Significance
Cohesin complexes tether replicated sister chromatids together from DNA replication until anaphase onset. This tethering promotes proper chromosome segregation, DNA damage repair, and the regulation of some genes. Cohesins are deposited on chromosomes by sister chromatid cohesion (Scc) proteins, Scc2 and Scc4, which form a heterodimeric cohesin deposition complex in budding yeast. Although Scc2/Scc4 is essential for cohesin deposition, its regulation is poorly understood. We demonstrate that dephosphorylation promotes constitutive Scc2 cleavage within its amino terminus, resulting in the loss of its interaction with Scc4. Moreover, an scc2 truncation mutant that mimics cleaved Scc2 is defective in cohesin deposition. We suggest that Scc2 cleavage, which is regulated by modulation of Scc2’s phosphorylation status, represents a previously unidentified mechanism that controls Scc2/Scc4 cohesin deposition activity.
Keywords: chromosome segregation, Scc2 phosphorylation, chromosome instability, NIPBL, MAU2
Abstract
Sister chromatid cohesion (SCC), efficient DNA repair, and the regulation of some metazoan genes require the association of cohesins with chromosomes. Cohesins are deposited by a conserved heterodimeric loading complex composed of the Scc2 and Scc4 proteins in Saccharomyces cerevisiae, but how the Scc2/Scc4 deposition complex regulates the spatiotemporal association of cohesin with chromosomes is not understood. We examined Scc2 chromatin association during the cell division cycle and found that the affinity of Scc2 for chromatin increases biphasically during the cell cycle, increasing first transiently in late G1 phase and then again later in G2/M. Inactivation of Scc2 following DNA replication reduces cellular viability, suggesting that this post S-phase increase in Scc2 chromatin binding affinity is biologically relevant. Interestingly, high and low Scc2 chromatin binding levels correlate strongly with the presence of full-length or amino-terminally cleaved forms of Scc2, respectively, and the appearance of the cleaved Scc2 species is promoted in vitro either by treatment with specific cell cycle-staged cellular extracts or by dephosphorylation. Importantly, Scc2 cleavage eliminates Scc2–Scc4 physical interactions, and an scc2 truncation mutant that mimics in vivo Scc2 cleavage is defective for cohesin deposition. These observations suggest a previously unidentified mechanism for the spatiotemporal regulation of cohesin association with chromosomes through cell cycle regulation of Scc2 cohesin deposition activity by Scc2 dephosphorylation and cleavage.
Multisubunit, ring-shaped cohesin complexes play key roles in chromosome morphogenesis that are required for faithful chromosome transmission to daughter cells. Newly replicated sister chromatids become tethered together by cohesins during S phase, which promotes chromosome biorientation on mitotic spindles (1). Cohesins also mediate efficient DNA double-strand break repair by homologous recombination (2, 3) and the formation or stabilization of chromatin loops that affect various nuclear processes, such as gene expression and Ig gene rearrangements (reviewed in refs. 4 and 5). Altered gene expression resulting from defective cohesin-mediated chromatin looping is likely responsible for the pathogenesis of Cornelia de Lange Syndrome (CdLS), a dominantly inherited human developmental disorder (6).
Sister chromatid cohesion (Scc) proteins form a heterodimeric cohesin deposition complex, but the complex's activity in deposition is not understood (7). Cohesins copurify with Scc2/Scc4, suggesting that Scc2/Scc4 plays a direct role in deposition (8–11). In the absence of either loader complex subunit, cohesin rings assemble, but fail to be deposited (7, 12, 13). ATP hydrolysis by cohesin’s structural maintenance of chromosome (SMC) subunits is required for cohesin loading, and deposition is inhibited when SMC hinge domains, which mediate Smc1/3 interactions within cohesin, are artificially tethered (8, 14, 15). Thus, Scc2/Scc4 may activate cohesin’s ATPase activity or facilitate a conformational change in cohesin structure that promotes its loading, perhaps by permitting transient hinge opening to allow chromatin to enter cohesin rings or by promoting cohesin oligomerization (14, 16).
Factors that regulate Scc2/Scc4 chromatin association are only beginning to be elucidated. Interactions of Scc2 and Scc4 orthologs from Xenopus and humans, and their stable association with chromatin, require the amino termini of both proteins (10, 13, 17, 18). In contrast, the fission yeast Scc2 ortholog alone binds nonchromatinized DNA, but does not exhibit an expected preference for sequences shown to associate with Scc2/Scc4 in vivo (19). Xenopus Scc2/Scc4 chromatin association requires prereplication complexes and Drf1-dependent kinase (DDK) activity (10, 12, 20), although this scenario is not the case in budding yeast (21). Scc2/Scc4 interactions with histone deacetylases and an ATP-dependent chromatin remodeler suggest that underlying chromatin structure also influences Scc2/Scc4 chromatin association (22–26). Whether Scc2/Scc4 plays a role in chromatin remodeling or merely deposits cohesins at remodeled sites is unknown, however.
The chromatin association of Scc2/Scc4 and its orthologs is also regulated temporally during the cell cycle, although the specifics of association vary across species. Scc2/Scc4 associates with chromatin in late mitosis of the previous cell cycle in metazoans (12, 13) and in late G1 in budding yeast, but in all cases, this association precedes DNA replication initiation so that cohesins are deposited in time to tether newly replicated sister chromatids together. Surprisingly, budding yeast Scc2/Scc4 chromatin association is more robust in mitotically arrested cells than in G1-staged cells. Reduced G1 Scc2/Scc4 chromatin association is not due to the absence of either loader subunit, because Scc2 and Scc4 protein levels vary little during the cell cycle, or by a lack of assembled cohesin complexes in G1, because Scc2/Scc4 chromatin association occurs independently of cohesins (18, 27, 28). Scc2/Scc4 removal from chromatin is also regulated and occurs during mitosis in Xenopus and, more specifically, during prophase in humans (12, 13). Although factors responsible for regulating Scc2/Scc4 chromatin association/dissociation during the cell cycle remain enigmatic, evidence that multiple Scc2 orthologs are phosphorylated suggests the intriguing possibility that Scc2 posttranslational modifications regulate Scc2/Scc4 chromatin association (29–31).
Here, we describe our efforts to understand how budding yeast Scc2/Scc4 chromatin binding is regulated during the cell cycle. Our results demonstrate the existence of multiple Scc2 protein species in vivo and that a specific cleaved form of Scc2 accumulates at cell cycle periods when Scc2 chromatin binding is weak. The appearance of this cleaved Scc2 species is strongly correlated with Scc2 dephosphorylation, suggesting that the phosphorylation state of Scc2 is critical for the regulation of its stability. Scc2 cleavage is also correlated with the loss of Scc2–Scc4 interactions, and an scc2 truncation mutant that mimics cleaved Scc2 is defective in cohesin deposition. These observations suggest that Scc2–Scc4 interactions and, therefore, the function of the complex in cohesin deposition, may be influenced by dephosphorylation-induced Scc2 cleavage.
Results
Scc2’s Chromatin Association Is Biphasic Through the Cell Cycle.
Previous immunoblot and chromatin immunoprecipitation (ChIP) analyses revealed that although Scc2 and Scc4 protein levels vary little during the cell cycle, their chromatin binding is higher in mitotically arrested cells than in G1-staged cells (27). To eliminate the possibility of epitope masking during ChIP in G1 cells, we used chromatin fractionation as an alternative method to assess Scc2/Scc4 chromatin association (32). Strains expressing Scc2 or Scc4 tagged with three tandem FLAG epitopes (hereafter referred to as Scc2-FLAG or Scc4-FLAG) (27) were arrested either in G11 by using α-factor (αF) mating pheromone or in mitosis by using a conditional cdc16 mutant. We observed that although substantial pools of soluble (SN1) Scc2 and Scc4 exist, chromatin-bound Scc2 and Scc4 levels are higher in chromatin bound pellet fractions prepared from mitotically arrested cells than in pellets from G1 cells (Fig. S1 A and B), consistent with previous ChIP results (27). Although both Scc2 and Scc4 exhibit this unexpected post-S phase increase in chromatin binding, we focus here on the characterization of Scc2 chromatin binding.
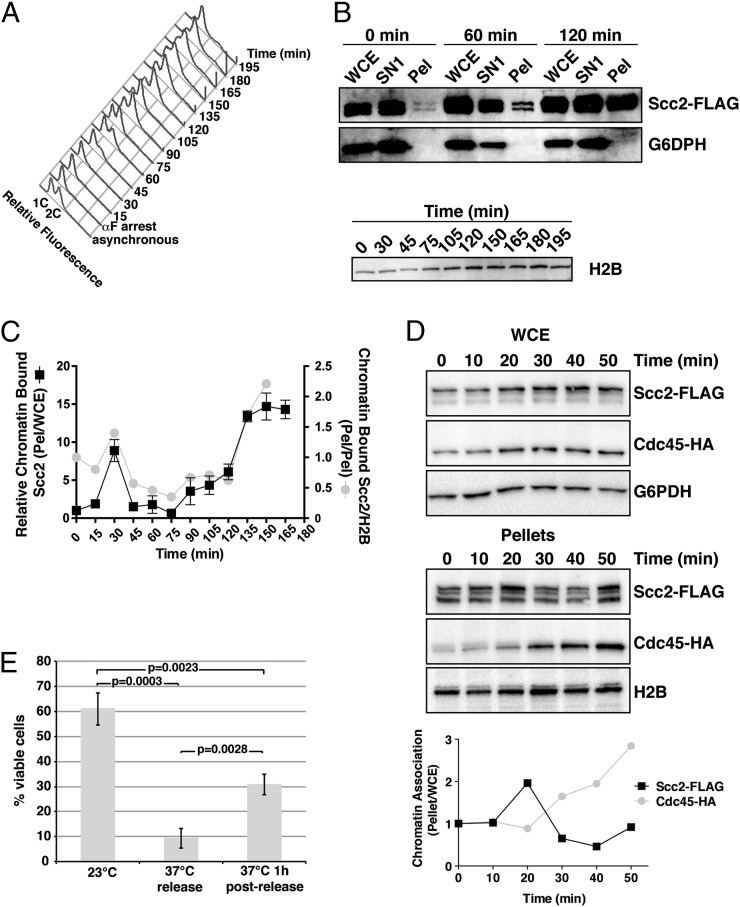
To better determine when Scc2 chromatin binding affinity increases during the cell cycle, we fractionated G1-staged cells and cells at 15-min intervals post-αF release. Ratios of the total amount of Scc2, present in two predominant forms (see below), in pellet fractions relative to the amount in their corresponding whole cell extracts (WCE) were then determined at each time point by using semiquantitative immunoblotting. As observed previously, Scc2 chromatin binding increased from G1 to mitosis (Fig. 1 B and C). Interestingly however, this increase occurred in two distinct intervals. The first increase occurred transiently at ∼30 min after G1 release before cells had entered S phase, as indicated by histograms of DNA content (Fig. 1A), but then quickly returned to levels observed in G1-staged cells. A second increase then began at ∼90 min after release, after most cells completed DNA replication, and persisted into mitosis. This biphasic nature of Scc2 recruitment to chromatin was also evident when chromatin-bound Scc2 was instead normalized to chromatin-bound H2B (Fig. 1C). Fractionation of cells containing epitope-tagged Cdc45, whose recruitment to prereplication complexes shortly precedes origin firing during DNA replication initiation (33), revealed that the peak of Scc2 chromatin association precedes Cdc45 recruitment (Fig. 1D), more precisely positioning this first peak of Scc2 chromatin association in late G1.
Fig. 1.
Scc2 chromatin association is biphasic. (A) DNA histograms of propidium iodide-stained (1891-32C) cells are shown in an asynchronous population and following αF-arrest and release into fresh media at 23 °C in 15-min intervals. Pre- and post-DNA replication DNA contents (1C and 2C, respectively) are indicated. (B) FLAG and G6PDH immunoblots of WCE, SN1, and pellet fractions and H2B immunoblots of pellet fractions are shown for a subset of the time course samples taken in A. (C) Scc2 protein levels in pellets were analyzed relative to their levels in WCEs (Pel:WCE, black squares), as determined by using semiquantitative immunoblotting, and this ratio was set equal to 1 for the 0 min time point. Subsequent time points were calculated relative to 0 min. Levels of Scc2-FLAG in time course pellet fractions were also normalized to chromatin-bound H2B (gray circles), as an additional control. (D) Scc2-FLAG Cdc45-HA (PMY715) cells were released from an αF arrest and sampled by chromatin fractionation at the time of release (0 min) and at 10-min intervals. FLAG, HA, G6PDH, and H2B immunoblots of WCE and pellet fractions are shown. Quantitation of ratios of Scc2 and Cdc45 in pellets as a function of their amounts in WCE is shown. (E) Viabilities of scc2-4 (1875-39B) cells under the indicated release conditions are shown with error bars indicating SD, n = 3. Student t tests determined significance, P.
Because synchrony through mitosis is poor following G1 release, we also examined Scc2 chromatin association in cells arrested at different stages of mitosis to determine when Scc2 chromatin association returns to G1 levels. Scc2 levels remained high in the pellet fractions of both metaphase- and anaphase-arrested cells (conditional cdc16 and cdc14 mutants, respectively), but were dramatically reduced in late anaphase/early telophase-arrested (cdc15) cells (Fig. S1B). Thus, Scc2 removal from chromatin in late mitosis requires progression through the cell cycle stage dependent on Cdc14, a protein phosphatase involved in the mitotic exit network and rDNA segregation that acts directly to reverse inhibitory Cdc15 phosphorylation (34, 35). Furthermore, Scc2-FLAG levels were indistinguishable in the chromatin pellets of otherwise isogenic strains carrying ∼190 or 25 rDNA repeats, suggesting that Scc2 chromatin binding is unlikely to be due to an association solely with rDNA at late cell cycle stages (Fig. S1C). Thus, we conclude that Scc2 chromatin binding affinity throughout the genome is cell cycle regulated.
Scc2 Inactivation in Post-S Phase Cells Reduces Cellular Viability.
Robust G2/M Scc2 chromatin binding suggests that Scc2 has a post-S phase function. To test this possibility, we determined whether Scc2 inactivation after S phase reduces conditional scc2-4 mutant cell viability (Fig. 1E). Briefly, scc2-4 cells released from G1 arrest for 1 h at 23 °C to allow for completion of S phase (Fig. S2) were then shifted to 37 °C for 2.5 h to inactivate Scc2-4. Control cultures were released from G1 and maintained at 23 °C or were shifted to 37 °C immediately following release. αF was readded to all cultures 1 h after release to prevent cells from entering the next S phase. Interestingly, scc2-4 cells shifted to 37 °C after DNA replication still exhibited a 50% reduction in viability compared with cells maintained at 23 °C (∼31% versus 61%, respectively), but were less severely affected than scc2-4 cells that had traversed S phase in the absence of Scc2 function (9% viability) (Fig. 1E). These data are in agreement with a reported viability reduction following Scc2 inactivation in post-S phase fission yeast cells (36). Taken together, these results suggest that Scc2 has a post-S phase function and are consistent with elevated Scc2 chromatin association in post-S phase cells.
The Appearance of Scc2 Species Is Cell Cycle Regulated.
Interestingly, two predominant Scc2 species were present in WCEs of both G1 and mitotic samples (Fig. 1 and Fig. S1). To determine whether these two Scc2 species are cell cycle regulated, Scc2-FLAG immunopurified from an extract of asynchronously growing cells was exposed to one of several extracts prepared from untagged Scc2 cells subjected to a G1 arrest/release time course. The effect of extract treatment on the ratio of slower to faster migrating forms of Scc2 was then determined by FLAG immunoblot. In general, we find that the ratio of slower to faster migrating Scc2 species decreased following treatment with extracts of cells from intervals with reduced chromatin Scc2 association, but this ratio remained higher following treatment with extracts of cells from intervals with robust Scc2 chromatin association (Fig. 2A). Thus, the appearance of different Scc2 species is cell cycle regulated, and slower migrating Scc2 predominates in cell cycle intervals in which Scc2 chromatin association is higher.
Fig. 2.
Scc2 dephosphorylation promotes amino-terminal cleavage. (A) Scc2-FLAG, immunopurified from asynchronously growing (1891-32C) cells, was incubated with WCE of untagged (1891-36D) cells staged at specific cell cycle intervals (indicated by min postαF release) or with pooled and boiled WCE. Following extensive washing, Scc2-FLAG species were analyzed via immunoblot (Upper). The ratios of slower:faster migrating forms of Scc2 were determined by using the immunoblot and plotted (Lower). (B) WCE of asynchronously growing Scc2-FLAG (1891-32C) cells was treated with lambda phosphatase or mock treated and analyzed by immunoblot using FLAG [Left (Upper and Lower are long and short exposures, respectively)] or a rabbit anti-Scc2 against the Scc2 amino terminus (Upper Right). Lower Right is a reprobing of the third image with FLAG. Carets, arrows, and asterisks indicate phosphorylated, full-length, and cleaved species of Scc2, respectively. (C) Scc2-FLAG immunopurified from WCE of asynchronously growing Scc2-FLAG cells (1891-32C) was treated with λ phosphatase before, or after, its isolation, and Scc2 species distributions were then determined by FLAG immunoblot.
Dephosphorylation Promotes Scc2 Instability.
Closer examination of WCEs of asynchronously growing cells revealed several other less prominent Scc2 species that migrate more slowly than the two predominant forms in SDS/PAGE (Fig. 2B). These species were detected by immunoblot with carboxyl-terminal–directed FLAG antibody or rabbit polyclonal serum that recognizes amino-terminal Scc2 residues spanning amino acids 40–200 (Fig. S3). The loss of these species following treatment of cell extracts with lambda phosphatase suggests that phosphorylation contributes to the existence of multiple Scc2 species (Fig. 2B), consistent with proteomics studies that document Scc2 ortholog phosphorylation (29–31).
Surprisingly, Scc2 detection with the polyclonal Scc2 serum was eliminated following phosphatase treatment (Fig. 2B). However, subsequent reprobing with FLAG antibody revealed that the faster migrating form of the two predominant Scc2 species remained (Fig. 2B, Right). Given that the anti-Scc2 serum is unlikely to recognize phosphorylated epitopes on a bacterially produced antigen, these observations instead suggest that dephosphorylation of Scc2 promotes its amino-terminal cleavage, producing the faster migrating of the two predominant Scc2 species. The slower migrating form is therefore likely to represent intact Scc2.
Dephosphorylation-mediated Scc2 instability may result from an inherent destabilization of Scc2, or an additional factor may mediate cleavage. To distinguish between these possibilities, Scc2-FLAG was treated with phosphatase either before or after its immunopurification from WCE, followed by immunoblot analysis. We observed that Scc2 cleavage occurred only when phosphatase treatment was performed in the WCE, suggesting that an additional cellular factor mediates cleavage (Fig. 2C). Scc2 was similarly cleaved following phosphatase treatment in a WCE prepared from cells that lack the major vacuolar proteases, Pep4, Prb1, and Prc1, which degrade proteins nonspecifically (reviewed in refs. 37 and 38), and in cells treated with the proteasome inhibitor, MG132 (Fig. S4). Thus, Scc2 cleavage in phosphatase-treated extracts is unlikely to be a consequence of nonspecific protein degradation or proteasome-mediated proteolysis, but is instead a result of Scc2 dephosphorylation and subsequent protein cleavage mediated by a factor present within the WCE.
That Scc2 is cleaved and processed in vivo is supported by mass spectrometric (MS) analyses. Scc2-FLAG was immunopurified from extracts prepared by cryogenic grinding of flash-frozen cells either staged in mitosis or taken 1 h after αF release (late S phase) to enhance detection of full-length and cleaved Scc2 species, respectively (Fig. 3A). Samples were then subjected to in-gel trypsin digestion, which cleaves peptides carboxyl-terminal to lysine and arginine residues. Notably, 52% of the total S phase peptides derived from Scc2 residues 144–166 lacked lysine or arginine residues immediately preceding their amino termini, resulting in a series of half-tryptic peptides that were progressively shorter from the amino terminus only, a phenomenon we refer to as laddering (Fig. 3B). In contrast, laddering was never observed in this region in the mitotic sample (Fig. 3B), an indication that these laddered peptides unique to S phase are forms of cleaved Scc2 processed in vivo. Cleaved and processed Scc2 species whose amino termini map within residues 144–155 are expected to be ∼15 kDa smaller than full-length Scc2, which is consistent with the electrophoretic mobility change we observe. The importance of this region in Scc2 function is evident by the fact that cells expressing a deletion of Scc2 residues 143–155 as the sole source of Scc2 are inviable, despite the mutant protein’s ability to associate with both Scc4 and chromatin (Fig. S5). Interestingly, Scc2∆143–155 is also subject to cleavage (Fig. S5), suggesting that the initial proteolytic event occurs outside Scc2 residues 143–155 and that subsequent processing produces the Scc2 cleavage product that we observe.
Fig. 3.
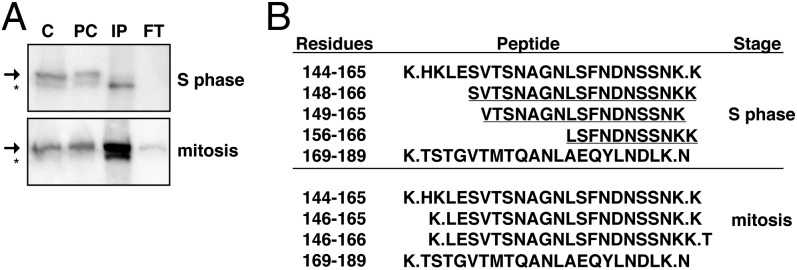
Cell cycle-specific cleavage and processing of Scc2. (A) Full-length and cleaved Scc2 forms immunoprecipitated from late S- or M-staged extracts (1891-32C) were subjected to MS. Crude (C), precleared extract (PC), immunoprecipitated (IP), and flow through (FT) samples are shown. (B) Scc2 peptides spanning residues 140–189 identified by MS in tryptic digests from late S phase or mitotic cells are shown. Full tryptic peptides are flanked by periods and the adjacent amino acids. Semitryptic peptides are underlined.
Phosphorylation Protects Scc2 from Cleavage.
Because Scc2 cleavage and reduced chromatin binding affinity appear to coincide in the cell cycle, we determined whether Scc2 phosphorylation varies in the cell cycle. We noted no gross changes in phosphorylated Scc2 species following αF synchronization and release, however, suggesting that the overall degree of Scc2 phosphorylation remains constant throughout the cell cycle (Fig. S6A). To determine whether the phosphorylation status of Scc2 affects its cleavage in the cell cycle, immunopurified Scc2-FLAG was treated with phosphatase before incubation with extracts prepared from cells at distinct cell cycle periods. Scc2 treated with phosphatase before incubation with extract yielded only the cleaved Scc2 species regardless of which cell cycle positioned extract was used (Fig. S6B). These observations contrasted with mock-treated Scc2-FLAG, which showed similar cell cycle stage-specific decreases in the proportions of full-length and cleaved Scc2 species. These results indicate that Scc2 cleavage occurs constitutively following dephosphorylation.
Scc2 Cleavage Eliminates Its Interactions with Scc4 and Reduces Its Cohesin Deposition Activity.
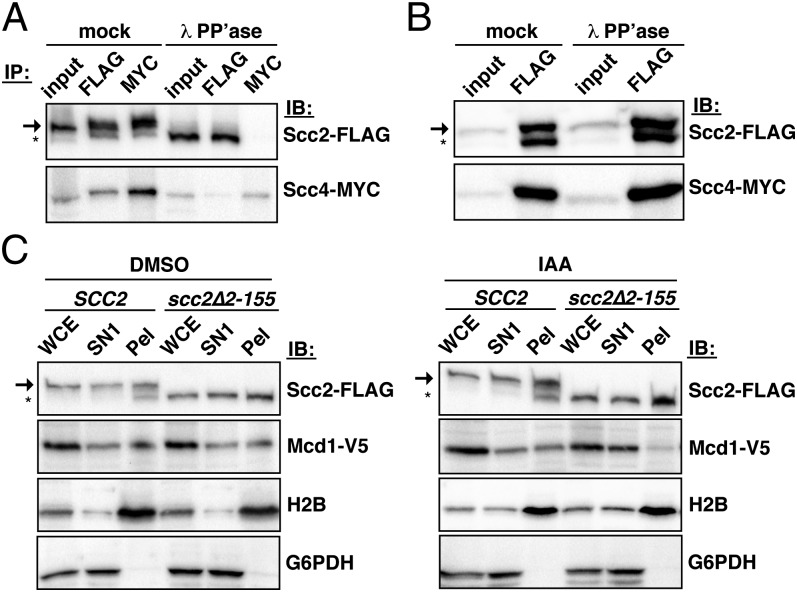
The amino termini of human and Xenopus Scc2 homologs are required for interactions with their respective Scc4 orthologs (10, 13, 17, 18). Whether this requirement is true in budding yeast is unclear, however, because S. cerevisiae Scc2 lacks the corresponding amino-terminal region present in multicellular eukaryotic homologs. To assess the basis of budding yeast Scc2–Scc4 interactions, we determined the effect of dephosphorylation-promoted Scc2 cleavage on its association with Scc4. WCE from Scc2-FLAG Scc4-6His-13Myc cells was first phosphatase treated to promote Scc2 cleavage or mock treated and then subjected to reciprocal coimmunoprecipitation. We find that whereas Scc4 and full-length Scc2, but not its cleaved form, are efficiently coimmunoprecipitated in mock-treated extracts, the reciprocal coimmunoprecipitations of Scc4 and Scc2 are dramatically reduced following dephosphorylation-promoted Scc2 cleavage (Fig. 4A and Fig. S3). In contrast, phosphatase treatment of Scc2-FLAG after it has been immunopurified, which does not promote Scc2 cleavage, does not alter its ability to coimmunoprecipitate Scc4, indicating that cleavage, rather than Scc2 dephosphorylation per se, is responsible for the reduction in Scc2–Scc4 interactions (Fig. 4B). We conclude that the budding yeast Scc2 amino terminus is required for its interaction with Scc4, as is the case for the human and Xenopus homologs of these two proteins.
Fig. 4.
Scc2 cleavage affects its interaction with Scc4 and Mcd1 chromatin association. (A) Reciprocal coimmunoprecipitations were performed in λ phosphatase- or mock-treated extracts of Scc2-FLAG Scc4-13MYC cells (1891-32C). The arrow and asterisk indicate full-length and cleaved Scc2, respectively. (B) Scc2-FLAG immunopurified from Scc2-FLAG Scc4-13MYC cells (1891-32C) was subsequently either mock treated or treated with phosphatase. Samples were then immunoblotted with FLAG or MYC to determine the ability of phosphatase-treated samples to coimmunoprecipitate Scc4-13MYC. (C) Cells expressing plasmid-borne Scc2-FLAG or Scc2∆2-155-FLAG and chromosomal Scc2 fused to an auxin-inducible domain (JWY214 and JWY215, respectively) were treated with auxin or vehicle only (DMSO) as described in the text and then subjected to fractionation. Immunoblots of WCE, SN1, and pellet fractions are shown for the indicated proteins.
An expected consequence of the loss of Scc2–Scc4 interactions following dephosphorylation-promoted Scc2 cleavage is reduced cohesin deposition. To test this prediction, we examined the chromatin association of the Mcd1 cohesin subunit in cells expressing as the sole source of Scc2 a truncation mutant that lacks residues 2–155, which corresponds to the most extensively cleaved and processed Scc2 species observed by mass spectrometry. Although Scc2∆2-155 is present at levels similar to a wild-type control in the WCE, this mutant is incapable of serving as the only source of Scc2 function (Fig. S5A). Therefore, Scc2-FLAG or Scc2∆2-155-FLAG was expressed from a plasmid in cells in which chromosomally derived Scc2 is rapidly degraded by addition of a plant auxin to the culture medium (SI Materials and Methods). αF-synchronized cells were treated with auxin, released from G1 in auxin-containing medium, and then subjected to chromatin fractionation after cells reached a mitotic arrest. As predicted, cells dependent solely on Scc2∆2-155 for Scc2 function had significantly lower levels of Mcd1-V5 in chromatin pellets and higher levels of Mcd1-V5 in the soluble SN1 fraction compared with cells that expressed wild-type Scc2 (Fig. 4C). Although cleaved Scc2 does not coimmunoprecipitate with Scc4, which likely assays interactions of soluble proteins rather than interactions within the context of chromatin, we note that cleaved Scc2 and Scc2∆2–155 remain in chromatin pellets (Fig. 4C), suggesting that the presence of chromatin may stabilize Scc2 and its interactions in vivo. Nevertheless, we demonstrate that cleaved Scc2 is compromised in its cohesin deposition activity.
Discussion
We demonstrate that Scc2 chromatin binding is regulated biphasically during the cell cycle, with increases that occur first transiently in late G1 when Scc2/Scc4-mediated cohesin deposition is required to tether together sister chromatids produced in the ensuing S phase, and then later, in post-S phase cells, for unknown reasons. Scc2 is a phospho-protein and is subject to amino-terminal cleavage in vivo, which can be faithfully recapitulated in vitro by treatment of immunopurified Scc2 with extracts of cells in stages of the cell cycle that exhibit poor Scc2 chromatin binding, and by dephosphorylation of cellular extracts. Although the appearance of amino terminally cleaved Scc2 in vivo correlates with decreased Scc2 chromatin binding, cleaved Scc2 is not immediately lost from chromosomes. Importantly, cleaved Scc2 does not interact with Scc4 and is likely inactive, as indicated by dramatically reduced cohesin association in a strain expressing only an amino terminally truncated Scc2. A model consistent with these data are that dephosphorylation, likely of Scc2 itself, promotes proteolysis within the amino terminus of Scc2, which disrupts Scc2–Scc4 interactions, resulting in the inactivation of Scc2/Scc4-mediated cohesin deposition. In this view, the regulation of Scc2 phosphorylation status is the critical event controlling its interaction with Scc4 and, therefore, its activity in cohesin deposition.
Biphasic Scc2 recruitment to chromatin was unexpected. The existence of mechanisms that target Scc2/Scc4 to key chromosomal regions, such as pericentromeres, may explain why the modest peak of Scc2/Scc4 in late G1 suffices in early cell cycle periods. Kinetochores directly mediate Scc2/Scc4 enrichment within pericentromeric chromatin, thereby ensuring the robust cohesion of sister chromatids that is vital for promoting chromosome biorientation, even under conditions in which cohesins are limiting (27, 39, 40). Perhaps more surprising given a previous report that Scc2 is not essential after S phase in the absence of DNA damage (7) are our observations that robust Scc2/Scc4 chromatin association is achieved in G2/M cells and that post-S phase cell viability is reduced following Scc2 inactivation. The reason for the discrepancy in these two studies is unclear, because both used the same conditional scc2-4 allele. Nevertheless, our results are consistent with a post-S phase role for Scc2/Scc4. Although this role is undefined, we note that a cohesin-independent role for the human Scc2 ortholog, NIPBL, in the transcriptional regulation of a subset of genes has recently been proposed (28), and several studies have demonstrated Scc2/Scc4-cohesin colocalization on chromosomes (27, 41, 42). It remains possible then that Scc2/Scc4 contributes to the stability, and therefore function, of cohesins on mitotic chromosomes or is involved in anchoring cohesins to particular chromosomal locations that are required for optimal function.
Our results also indicate that passage through mitosis resets high levels of Scc2 chromatin association observed through midanaphase to the lower levels observed in late anaphase/early telophase and G1-arrested cells, suggesting that this transition depends on the activity of Cdc14, a protein phosphatase with an important role in the inactivation of cyclin-dependent kinases necessary for mitotic exit (34). Although Cdc14 is largely sequestered in the nucleolus until anaphase onset, Dsn1 kinetochore subunit dephosphorylation in metaphase is Cdc14-dependent, suggesting that sufficient levels of Cdc14 may escape nucleolar sequestration and could promote Scc2 dephosphorylation and subsequent cleavage during S phase (43). Further experimentation will be required, however, to determine whether Scc2 is a direct substrate of Cdc14 in vitro. Furthermore, our finding that Scc2 remains chromatin associated until early telophase contradicts a recent suggestion that Scc2/Scc4 chromatin association is cohesin dependent, because cohesins are removed from chromosomes at the metaphase/anaphase transition by separase-dependent proteolysis (44, 45).
Vertebrate Scc2 and Scc4 orthologs interact physically through their amino termini (10, 13, 17, 18), but budding yeast Scc2 appears to lack the corresponding amino-terminal region. Nevertheless, we found that Scc2 cleavage prevents its coimmunoprecipitation with Scc4, indicating that the budding yeast Scc2 amino-terminal domain is required for stable interactions with Scc4. Moreover, disrupted Scc2–Scc4 interactions resulting from Scc2 cleavage strongly suggest that, despite its relative stability, the carboxyl-terminal cleavage product of Scc2 is unable to mediate cohesin deposition. It will be of interest to explore additional regions of the Scc2 and Scc4 proteins to identify which domains are required for the different functional activities of the deposition factor complex.
That Scc2 is constitutively cleaved in vitro following its dephosphorylation suggests that Scc2’s phosphorylation status ultimately regulates its proteolysis and, consequently, its ability to associate with Scc4 and mediate cohesin deposition. Notably, we did not detect gross cell cycle-specific alterations in Scc2 electrophoretic migration patterns, sometimes indicative of changes in phosphorylation state, suggesting that the phosphorylation status of a small number of key Scc2 residues may determine its susceptibility to cleavage. Importantly, we also find that in vitro Scc2 dephosphorylation per se is insufficient to disrupt Scc2–Scc4 interactions, suggesting that dephosphorylation instigates in vivo events that culminate in Scc2 proteolytic cleavage and inactivation. One scenario is that chromatin-bound Scc2 is first targeted by a protein phosphatase at specific cell cycle intervals. Dephosphorylated Scc2 then becomes susceptible to cleavage, disrupting its interaction with Scc4 and, importantly, its cohesin deposition activity. Interestingly, our observation that cleaved Scc2 maintains an association with chromatin strongly suggests that chromatin-bound Scc2 is a suitable cleavage substrate. We note, however, that immunopurified Scc2 is susceptible to cleavage, indicating that residence on chromatin is not essential for Scc2 proteolysis. Therefore, it is possible that Scc2 dephosphorylation reduces its chromatin binding affinity directly, or indirectly promotes its eviction from chromatin through the activity of an unknown factor, and once removed, Scc2 is then susceptible to cleavage by proteases. This scenario seems unlikely, however, given that only a small fraction of the substantial pool of Scc2 is cleaved in vivo. In any case, these data suggest that the phospho-regulation of Scc2 stability represents another in a growing list of mechanisms that regulate the spatiotemporal association of cohesin with chromosomes.
Materials and Methods
See SI Materials and Methods for detailed materials and methods. Relevant strain genotypes are listed in Table S1. G1 or mitotic arrests using αF mating pheromone or nocodazole, respectively, were done as described (46). To determine viability, cultures were serially diluted and plated in triplicate at a density of ∼200 cells per plate. Colonies were counted after 3 d at 23 °C, and the percent viable cells was calculated by using cell numbers in the original culture at the time of dilution. Chromatin fractionation was performed as described (32) with minor modification. Chromatin-bound proteins were quantitated relative to the amount of protein present in the corresponding WCE by using semiquantitative immunoblotting. Unless stated otherwise, full-length and cleaved forms of Scc2, whose resolution required 6% (vol/vol) PAGE gels, were included in computations of chromatin binding. In the in vitro Scc2 cleavage assay, beads containing immunopurified Scc2-FLAG were incubated with a second WCE lacking FLAG-tagged proteins for 2 h at 4 °C. Where indicated, WCEs (∼40 μg/mL total protein) were treated with 200 U of lambda phosphatase (New England Biolabs) in a 100-μL reaction incubated at 30 °C for 0.5–1 h. Polypeptides corresponding to amino acids 40–200 of Scc2, either free of, or fused to GST, were purified and used to inoculate rabbits to raise polyclonal sera against Scc2 (Covance Research Products). Scc2-FLAG was immunopurified as described (47) and, following electrophoresis, was subjected to in-gel trypsin digestion in preparation for mass spectrometry. Peptides were identified by using nanoelectrospray liquid chromatography tandem mass spectrometry.
Supplementary Material
Acknowledgments
We thank Drs. D. Bentley, D. Schneider, R. Sclafani, and R. Zhao for strains or reagents; S. Johnson and A. Johnson for advice on antibody production and mutagenesis, respectively; J. Ramey for advice with figures; and Drs. Chad Pearson and Robert Sclafani for helpful comments on the manuscript. This work was supported by National Institutes of Health Grants R01 GM066213 (to P.C.M.) and T-32 GM008730 (to J.W.), and a fellowship from the Cornelia de Lange Syndrome Foundation (to J.W.). This work was also supported in part by Flow Cytometry Shared Resource of the University of Colorado Cancer Center Support Grant P30CA046934.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321722111/-/DCSupplemental.
References
- 1.Eckert CA, Gravdahl DJ, Megee PC. The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 2007;21(3):278–291. doi: 10.1101/gad.1498707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16(6):1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 3.Ünal E, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16(6):991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 4.Dorsett D. Cohesin: Genomic insights into controlling gene transcription and development. Curr Opin Genet Dev. 2011;21(2):199–206. doi: 10.1016/j.gde.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ribeiro de Almeida C, Stadhouders R, Thongjuea S, Soler E, Hendriks RW. DNA-binding factor CTCF and long-range gene interactions in V(D)J recombination and oncogene activation. Blood. 2012;119(26):6209–6218. doi: 10.1182/blood-2012-03-402586. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Krantz ID. Cornelia de Lange syndrome, cohesin, and beyond. Clin Genet. 2009;76(4):303–314. doi: 10.1111/j.1399-0004.2009.01271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciosk R, et al. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5(2):243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 8.Arumugam P, et al. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr Biol. 2003;13(22):1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 9.D’Ambrosio C, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22(16):2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi TS, Basu A, Bermudez V, Hurwitz J, Walter JC. Cdc7-Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev. 2008;22(14):1894–1905. doi: 10.1101/gad.1683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gause M, et al. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma. 2008;117(1):51–66. doi: 10.1007/s00412-007-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14(17):1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 13.Watrin E, et al. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol. 2006;16(9):863–874. doi: 10.1016/j.cub.2006.03.049. [DOI] [PubMed] [Google Scholar]
- 14.Gruber S, et al. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127(3):523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 15.Hu B, et al. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr Biol. 2011;21(1):12–24. doi: 10.1016/j.cub.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang CE, Milutinovich M, Koshland D. Rings, bracelet or snaps: Fashionable alternatives for Smc complexes. Philos Trans R Soc Lond B Biol Sci. 2005;360(1455):537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braunholz D, et al. Isolated NIBPL missense mutations that cause Cornelia de Lange syndrome alter MAU2 interaction. Eur J Hum Genet. 2012;20(3):271–276. doi: 10.1038/ejhg.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bermudez VP, et al. In vitro loading of human cohesin on DNA by the human Scc2-Scc4 loader complex. Proc Natl Acad Sci USA. 2012;109(24):9366–9371. doi: 10.1073/pnas.1206840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murayama Y, Uhlmann F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature. 2014;505(7483):367–371. doi: 10.1038/nature12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6(10):991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- 21.Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8(20):1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- 22.Jahnke P, et al. The Cohesin loading factor NIPBL recruits histone deacetylases to mediate local chromatin modifications. Nucleic Acids Res. 2008;36(20):6450–6458. doi: 10.1093/nar/gkn688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakimi MA, et al. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418(6901):994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 24.Ritchie K, et al. Loss of ATRX leads to chromosome cohesion and congression defects. J Cell Biol. 2008;180(2):315–324. doi: 10.1083/jcb.200706083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baetz KK, Krogan NJ, Emili A, Greenblatt J, Hieter P. The ctf13-30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol Cell Biol. 2004;24(3):1232–1244. doi: 10.1128/MCB.24.3.1232-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, Hsu JM, Laurent BC. The RSC nucleosome-remodeling complex is required for Cohesin’s association with chromosome arms. Mol Cell. 2004;13(5):739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 27.Kogut I, Wang J, Guacci V, Mistry RK, Megee PC. The Scc2/Scc4 cohesin loader determines the distribution of cohesin on budding yeast chromosomes. Genes Dev. 2009;23(19):2345–2357. doi: 10.1101/gad.1819409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuin J, et al. A cohesin-independent role for NIPBL at promoters provides insights in CdLS. PLoS Genet. 2014;10(2):e1004153. doi: 10.1371/journal.pgen.1004153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson-Grady JT, Villén J, Gygi SP. Phosphoproteome analysis of fission yeast. J Proteome Res. 2008;7(3):1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- 30.Villén J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci USA. 2007;104(5):1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dephoure N, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105(31):10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11(24):3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280(5363):593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 34.Visintin R, et al. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2(6):709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 35.Jaspersen SL, Morgan DO. Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr Biol. 2000;10(10):615–618. doi: 10.1016/s0960-9822(00)00491-7. [DOI] [PubMed] [Google Scholar]
- 36.Furuya K, Takahashi K, Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 1998;12(21):3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones EW. Three proteolytic systems in the yeast saccharomyces cerevisiae. J Biol Chem. 1991;266(13):7963–7966. [PubMed] [Google Scholar]
- 38.Wolf DH. From lysosome to proteasome: The power of yeast in the dissection of proteinase function in cellular regulation and waste disposal. Cell Mol Life Sci. 2004;61(13):1601–1614. doi: 10.1007/s00018-004-4134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol. 2010;20(10):957–963. doi: 10.1016/j.cub.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Natsume T, et al. Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol Cell. 2013;50(5):661–674. doi: 10.1016/j.molcel.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin W, Jin H, Liu X, Hampton K, Yu HG. Scc2 regulates gene expression by recruiting cohesin to the chromosome as a transcriptional activator during yeast meiosis. Mol Biol Cell. 2011;22(12):1985–1996. doi: 10.1091/mbc.E10-06-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onn I, Koshland D. In vitro assembly of physiological cohesin/DNA complexes. Proc Natl Acad Sci USA. 2011;108(30):12198–12205. doi: 10.1073/pnas.1107504108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akiyoshi B, Biggins S. Cdc14-dependent dephosphorylation of a kinetochore protein prior to anaphase in Saccharomyces cerevisiae. Genetics. 2010;186(4):1487–1491. doi: 10.1534/genetics.110.123653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernius J, et al. Cohesin-dependent association of scc2/4 with the centromere initiates pericentromeric cohesion establishment. Curr Biol. 2013;23(7):599–606. doi: 10.1016/j.cub.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103(3):375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 46.Weber SA, et al. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2(9):E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cristea IM, Chait BT. Affinity purification of protein complexes. Cold Spring Harbor Protocols. 2011;2011(5):pdb prot5611. doi: 10.1101/pdb.prot5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.