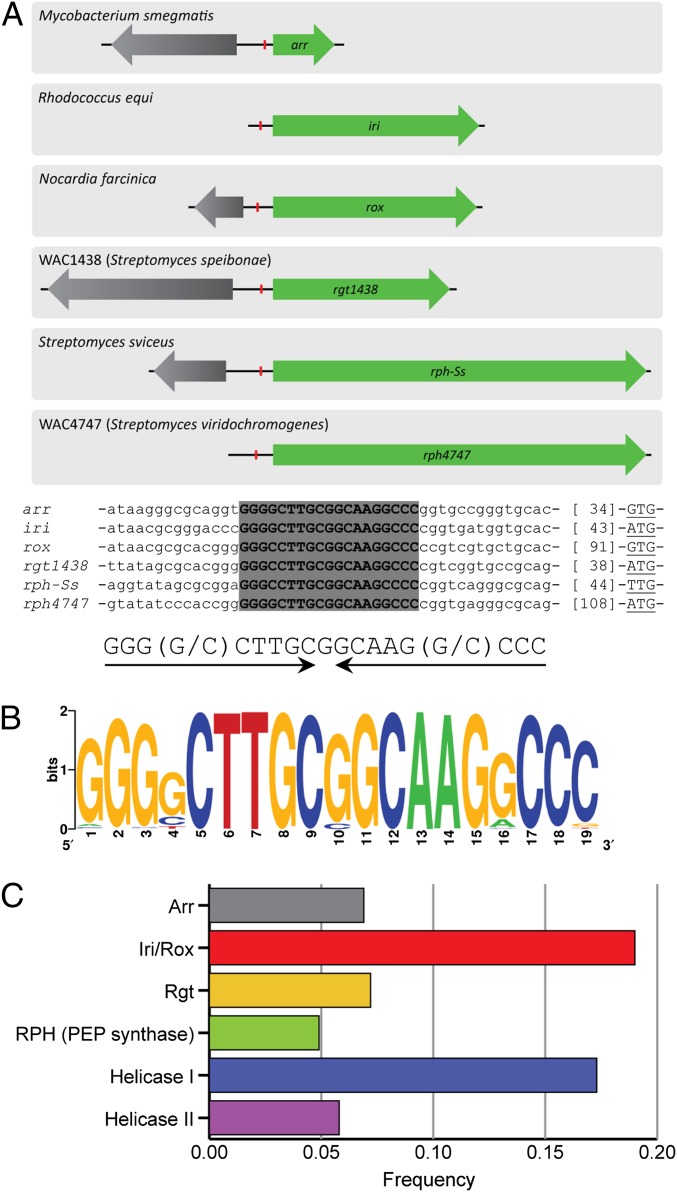
A diverse collection of RIF-inactivating mechanisms has been described from various pathogenic and nonpathogenic actinomycetes. We recently reported the identification and characterization of a RIF-inactivating glycosyltransferase (rgt1438) from a screen of soil actinomycetes (23). Examination of the intergenic DNA sequence upstream of rgt1438 identified a 19-bp inverted repeat motif (9-bp repeats separated by a single nucleotide spacer) (Fig. 2A). We hypothesized that this element could be involved in gene regulation. The conservation of this putative RIF-associated element (RAE) was examined by scanning the upstream intergenic regions from various characterized genes encoding RIF-inactivating enzymes (20, 21, 23, 24). This collection included three mechanisms of RIF inactivation (glycosylation, ADP ribosylation, and monooxygenation) from four divergent genera of actinomycetes (Streptomyces, Mycobacteria, Rhodococcus, and Nocardia) (Fig. 2A). The RAE was conserved upstream of all these genes with near-perfect nucleotide identity (Fig. 2A). A BLASTn search of the RAE was conducted from intergenic regions of the nucleotide collection and whole-genome shotgun contigs from the National Center of Biotechnology Information (25). The RAE was found over 400 times and predominantly associated with actinobacterial genomes (Dataset S1). The RAE was not associated with rifamycin-producing bacteria. A sequence logo with this RAE collection shows the remarkable conservation of this element (Fig. 2B). To determine if there is a correlation between the RAE and specific genes, we compiled a list of available divergent ORFs flanking each RAE and loosely binned orthologous proteins (Dataset S2). The RAE was strongly associated with ORFs resembling previously characterized RIF-inactivating enzymes (Fig. 2C). The RAE is observed accompanying an ORF encoding a RIF ADP ribosyltransferase (Arr), RIF glycosyltransferase (Rgt), or RIF monooxygenase (Iri/Rox) enzyme at a frequency of 0.33. Furthermore, the RAE is strongly associated with a number of uncharacterized ORFs. The RAE is found upstream of ORFs annotated as phosphoenolpyruvate (PEP) synthases and two distinct helicases (Fig. 2C). This survey revealed that the RAE is highly conserved across many actinobacterial genomes and has coevolved with a repertoire of genes, including RIF-inactivating genes and several other genes that are presumably expressed in response to RIF.
Fig. 2.
The RAE is associated with RIF-inactivating genes. (A) The RAE is conserved upstream of various characterized RIF-inactivating genes from diverse actinobacteria. arr, RIF ADP ribosyltransferase; iri and rox, RIF monooxygenase; rgt1438, RIF glycosyltransferase; rph-Ss and rph4747, RIF phosphotransferase. The red line indicates the position of the RAE. An alignment of the RAE identified a conserved 19-nt inverted repeat boxed in gray. (B) Sequence logo from over 400 RAEs found in the nucleotide collection and whole-genome shotgun contig databases. Sequence logos were generated using WebLogo. (C) RAE gene associations.
The rph Gene Is Responsible for Phosphorylating RIF.
A number of actinomycetes capable of inactivating RIF by phosphorylation have been described; however, the gene responsible for this phenotype, rph, has yet to be identified (22, 23). The strong correlation of the RAE with genes encoding RIF-inactivating enzymes led us to apply this link to identify the rph gene. In a previous phenotypic screen, we identified a collection of RIF-phosphorylating soil actinomycetes, and thus, we selected one of these strains for additional investigation (23). The draft genome of WAC4747 (most closely resembling Streptomyces viridochromogenes based on 16S rRNA analysis) was determined. We also biochemically screened our in-house collection of genome-sequenced actinomycetes for their ability to phosphorylate RIF and identified Streptomyces sviceus ATCC 29083 as a candidate (Fig. S1). Putative rph genes were identified by probing the genome sequences of WAC4747 and S. sviceus for the RAE and examining associated downstream genes. The RAE was not found to be flanking any genes resembling known antibiotic phosphotransferases (26). The RAE was found on two contigs within the WAC4747 genome: on a 3-kb contig upstream of an ORF encoding a putative PEP synthase (ORF1) and an 8.5-kb contig upstream of a putative helicase (ORF2) (Table S1). Similarly, the RAE was found upstream of predicted PEP synthase and helicase ORFs within the S. sviceus genome in addition to a candidate RIF monooxygenase (Table S1). PEP synthase and helicase proteins have not been previously associated with antibiotic resistance. Helicases harness the energy of ATP hydrolysis for separation of nucleotide strands, whereas PEP synthases catalyze the conversion of ATP and pyruvate to PEP, AMP, and Pi (27, 28). Orthologous genes of ORF1 and ORF2 are conserved in other actinomycete genomes and colocalized with the RAE (Fig. 2C).
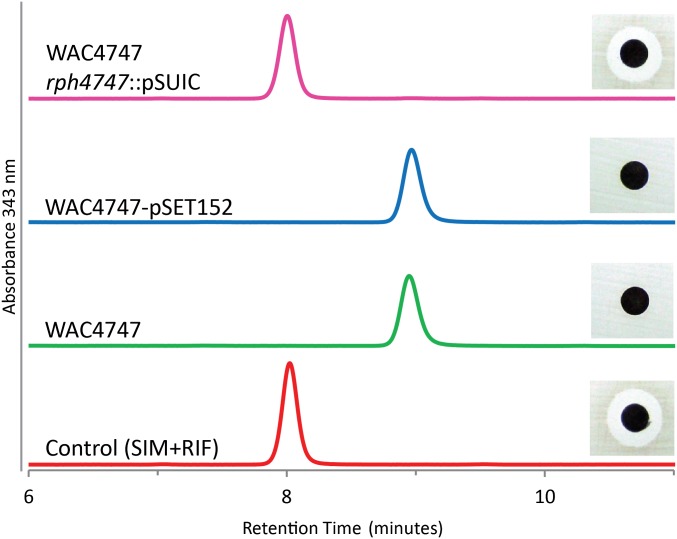
We individually disrupted ORF1 and ORF2 from WAC4747 and evaluated the ability of the mutant strains to phosphorylate RIF. Analysis of conditioned media from WAC4747 grown in the presence of RIF showed complete inactivation of RIF within 48 h (Fig. 3). In contrast, a mutant strain of WAC4747 with a disrupted ORF1 (putative PEP synthase) failed to inactivate RIF (Fig. 3). Disruption of ORF2 (putative helicase) in WAC4747 had no effect on RIF inactivation. These findings suggest that ORF1 (named rph4747) encodes the RPH enzyme. Inactivation of the orthologous putative PEP synthase gene from S. sviceus (rph-Ss) generated identical results (Fig. S2). Mutant strains of WAC4747 and S. sviceus with a disrupted rph gene displayed a modest twofold decrease in RIF minimum inhibitory concentrations (MICs) compared with the WT strains (Table S2). This result suggests, similar to other actinomycetes, that WAC4747 and S. sviceus harbor multiple RIF resistance mechanisms (21, 23).
Fig. 3.
The rph4747 gene from WAC4747 is responsible for the inactivation of RIF. Strains of WAC4747 were grown in the presence of RIF for 48 h, and conditioned media were analyzed for residual RIF activity using an RIF-sensitive indicator organism (B. subtilis). Samples were additionally analyzed using HPLC. WAC4747 and the control, WAC4747-pSET152 (empty vector), completely inactivate RIF within 48 h. The rph4747 mutant strain, WAC4747 rph4747::pSUIC, does not inactivate RIF and is comparable with the control of media and drug (Streptomyces isolation media (SIM)+RIF). Chromatograms are displayed at an absorbance of 343 nm.
We assessed the ability of the rph gene to contribute to RIF resistance by expression in a heterologous host. Introduction of rph4747 or rph-Ss into RIF-sensitive Escherichia coli conferred a 64-fold increase in RIF MIC compared with the control (Table 1). E. coli-expressing rph displayed high-level resistance up to or over 512 µg/mL to a panel of natural product and clinically used semisynthetic rifamycin antibiotics (Table 1). This result highlights the ability of rph to confer broad high-level resistance to rifamycin antibiotics.
Table 1.
Rifamycin MIC determinations of various rph genes expressed in E. coli
| Construct*/IPTG | MIC (µg/mL) | |||
| RIF | Rifamycin SV | Rifabutin | Rifaximin | |
| pET22b | ||||
| − | 8 | 64 | 8 | 16 |
| + | 8 | 64 | 8 | 16 |
| pET22b-rph4747 | ||||
| − | 512 | 512 | 512 | 64 |
| + | >512 | >512 | 512 | 512 |
| pET28b | ||||
| − | 8 | 64 | 8 | 16 |
| + | 8 | 64 | 8 | 16 |
| pET28b-rph-Ss | ||||
| − | 512 | >512 | 512 | 512 |
| + | >512 | >512 | 512 | 512 |
| pET19Tb | ||||
| − | 8 | 64 | 8 | 8 |
| + | 8 | 64 | 8 | 8 |
| pET19Tb-rph-Bc | ||||
| − | 128 | 256 | 64 | 64 |
| + | >512 | >512 | 512 | 512 |
| pET19Tb-rph-Lm | ||||
| − | 128 | 128 | 32 | 32 |
| + | >512 | >512 | 512 | 512 |
IPTG, isopropyl β-d-1-thiogalactopyranoside.
E. coli Rosetta(DE3)pLysS host.
RAE Responds to RIF.
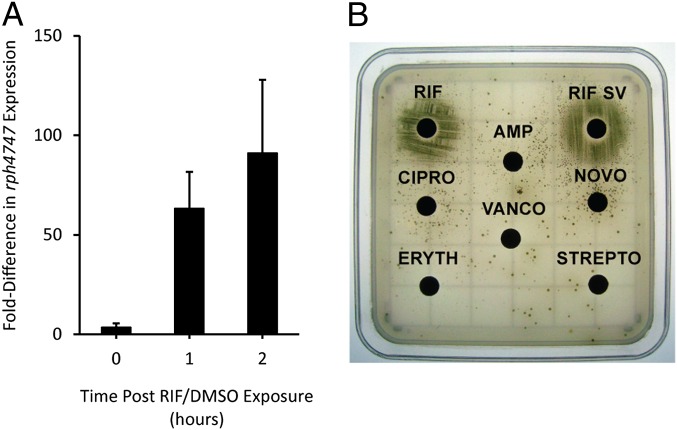
Because of the strong association of the RAE and genes encoding RIF-inactivating enzymes, we hypothesized that this DNA motif is likely involved in gene expression in response to RIF. Changes in transcript levels of rph4747 expression were monitored using quantitative RT-PCR with growth of WAC4747 in the presence or absence of RIF. In the presence of RIF, the rph4747 transcript was up-regulated 63- and 91-fold after 1 and 2 h of growth, respectively, compared with the control, DMSO (Fig. 4A). This finding confirms that genes paired with the RAE are up-regulated in response to RIF.
Fig. 4.
The RAE is involved in gene regulation in response to RIF. (A) Quantitative RT-PCR analysis of rph4747 transcripts. WAC4747 was grown in the presence of RIF or DMSO. Data are presented as fold difference of rph4747 transcripts from three independent biological replicates, and error bars represent SDs. (B) A WAC4747 KAN resistance reporter assay. The RAE from WAC4747 was fused to a promoterless KAN resistance cassette and integrated in the genome of WAC4747. Growth on KAN-containing media were only achieved in the presence of RIF and RIF SV. Subinhibitory concentrations of antibiotics were used: AMP, 5 µg; ciprofloxacin (CIPRO), 0.5 µg; erythromycin (ERYTH), 0.125 µg; novobiocin (NOVO), 0.05 µg; RIF, 2.5 µg; RIF SV, 5 µg; streptomycin (STREPTO), 0.05 µg; vancomycin (VANCO), 0.05 µg.
To validate the role of the RAE in RIF-specific gene regulation, we used a kanamycin (KAN) resistance reporter assay. The reporter was constructed by fusing the intergenic DNA upstream of the rph4747 gene ahead of a promoter-less KAN resistance cassette followed by integration into the genome of WAC4747, creating the strain WAC4747-PRAE/neo-pSET152. This strain was not KAN-resistant; showing gene expression downstream of the RAE is tightly regulated. However, exposure to subinhibitory concentrations of rifamycin antibiotics (RIF and the parent natural product rifamycin SV) induced growth on KAN-containing media, confirming that RAE is involved in rifamycin-responsive gene regulation (Fig. 4B). A number of diverse antibiotics, from natural and synthetic origins, inhibiting various biological targets failed to induce KAN resistance, confirming that gene regulation from the RAE is rifamycin-specific. Additionally, a reporter construct was made where the RAE was replaced with a random sequence of nucleotides. This construct failed to respond to RIF (Fig. S3) and established that the RAE nucleotide sequence is critical for gene expression.
In some instances, antibiotic resistance gene expression is dependent on the antibiotic successfully binding and inhibiting its target (29, 30). We generated RIF-resistant rpoB mutants of WAC4747-PRAE/neo-pSET152 to determine if inhibition of RNAP is a prerequisite for RAE-mediated gene induction. This mutant strain gained KAN resistance in response to RIF (Fig. S3), indicating that gene expression mediated by the RAE is independent of RNAP inhibition.
Diversity of the RPH Protein Family.
A BLASTp analysis of the RPH enzyme reveals that it is related to enzymes annotated as PEP synthases (EC 2.7.9.2). The mechanism of PEP synthase proceeds through a phosphoenzyme intermediate, where the β-phosphate of ATP is transferred to a histidine (His) residue, which in turn, is transferred to pyruvate (31). PEP synthase is composed of three domains: the N-terminal domain is responsible for nucleotide binding, the C-terminal domain binds pyruvate, and the central segment contains the catalytic His domain.
A global alignment of RPH4747 and PEP synthase from E. coli indicates low overall amino acid similarity but relatively high similarity near the N terminus. A search of the Conserved Domain Database (32) with RPH4747 showed that the N terminus consists of an ATP binding domain, similar to PEP synthase. In contrast to PEP synthase, the catalytic His domain of RPH is located at the C terminus (Fig. S4). The central region of RPH4747 shows no similarity to any previously characterized proteins. Therefore, RPH4747 shares some similarities with PEP synthase, but the overall architectures of these two enzymes differ. A search of the RPH using the Conserved Domain Architecture Retrieval Tool (33) showed that this domain architecture is found in over 1,600 proteins and primarily restricted to bacterial genomes. Although the majority of these enzymes is annotated as PEP synthases (or PEP binding proteins), there is no biochemical evidence confirming this activity, and therefore, it represents an unexplored protein family.
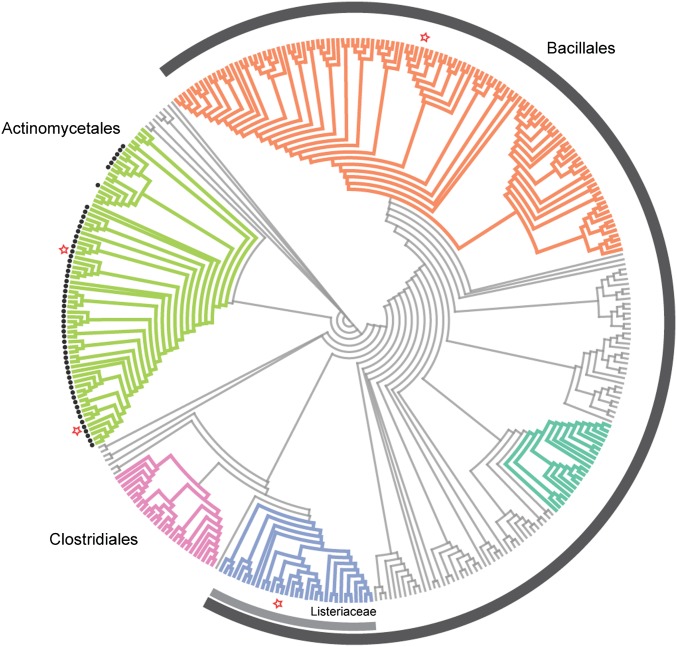
We performed a phylogenetic analysis of the RPH protein family to explore the relationship of the sequences in this family (Dataset S3). The RPH cladogram is shown in Fig. 5. We can identify several clades with high bootstrap support that correspond to well-defined taxonomic groups, even if resolution within and between these groups remains unclear (Fig. S5). RPH proteins are widely distributed, predominately in Gram-positive bacteria. Although many RPH orthologs are found within Actinobacteria, the majority is associated with Bacilli and Clostridia. We examined the intergenic DNA sequence upstream of RPH ORFs and discovered that the majority of ORFs from Actinobacteria is associated with the RAE (Fig. 5). This finding is consistent with our previous survey of the RAE and gene associations (Fig. 2C). The RAE was not associated with RPHs outside Actinobacteria. It is, therefore, uncertain whether these enzymes possess RPH activity.
Fig. 5.
Cladogram of the RPH protein family. Colored clades represent groups with over 85% bootstrap support based on maximum likelihood analysis using RAxML. Black circles indicate RPHs that are paired with the RAE. Red stars indicate RPHs selected for characterization: RPH4747, WAC4747; RPH-Bc, B. cereus ATCC 14579; RPH-Lm, L. monocytogenes str. 4b. F2365; RPH-Ss, S. sviceus ATCC 29083. The gray bar represents RPHs from bacteria of the Bacillales order. The light gray bar represents RPHs from the Listeriaceae family. A comprehensive cladogram with labels can be found in Fig. S5.
A previous biochemical screen by Dabbs et al. (34) showed that a number of bacteria from the genus Bacillus inactivated RIF by phosphorylation. This report was intriguing considering that Bacillus spp. are highly susceptible to RIF (35). Our phylogenetic analysis of the RPH protein family revealed orthologous enzymes from a number of Bacilli. This list also encompassed a number of pathogenic bacteria, including Bacillus cereus, Bacillus anthracis, and Listeria monocytogenes. These pathogens have been previously reported to be highly susceptible to RIF (36, 37). Indeed, this finding was confirmed by performing RIF susceptibility testing with B. cereus and L. monocytogenes, which showed a RIF MIC of 62.5 ng/mL. RPH orthologs from B. cereus (RPH-Bc) and L. monocytogenes (RPH-Lm) displayed 55% and 52% identity and 74% and 71% similarity, respectively, compared with RPH4747. We selected these two RPH orthologs as representatives from our phylogenetic analysis and evaluated their ability to confer RIF resistance when expressed in a heterologous host. E. coli expressing rph-Bc or rph-Lm exhibited a 64-fold increase in RIF MIC and also conferred high-level resistance to a variety of rifamycin analogs (Table 1). This result confirms that RPH orthologs are found in RIF-sensitive pathogenic bacteria and will confer high-level and broad rifamycin resistance when expressed in a heterologous host.
Biochemical Characterization of RPH Activity.
Recombinant RPH enzymes were overexpressed from E. coli and purified for characterization. Initially, RPH activity was monitored using a TLC assay, which confirmed that ATP served as a cosubstrate (Fig. S6). RIF-phosphate product from various RPHs was purified and analyzed using high-resolution MS and NMR spectroscopy (Fig. S6). This analysis confirmed that RPHs share the same regiospecificity and phosphorylate RIF at the hydroxyl group attached to the C21 of the ansa-chain, consistent with precedent using whole-cell inactivation (Fig. 1) (22).
Liquid chromatography electrospray ionization MS coupled with multiple reaction monitoring were used to quantify RPH activity. Steady-state kinetics were performed for three RPH enzymes from diverse bacterial hosts; RPH-Ss from S. sviceus, RPH-Bc from B. cereus, and RPH-Lm from L. monocytogenes. The gene encoding RPH-Ss is associated with the RAE, whereas the genes encoding RPH-Bc and RPH-Lm are not. All RPHs showed comparable kinetic constants regardless of the host source and RAE association (Table S3). RPHs exhibited very low Km values for RIF within the high-nanomolar range, indicating a high affinity for RIF. The Km values for RIF only varied twofold among the different RPH enzymes. Similarly, all RPHs displayed comparable kcat constants (catalytic rate constant).
Although PEP synthase and RPH share aspects of function and amino acid structure, it is unclear whether these two protein families have similar catalytic features. In addition to the liquid chromatography electrospray ionization MS assay, we further monitored RPH-Bc activity by measuring the generation of free Pi. Steady-state kinetic constants generated using this orthogonal assay were analogous to values obtained with the MS method described above (Table S3), confirming stoichiometric production of RIF-phosphate and Pi by RPH-Bc. With this assay, RPHs were also assayed for PEP synthase activity. All RPHs failed to phosphorylate pyruvate and showed no PEP synthase activity.
A sequence alignment of the region surrounding the catalytic His residue from PEP synthase and structurally and mechanistically related enzymes identified a conserved motif T-X-X-G-G-X-X-X-H; the conserved His is required for formation of a phosphoenzyme intermediate (Fig. S7). A similar motif was found in the RPH family of enzymes fewer than 50 aa away from the C terminus of the protein. An His-827-Ala mutant of RPH-Bc failed to confer RIF resistance in E. coli, and the enzyme had no in vitro enzymatic activity (Fig. S7). Together, these results suggest that RPH is mechanistically related to PEP synthase, likely using a phosphohistidine intermediate during catalysis, despite having architectural differences with the metabolic enzyme.
Discussion
The genetic reservoirs of antibiotic resistance and their regulation are poorly understood. We and others have shown that nonpathogenic environmental bacteria harbor an extensive and ancient resistome that is the likely source of resistance elements circulating in pathogens today (4–6, 9). The regulation of resistance genes by target antibiotics has been shown in only a few cases (for example, the induction of the efflux protein TetA by tetracycline binding to TetR and the complex regulation of β-lactam resistance by these antibiotics in Staphylococcus aureus) (38, 39). In this study, we characterize a new family of rifamycin resistance enzymes along with a conserved RIF-associated regulatory element, the RAE, which is shared among genes encoding various RIF-inactivating enzymes from actinomycetes. The RAE is composed of a 19-nt inverted repeat that is essential for modulating gene expression. Such inverted repeat DNA motifs have long been linked to gene regulation and often interact with protein regulators. Additional work is required to reveal the details of the mechanism of induction associated with this RAE and determine if any additional regulators are involved.
For over two decades, a number of actinomycetes has been reported to inactivate RIF by phosphorylation; however, the gene encoding the RPH was not known. Our observation of the strong correlation of the RAE with various actinomycete genes known to encode RIF-inactivating enzymes suggested a common regulatory mechanism and an entry point to discover the rph gene. Indeed, we successfully identified and validated that a gene predicted to encode a PEP synthase was, in fact, the rph. The RPH enzyme does not show similarity to any previously identified antibiotic phosphotransferases. Instead, RPH is related to the multidomain gluconeogenic enzyme PEP synthase. Although RPH and PEP synthase differ in overall domain architectures (Fig. S4), RPH enzymes have been routinely annotated as PEP synthases from sequenced genomes. Therefore, RPH represents a previously unexplored and misannotated protein family. Kinetic analysis and site-directed mutagenesis confirmed that the RPH enzyme is mechanistically related to PEP synthase and does not use pyruvate as a substrate. A Conserved Domain Architecture Retrieval Tool search has shown that this protein family is widely distributed across bacteria and that these enzymes are likely not involved in gluconeogenesis and may have diverse functions, including antibiotic resistance. Additional work is required to decipher their substrates and functions.
RPH orthologs from actinomycetes are colocalized with the RAE. However, the majority of RPHs is found in other Gram-positive genera (Fig. 5). Interestingly, many of these Gram positives have an RIF-sensitive phenotype. Of particular concern are the orthologs of rph that are found within the genomes of pathogens, such as B. anthracis, B. cereus , and L. monocytogenes. Heterologous expression and in vitro enzyme assays confirmed they are bona fide RPHs and not PEP synthases and confer high-level and broad-spectrum resistance with equivalent competence to their Streptomycete counterparts. This unexpected diversity and prevalence of rph genes in a number of bacterial genera are troubling given the renewed interest in expanding the application of these antibiotics outside their use in mycobacterial infection (12, 14–16).
Antibiotic resistance genes that do not normally confer resistance for their native host but are proficient at conferring resistance when expressed in an alternate host are called silent resistance genes (40). The majority of rph genes is not from actinomycetes and not associated with the RAE. Therefore, these bacteria cannot respond to RIF and activate gene expression accordingly. We speculate that these nonactinomycete rph genes are evolutionary related to rphs from actinomycetes but have an alternate function in their native host, which is not associated with antibiotic resistance. Their ability to bind and phosphorylate rifamycins is possibly caused by structural similarities with their natural substrates. Silent resistance genes have, thus far, been largely underappreciated, likely because of their inability to confer an antibiotic resistance phenotype in their native host. Recently, functional metagenomics have revealed numerous novel resistance genes, and continued research in this area is likely to uncover additional genes (41–43).
Antibiotic resistance mediated by enzymes is of particular clinical concern because of their potential to become mobilized and distributed horizontally between diverse species of bacteria. Examples of this mobilization are prevalent for many families of antibiotics, and this trend has begun to emerge with RIF-inactivating enzymes. The Arr enzyme, which is responsible for the inactivation of RIF by ADP ribosylation, was first described from the normally benign Mycobacterium smegmatis; however, the gene encoding this enzyme is now mobile and found on transmissible plasmids and integrons carrying multiple antibiotic resistance determinants (24, 44, 45). These mobile elements are now found in a variety of pathogens. The dissemination of Arr and potentially other RIF-inactivating enzymes in the near future threatens the efficacy of the entire rifamycin family of antibiotics.
This work has established that RPH enzymes do not discriminate between various rifamycins and effectively inactivate natural product and semisynthetic derivatives of this family. Understanding the molecular interactions between RPH and rifamycin substrates will provide the basis for generating rifamycin analogs that are not susceptible to these enzymes. The possibility of resistance-proof rifamycins has been exemplified with the recent development of C25 carbamate rifamycin derivatives that retain activity against Arr-containing bacteria (46).
Materials and Methods
Additional details are available in SI Text.
Inactivation of the rph Gene from WAC4747 and S. sviceus.
Disruption of the rph gene was accomplished by insertional inactivation using a previously described strategy (23). A partial fragment (∼1,000 bp) of the rph4747 gene from WAC4747 was amplified by PCR using primers rph4747-part-F and rph4747-part-R and cloned into pSUIC (a suicide version of pSET152 that lacks attP and int). The rph-Ss gene from S. sviceus ATCC 29083 was amplified using primers rph-Ss-part-F and rph-Ss-part-R and cloned as described above. The helicase gene from WAC4747 was amplified using primers hel4747-part-F and hel4747-part-R. Conjugations were performed as previously described (23). A list of primers can be found in Table S4.
RIF Inactivation Assay.
Fresh spores were used to inoculate 5 mL SIM media (5). Cultures were grown for 4 d at 30 °C with shaking at 250 rpm; 0.5 mL culture was then used to inoculate 4.5 mL fresh SIM containing RIF at 20 µg/mL and represented the zero time point. At 48 h, conditioned media samples were taken and analyzed for residual RIF activity using the Kirby–Bauer disk diffusion assay with Bacillus subtilis as the RIF-sensitive indicator microorganism. Additionally, supernatant samples were extracted with an equal volume of methanol, clarified by centrifugation, and analyzed using a Waters e2695 HPLC system equipped with an XSelect CSH C18 5 µM column (4.6 × 100 mm) with the following method: linear gradient of 37–55% (vol/vol) acetonitrile in water with 0.05% TFA at 1 mL/min over 12 min.
Supplementary Material
Acknowledgments
We thank Andrew G. McArthur for assistance with the RPH phylogenetic analysis and Christine King for help with sequencing of the WAC4747 genome. We thank Michael Fischbach for providing Streptomyces sviceus ATCC 20983 and Lori Burrows for the gift of Listeria monocytogenes str. 4b. F2365. We also thank Georgina Cox and Grace Yim for helpful discussions. This research was funded by Canadian Institutes of Health Research Grant MT-13536 and a Canada Research Chair in Antibiotic Biochemistry (G.D.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The DNA sequence reported in this paper has been deposited in the NCBI database (accession no. KJ151292).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402358111/-/DCSupplemental.
References
- 1.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325(5944):1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper MA, Shlaes D. Fix the antibiotics pipeline. Nature. 2011;472(7341):32. doi: 10.1038/472032a. [DOI] [PubMed] [Google Scholar]
- 3.Bérdy J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J Antibiot (Tokyo) 2012;65(8):441. doi: 10.1038/ja.2012.54. [DOI] [PubMed] [Google Scholar]
- 4.D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477(7365):457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 5.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311(5759):374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 6.Bhullar K, et al. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE. 2012;7(4):e34953. doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benveniste R, Davies J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc Natl Acad Sci USA. 1973;70(8):2276–2280. doi: 10.1073/pnas.70.8.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall CG, Broadhead G, Leskiw BK, Wright GD. D-Ala-D-Ala ligases from glycopeptide antibiotic-producing organisms are highly homologous to the enterococcal vancomycin-resistance ligases VanA and VanB. Proc Natl Acad Sci USA. 1997;94(12):6480–6483. doi: 10.1073/pnas.94.12.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsberg KJ, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337(6098):1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright GD. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5(3):175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 11.Sensi P, Margalith P, Timbal MT. Rifomycin, a new antibiotic; preliminary report. Farmaco Sci. 1959;14(2):146–147. [PubMed] [Google Scholar]
- 12.Aristoff PA, Garcia GA, Kirchhoff PD, Hollis Showalter HD. Rifamycins—obstacles and opportunities. Tuberculosis (Edinb) 2010;90(2):94–118. doi: 10.1016/j.tube.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Campbell EA, et al. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104(6):901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 14.Huang DB, DuPont HL. Rifaximin—a novel antimicrobial for enteric infections. J Infect. 2005;50(2):97–106. doi: 10.1016/j.jinf.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 15.Cremonini F, Lembo A. Rifaximin for the treatment of irritable bowel syndrome. Expert Opin Pharmacother. 2012;13(3):433–440. doi: 10.1517/14656566.2012.651458. [DOI] [PubMed] [Google Scholar]
- 16.Rothstein DM, van Duzer J, Sternlicht A, Gilman SC. Rifalazil and other benzoxazinorifamycins in the treatment of chlamydia-based persistent infections. Arch Pharm (Weinheim) 2007;340(10):517–529. doi: 10.1002/ardp.200700080. [DOI] [PubMed] [Google Scholar]
- 17.Tupin A, et al. Resistance to rifampicin: At the crossroads between ecological, genomic and medical concerns. Int J Antimicrob Agents. 2010;35(6):519–523. doi: 10.1016/j.ijantimicag.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 18.Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79(1):3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 19.Floss HG, Yu TW. Rifamycin-mode of action, resistance, and biosynthesis. Chem Rev. 2005;105(2):621–632. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- 20.Andersen SJ, Quan S, Gowan B, Dabbs ER. Monooxygenase-like sequence of a Rhodococcus equi gene conferring increased resistance to rifampin by inactivating this antibiotic. Antimicrob Agents Chemother. 1997;41(1):218–221. doi: 10.1128/aac.41.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoshino Y, et al. Monooxygenation of rifampicin catalyzed by the rox gene product of Nocardia farcinica: Structure elucidation, gene identification and role in drug resistance. J Antibiot (Tokyo) 2010;63(1):23–28. doi: 10.1038/ja.2009.116. [DOI] [PubMed] [Google Scholar]
- 22.Yazawa K, Mikami Y, Maeda A, Morisaki N, Iwasaki S. Phosphorylative inactivation of rifampicin by Nocardia otitidiscaviarum. J Antimicrob Chemother. 1994;33(6):1127–1135. doi: 10.1093/jac/33.6.1127. [DOI] [PubMed] [Google Scholar]
- 23.Spanogiannopoulos P, Thaker M, Koteva K, Waglechner N, Wright GD. Characterization of a rifampin-inactivating glycosyltransferase from a screen of environmental actinomycetes. Antimicrob Agents Chemother. 2012;56(10):5061–5069. doi: 10.1128/AAC.01166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quan S, Venter H, Dabbs ER. Ribosylative inactivation of rifampin by Mycobacterium smegmatis is a principal contributor to its low susceptibility to this antibiotic. Antimicrob Agents Chemother. 1997;41(11):2456–2460. doi: 10.1128/aac.41.11.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morar M, Wright GD. The genomic enzymology of antibiotic resistance. Annu Rev Genet. 2010;44:25–51. doi: 10.1146/annurev-genet-102209-163517. [DOI] [PubMed] [Google Scholar]
- 27.Cooper RA, Kornberg HL. The mechanism of the phosphoenolpyruvate synthase reaction. Biochim Biophys Acta. 1967;141(1):211–213. doi: 10.1016/0304-4165(67)90269-3. [DOI] [PubMed] [Google Scholar]
- 28.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 29.Narayanan CS, Dubnau D. Evidence for the translational attenuation model: Ribosome-binding studies and structural analysis with an in vitro run-off transcript of ermC. Nucleic Acids Res. 1985;13(20):7307–7326. doi: 10.1093/nar/13.20.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexieva Z, Duvall EJ, Ambulos NP, Jr, Kim UJ, Lovett PS. Chloramphenicol induction of cat-86 requires ribosome stalling at a specific site in the leader. Proc Natl Acad Sci USA. 1988;85(9):3057–3061. doi: 10.1073/pnas.85.9.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narindrasorasak S, Bridger WA. Phosphoenolypyruvate synthetase of Escherichia coli: Molecular weight, subunit composition, and identification of phosphohistidine in phosphoenzyme intermediate. J Biol Chem. 1977;252(10):3121–3127. [PubMed] [Google Scholar]
- 32.Marchler-Bauer A, et al. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41(Database issue):D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: Protein homology by domain architecture. Genome Res. 2002;12(10):1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dabbs ER, et al. Rifampicin inactivation by Bacillus species. J Antibiot (Tokyo) 1995;48(8):815–819. doi: 10.7164/antibiotics.48.815. [DOI] [PubMed] [Google Scholar]
- 35.Dey A, Chatterji D. Tracing the variation in physiological response to rifampicin across the microbial spectrum. J Bacteriol Virol. 2012;42(2):87–100. [Google Scholar]
- 36.Vogler AJ, et al. Molecular analysis of rifampin resistance in Bacillus anthracis and Bacillus cereus. Antimicrob Agents Chemother. 2002;46(2):511–513. doi: 10.1128/AAC.46.2.511-513.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawkins AE, Bortolussi R, Issekutz AC. In vitro and in vivo activity of various antibiotics against Listeria monocytogenes type 4b. Clin Invest Med. 1984;7(4):335–341. [PubMed] [Google Scholar]
- 38.Hillen W, Berens C. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 39.Llarrull LI, Fisher JF, Mobashery S. Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new beta-lactams that meet the challenge. Antimicrob Agents Chemother. 2009;53(10):4051–4063. doi: 10.1128/AAC.00084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dantas G, Sommer MO. Context matters - the complex interplay between resistome genotypes and resistance phenotypes. Curr Opin Microbiol. 2012;15(5):577–582. doi: 10.1016/j.mib.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Torres-Cortés G, et al. Characterization of novel antibiotic resistance genes identified by functional metagenomics on soil samples. Environ Microbiol. 2011;13(4):1101–1114. doi: 10.1111/j.1462-2920.2010.02422.x. [DOI] [PubMed] [Google Scholar]
- 42.Pehrsson EC, Forsberg KJ, Gibson MK, Ahmadi S, Dantas G. Novel resistance functions uncovered using functional metagenomic investigations of resistance reservoirs. Front Microbiol. 2013;4(145):1–11. doi: 10.3389/fmicb.2013.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGarvey KM, Queitsch K, Fields S. Wide variation in antibiotic resistance proteins identified by functional metagenomic screening of a soil DNA library. Appl Environ Microbiol. 2012;78(6):1708–1714. doi: 10.1128/AEM.06759-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arlet G, et al. Plasmid-mediated rifampin resistance encoded by an arr-2-like gene cassette in Klebsiella pneumoniae producing an ACC-1 class C beta-lactamase. Antimicrob Agents Chemother. 2001;45(10):2971–2972. doi: 10.1128/AAC.45.10.2971-2972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tribuddharat C, Fennewald M. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43(4):960–962. doi: 10.1128/aac.43.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Combrink KD, et al. New C25 carbamate rifamycin derivatives are resistant to inactivation by ADP-ribosyl transferases. Bioorg Med Chem Lett. 2007;17(2):522–526. doi: 10.1016/j.bmcl.2006.10.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.