Significance
Few things are excluded in biology if they are not physically impossible, but some have been advocated to be, for instance, the absence of reverse flow of information from phenotype to genotype, often assimilated to Lamarckism. Regardless of the question of whether this prohibition is universally true or not, could any such prohibition logically result from natural selection? We introduce a mathematical model to examine this question and, more generally, the conditions under which various mechanisms for generating and transmitting variations in evolving populations may be favored or suppressed by natural selection.
Keywords: Darwinian evolution, Lamarckism, heredity, acquired characteristics, epigenetics
Abstract
The inheritance of characteristics induced by the environment has often been opposed to the theory of evolution by natural selection. However, although evolution by natural selection requires new heritable traits to be produced and transmitted, it does not prescribe, per se, the mechanisms by which this is operated. The mechanisms of inheritance are not, however, unconstrained, because they are themselves subject to natural selection. We introduce a schematic, analytically solvable mathematical model to compare the adaptive value of different schemes of inheritance. Our model allows for variations to be inherited, randomly produced, or environmentally induced, and, irrespectively, to be either transmitted or not during reproduction. The adaptation of the different schemes for processing variations is quantified for a range of fluctuating environments, following an approach that links quantitative genetics with stochastic control theory.
Three principles underlie the explanation of adaptations by natural selection: (i) individuals in a population have varied characteristics; (ii) their reproductive success correlates with these characteristics; (iii) the characteristics are inherited. The last principle, of inheritance, has always been the most contentious. At the time of Darwin and Wallace, its mechanisms were unknown, and fundamental questions, such as the role of the environment in the production of new, adaptive traits, were unsettled. Adaptation by natural selection does not, indeed, require any causal relation between the environment and newly generated traits, but neither does it exclude it; Darwin, for instance, included as potential sources of variations the direct and indirect effects of the environment, as well as the use and disuse of organs, in line with ideas previously propounded by Lamarck (1, 2).
Prominent followers of Darwin, however, came to exclude the possibility of inheritance of acquired characteristics. This viewpoint was notably formulated by Weismann in his theory of continuity of the germplasm (3). Experiments of amputations, which showed no incidence on the progeny, supported it. At the end of the nineteen century, it had became a central tenet of “neo-Darwinism” (4). Half a century later, the “Modern Synthesis,” which produced a synthesis between evolution theory and Mendel’s laws of inheritance (5), reached the same conclusion: it promoted a clear distinction between genotypes, inherited but only subject to random variations, and phenotypes, affected by the environment but not directly transmitted. These conclusions were based on studies in multicellular organisms, but subsequent experiments with microorganisms, which found that adaptive variations can precede changes of environmental conditions (6), further reinforced the conviction that biological evolution is mainly fueled by random variations. At a molecular level, finally, once prevalent instructional theories of enzymatic adaptation or antibody formation also came to be discarded in the 1950s and 1960s (7, 8). At this time, the successes of molecular biology in unraveling the mechanisms of heredity elevated a molecular refutation of Lamarckism, the unidirectional flow of information from DNA to proteins, as its “central dogma” (9).
Concurrent views, emphasizing the role of environmentally induced variations in evolution, have had several insightful proponents (10–13), but were also endorsed by dubious yet influential supporters (14). Examples of inherited acquired characteristics have, however, been long known, from the transmission of culture in humans to the uptake of extracellular DNA by bacteria. However, only recently have we gained a fuller recognition of the diversity of mechanisms for generating and transmitting variations (15). In addition to the well-recognized roles of mutations and recombinations of chromosomal DNA, a nonexhaustive list would include the transmission of acquired chromatin marks such as DNA methylation, the transmission of small interfering RNAs, the transmission of conformational states of molecules such as prions, or, at the cellular level, the transmission of self-sustaining states of gene regulation, and, at the organismal level, so-called parental effects (16).
Inheritance, long treated as an autonomous and universal mechanism to be experimentally characterized and then integrated to evolutionary theory, thus appears to consist of multiple and parallel systems whose origins and implications are to be explained within an evolutionary framework. The problem of a synthesis of inheritance with evolution is thus now doubled by the problem of the synthesis of inheritance by evolution. With this problem in view, we propose here a mathematical model where the adaptive values of different schemes for generating and transmitting variations can be compared. The model treats inheritance as a trait on which selection can act, although not in a direct way: systems of inheritance indeed pertain to the transmission of traits between individuals, and estimating their adaptive value therefore requires analyzing the dynamics of a population over several generations. In this sense, the adaptation of a mode of inheritance is necessarily of “second order”—a form of “evolvability.”
Our model thus quantifies the adaptation of different modes of inheritance by considering the long-term growth rate of populations. The model is schematic: it relies on an abstraction from physical implementations, along the example of Shannon’s model of communication (17). The model thus defines the “genotype” as what is transmitted between successive generations, and the “phenotype” as what determines the survival and reproduction of an individual, with no reference to their material support. As a consequence, our distinction between genotypic and phenotypic variations is not equivalent to the often-made distinction between genetic and epigenetic inheritance; any transmitted character, whether DNA encoded or not, will belong, from the standpoint of our model, to the genotype.
In general, few things are excluded in biology if they are not physically impossible—but some have been proposed to be, for instance, the reverse flow of information from phenotype to genotype. Regardless of the question of whether this prohibition is indeed universally true or not, it is interesting to consider whether any such prohibition could logically result from natural selection. More generally, under what conditions various mechanisms for generating and transmitting variations may be favored or suppressed by natural selection itself? We illustrate the versatility of our model by examining this question in the context of three biological phenomena that are often considered to be either irrelevant to evolution, or absent because “forbidden”:
-
i)
Noninherited variations, sometimes also referred to as phenotypic “noise,” and commonly thought to have no evolutionary implications; for instance, in the first chapter of The Origin of Species (1), Darwin states: “Any variation which is not inherited is unimportant for us.” Considering that new variations may generally be introduced at the genotypic and/or at the phenotypic level, and may thus be transmitted to future generations either totally, in part or not at all, what scheme is most conductive to adaptation? Does natural selection generically favor “developmental canalization,” i.e., a reduction of phenotypic differences between individuals inheriting a common genotype? Our model will highlight how the answer depends on the statistical structure of the environment, and how nontransmitted variations may under some conditions be more beneficial than transmitted variations.
-
ii)
The absence of reverse flow of information from phenotype to genotype, advocated by Weismann, but now challenged even in the species where it is best established (18). Given that isolating the transmitted genotype from the phenotype may involve dedicated mechanisms, can we characterize the conditions under which natural selection favors their presence?
-
iii)
The nondirected nature of new adaptive variations, associated with the refutation of any “Lamarckian” mechanism. Nothing in principle prevents the environment from inducing the generation of new traits, either at the phenotypic level, or at the genotypic level: the first effect, known as “plasticity,” has long been recognized (19), and the second one, long thought to be forbidden, is also observed (20). Are there nevertheless conditions under which direct integration of information into the transmitted genotype is logically excluded as a consequence of natural selection?
Despite the fundamental nature of these questions, no previous formal model exists, to our knowledge, that addresses them in a common and analytically tractable framework. Our model, however, is not without precedents: it is in line with traditional models of quantitative genetics (21) and relates to models of stochastic control in engineering (22). Processing unreliable informations from the past and present to confront an uncertain future, which may be seen as the fundamental “function” of systems of inheritance, is indeed at its core a question of control. We thus proposed previously that evolution in fluctuating environments could be viewed as a problem of stochastic control (23); this view is supported here by a formal analogy between our model and a basic algorithm in stochastic control theory, the Kalman filter (24).
Model
We provide in this section a general presentation of the model and derive its solution in a simple case. The general solution follows the same principles and its details are included in SI Appendix.
Definition.
The model considers a population of asexually reproducing individuals where each genotype is characterized by an “attribute” γ. The genetic or epigenetic nature of this attribute is irrelevant: we are only concerned with the origins of the transmitted information, either inherited, randomly produced, or environmentally induced, irrespective of its material support. In the parlance of Weismann, γ would be termed the “germplasm,” and, in the parlance of population genetics, the “breeding value.” At each time step, corresponding to a generation, each individual with attribute γ reproduces and is replaced by ξ offsprings sharing as common attribute  , with possibly
, with possibly 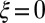 , in which case we conventionally define
, in which case we conventionally define 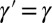 . (The model could be generalized to produce offsprings with different attributes, but we purposely ignore this unessential complication.) The individuals are noninteracting and the generations nonoverlapping. The values of ξ and
. (The model could be generalized to produce offsprings with different attributes, but we purposely ignore this unessential complication.) The individuals are noninteracting and the generations nonoverlapping. The values of ξ and 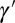 can depend on γ and on the current environmental state, which fluctuates independently of the population and is characterized by a variable
can depend on γ and on the current environmental state, which fluctuates independently of the population and is characterized by a variable 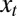 . This dependency can be stochastic and is generally given by a stochastic kernel
. This dependency can be stochastic and is generally given by a stochastic kernel 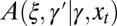 with the following properties:
with the following properties: 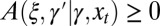 and
and 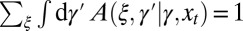 for all
for all 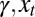 .
.
Population Dynamics.
For a given series of environmental states  , we define
, we define 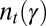 as the expected probability density function of the attribute γ in the population at time (generation) t, normalized to
as the expected probability density function of the attribute γ in the population at time (generation) t, normalized to 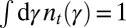 at any t. It satisfies a recursive equation which, more formally, is the recursion for the first moment of the branching process:
at any t. It satisfies a recursive equation which, more formally, is the recursion for the first moment of the branching process:
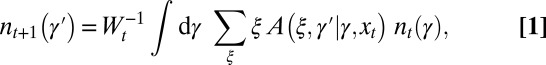 |
where 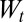 is a normalization ensuring
is a normalization ensuring 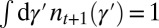 ,
,
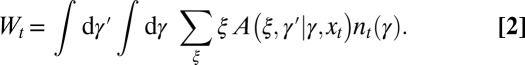 |
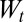 represents the factor by which the population increases, on average, between generations t and
represents the factor by which the population increases, on average, between generations t and 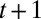 .
.
Starting from a large number 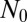 of individuals at time
of individuals at time 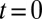 , the expected total number
, the expected total number 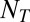 of individuals at time T is thus the following:
of individuals at time T is thus the following:
 |
Over T generations, this results in the following growth rate:
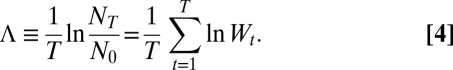 |
We are interested here in the long-term limit 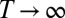 , under the assumption that the population does not go extinct. (We assume that the branching process is supercritical and ignore the fluctuations associated with small populations, which is justified in the large t limit when the population is exponentially growing.) This limit is mathematically well defined when the environment follows an ergodic process, in which case we have the following:
, under the assumption that the population does not go extinct. (We assume that the branching process is supercritical and ignore the fluctuations associated with small populations, which is justified in the large t limit when the population is exponentially growing.) This limit is mathematically well defined when the environment follows an ergodic process, in which case we have the following:
where 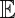 indicates an expectation with respect to the environmental fluctuations (25) (Λ is also known as the quenched Lyapunov exponent for the underlying branching process in a random environment).
indicates an expectation with respect to the environmental fluctuations (25) (Λ is also known as the quenched Lyapunov exponent for the underlying branching process in a random environment).
The long-term growth rate Λ is a “group-level” property, attached to the population as a whole rather than to any particular individual. It is relevant for the comparison of different schemes of inheritance because of the following property (23): given two populations, characterized by kernels 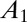 and
and 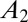 , and given an ergodic environmental process, if
, and given an ergodic environmental process, if 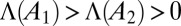 , then, almost surely,
, then, almost surely, 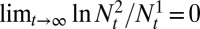 , where
, where 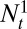 and
and 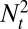 represent the respective sizes of the two populations at time t. In other words, Λ predicts the long-term outcome of a competition between populations characterized by different kernels A. This argument assumes an exponentially growing population, but its conclusions are equally valid in presence of a constraint on the total population size, in which case the population with smallest growth rate almost surely becomes extinct.
represent the respective sizes of the two populations at time t. In other words, Λ predicts the long-term outcome of a competition between populations characterized by different kernels A. This argument assumes an exponentially growing population, but its conclusions are equally valid in presence of a constraint on the total population size, in which case the population with smallest growth rate almost surely becomes extinct.
Long-Term Growth Rate.
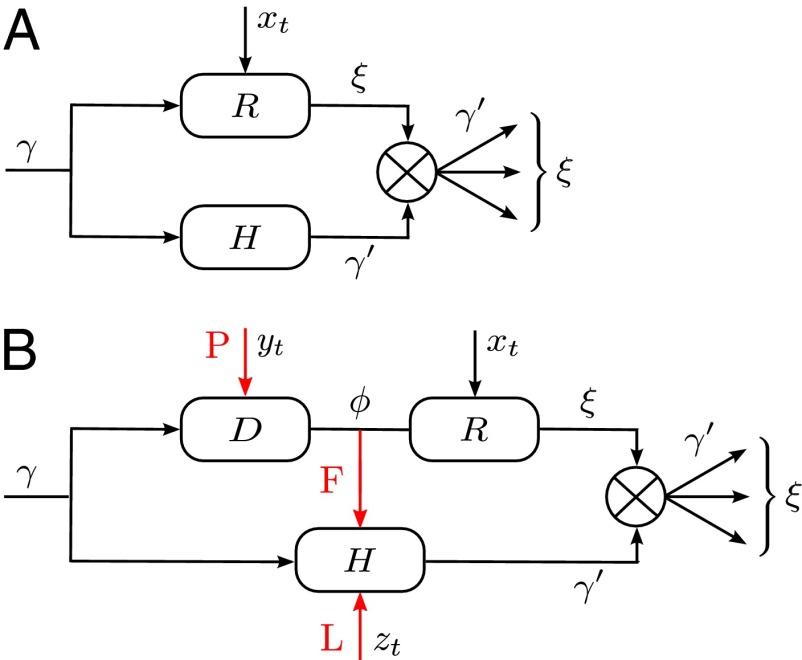
Different systems of inheritance are represented in the model by different kernels A. In the simplest case, schematically represented in Fig. 1A, the environment affects reproduction but not transmission, and the kernel A is factorized into the product of a reproduction kernel R and an heredity kernel H,
From now on, for simplicity, we also assume a single continuous attribute 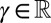 , although the model remains analytically solvable in the multidimensional case.
, although the model remains analytically solvable in the multidimensional case.
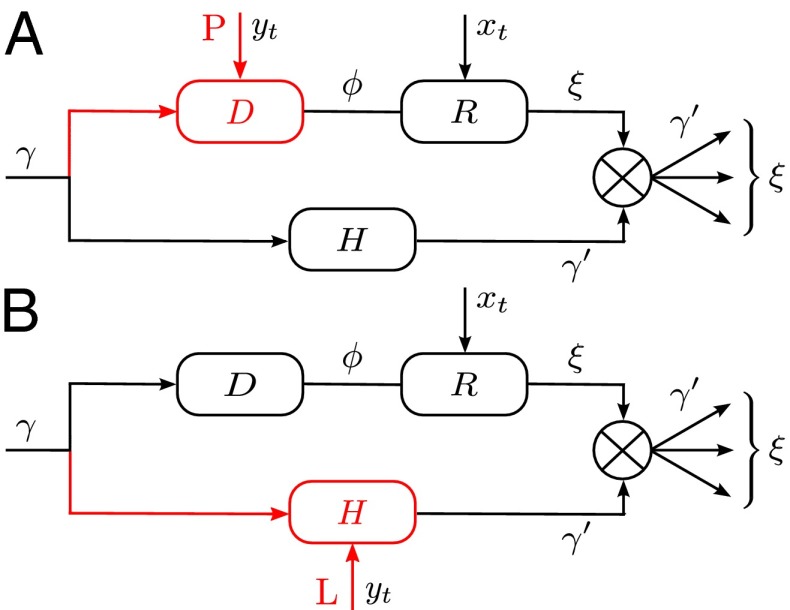
Fig. 1.
(A) Simple model. In this model, no distinction is made between inherited genotype and phenotype: both are characterized by a single attribute γ. The number ξ of offsprings and the attribute 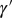 that they transmit are independently specified by two stochastic kernels, the “reproduction kernel”
that they transmit are independently specified by two stochastic kernels, the “reproduction kernel” 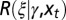 and the “heritability kernel”
and the “heritability kernel” 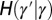 , where γ represents the attribute inherited by the individual and
, where γ represents the attribute inherited by the individual and 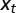 the current state of the environment. (B) General model. This model includes explicitly a phenotype ϕ derived from the inherited genotype γ through a stochastic kernel
the current state of the environment. (B) General model. This model includes explicitly a phenotype ϕ derived from the inherited genotype γ through a stochastic kernel 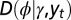 representing “development.” This kernel, and the kernels
representing “development.” This kernel, and the kernels 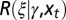 and
and 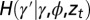 , can depend on external, environmental factors,
, can depend on external, environmental factors, 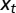 ,
, 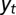 ,
, 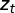 . The three red arrows represent respectively developmental plasticity (P), phenotype-to-genotype feedback (F), and direct Lamarckian effects (L).
. The three red arrows represent respectively developmental plasticity (P), phenotype-to-genotype feedback (F), and direct Lamarckian effects (L).
We take the heredity kernel to be Gaussian,
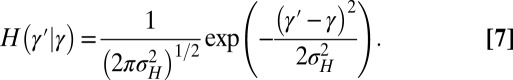 |
This corresponds to a standard assumption of additivity in population genetics: 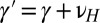 , with νH a normally distributed random variable with variance
, with νH a normally distributed random variable with variance 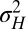 ,
, 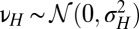 (this relation is extended below to include a possible reversion toward a mean, i.e.,
(this relation is extended below to include a possible reversion toward a mean, i.e., 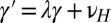 with
with 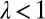 ).
).
To compute 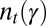 , as in Eq. 1, we only need to specify the first moment of the selection kernel
, as in Eq. 1, we only need to specify the first moment of the selection kernel 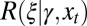 . We assume here that the expected number of offsprings of an individual with attribute γ in environment
. We assume here that the expected number of offsprings of an individual with attribute γ in environment 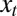 is the following:
is the following:
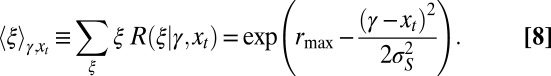 |
Here, the difference 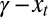 captures the “fitness” of an individual with attribute γ to the current criterion of selection
captures the “fitness” of an individual with attribute γ to the current criterion of selection 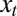 . Because in the simplest version of the model, depicted in Fig. 1A, there is no distinction between inherited genotype and phenotype, the attribute γ can be thought as a phenotypic trait and the “state of the environment”
. Because in the simplest version of the model, depicted in Fig. 1A, there is no distinction between inherited genotype and phenotype, the attribute γ can be thought as a phenotypic trait and the “state of the environment” 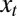 is thus being defined relative to the population, as the value of the trait that is optimal in this environment (for instance, in a classical simplistic picture of adaptation,
is thus being defined relative to the population, as the value of the trait that is optimal in this environment (for instance, in a classical simplistic picture of adaptation, 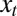 would be associated with the height of acacias and γ with the length of the giraffe neck). The variance
would be associated with the height of acacias and γ with the length of the giraffe neck). The variance 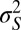 describes the selectivity of the environment;
describes the selectivity of the environment; 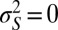 means that only one phenotype can survive at any given time, whereas
means that only one phenotype can survive at any given time, whereas 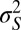 large means that many different phenotypes can survive. Finally,
large means that many different phenotypes can survive. Finally, 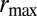 is the maximal reproductive rate per generation for the species; in particular,
is the maximal reproductive rate per generation for the species; in particular, 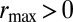 is a necessary condition for the population not to go extinct. [
is a necessary condition for the population not to go extinct. [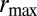 affects the growth rate Λ only through an additive constant, i.e.,
affects the growth rate Λ only through an additive constant, i.e., 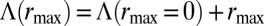 , and therefore plays no role when comparing different inheritance schemes.]
, and therefore plays no role when comparing different inheritance schemes.]
Starting at 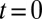 from a large population with a normally distributed attribute,
from a large population with a normally distributed attribute, 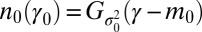 , the distribution of γ remains normally distributed at all times, i.e.,
, the distribution of γ remains normally distributed at all times, i.e., 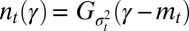 for all t; more generally, starting from any distribution
for all t; more generally, starting from any distribution 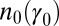 , the distribution of the trait in the population will converge to a Gaussian. Assuming that it does not go extinct, the long-term evolution of the population can thus be described in terms of just two parameters, the mean
, the distribution of the trait in the population will converge to a Gaussian. Assuming that it does not go extinct, the long-term evolution of the population can thus be described in terms of just two parameters, the mean 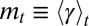 and variance
and variance 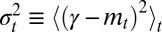 of γ at time t.
of γ at time t.
From Eq. 1, it follows that these two parameters satisfy the following:
where the so-called heritability 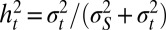 represents the contribution of the genotype to the total phenotypic variance (26) and satisfies here the following recursion:
represents the contribution of the genotype to the total phenotypic variance (26) and satisfies here the following recursion:
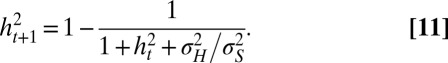 |
Eq. 1 also yields in terms of these variables 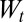 , the factor by which the population size increases between times t and
, the factor by which the population size increases between times t and 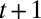 ,
,
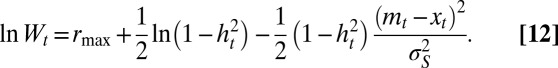 |
Stochastically Fluctuating Environments.
Up to here, the equations are valid for any kind of environmental process. If we now assume an ergodic environment, we obtain, by taking the 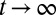 limit, a formal expression for the long-term growth rate,
limit, a formal expression for the long-term growth rate,
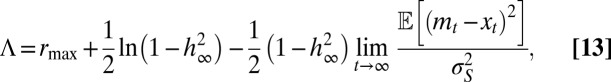 |
where 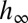 represents the fixed point of
represents the fixed point of 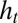 in Eq. 11. Λ is the sum of two terms: the first can be interpreted as the “genetic load” due to stabilizing selection and the second as the “evolutionary load” due to the lag between the mean phenotype
in Eq. 11. Λ is the sum of two terms: the first can be interpreted as the “genetic load” due to stabilizing selection and the second as the “evolutionary load” due to the lag between the mean phenotype 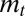 and the optimal phenotype
and the optimal phenotype 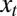 . The dynamics of
. The dynamics of 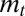 has generally no fixed point (unless the environment is constant), but, if the environment is ergodic, it has a stationary distribution and hence
has generally no fixed point (unless the environment is constant), but, if the environment is ergodic, it has a stationary distribution and hence 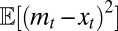 has a limit for
has a limit for 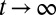 .
.
Λ may be explicitly computed for several stationary processes. Two aspects of the environment are particularly relevant when studying the adaptive value of mechanisms of heredity: the amplitude 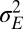 of the environmental fluctuations, and the scale
of the environmental fluctuations, and the scale 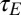 of their temporal correlations. As a simple dynamical process that encapsulates these two elements, we consider the following:
of their temporal correlations. As a simple dynamical process that encapsulates these two elements, we consider the following:
It corresponds to 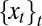 being generated by a stationary Gaussian Markov process with transition kernel
being generated by a stationary Gaussian Markov process with transition kernel 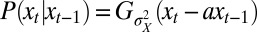 . The parameter
. The parameter 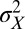 controls the degree of stochasticity and the parameter a the degree of correlation between successive environments; assuming
controls the degree of stochasticity and the parameter a the degree of correlation between successive environments; assuming 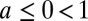 , the process followed by
, the process followed by 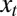 has a stationary distribution, namely
has a stationary distribution, namely 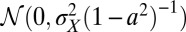 . The amplitude
. The amplitude 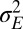 and relaxation time
and relaxation time 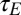 of this process are thus defined by the following:
of this process are thus defined by the following:
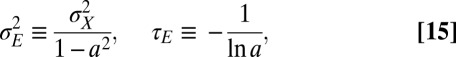 |
such that
This follows from the definition 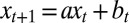 with
with 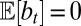 and
and 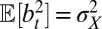 , and the assumption that
, and the assumption that 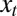 has a stationary distribution:
has a stationary distribution: 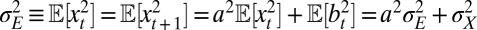 , and therefore
, and therefore 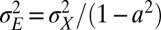 . Similarly, from
. Similarly, from 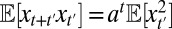 , it follows that
, it follows that 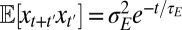 with
with 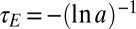 . This stochastic process is known as an autoregressive AR(1) model in signal processing and corresponds in physics to a discrete-time Ornstein–Uhlenbeck process.
. This stochastic process is known as an autoregressive AR(1) model in signal processing and corresponds in physics to a discrete-time Ornstein–Uhlenbeck process.
With this choice for the environmental temporal dynamics, we can show that 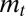 itself is normally distributed and we can compute
itself is normally distributed and we can compute 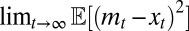 (SI Appendix). The calculation leads to a long-term growth rate Λ of the following form:
(SI Appendix). The calculation leads to a long-term growth rate Λ of the following form:
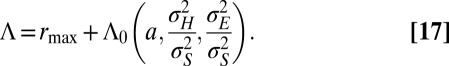 |
Without loss of generality, we can therefore assume that 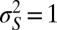 . Under this assumption, the expression for
. Under this assumption, the expression for 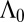 is given by the following:
is given by the following:
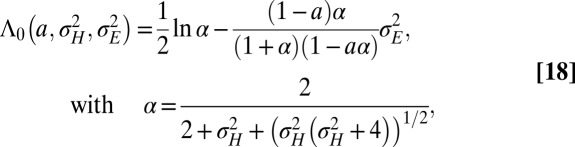 |
where the relation between α and  can also be inverted to give
can also be inverted to give 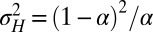 .
.
Not considering the additive parameter  , only three parameters, a,
, only three parameters, a, 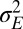 ,
, 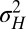 , are needed to characterize this simple model. The first two parameters pertain to the environmental process, with a representing temporal correlations
, are needed to characterize this simple model. The first two parameters pertain to the environmental process, with a representing temporal correlations 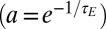 and
and 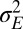 its stationary variance (in units of
its stationary variance (in units of 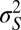 ), whereas
), whereas 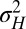 (in the same units) describes the stochasticity of inheritance (akin to a mutation rate).
(in the same units) describes the stochasticity of inheritance (akin to a mutation rate).
How Much Variation to Introduce?
This model allows us to address a first question pertaining to the evolution of heredity, the evolution of mutation rates: what is optimal degree of variation for a population adapting to a varying environment? This problem has been examined in many previous theoretical studies (27–30) and fueled by observations and experiments with microbial populations showing strikingly variable mutation rates (31). Within our model, the problem amounts to determining the value 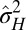 of
of 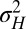 that optimizes the growth rate Λ for given environmental parameters
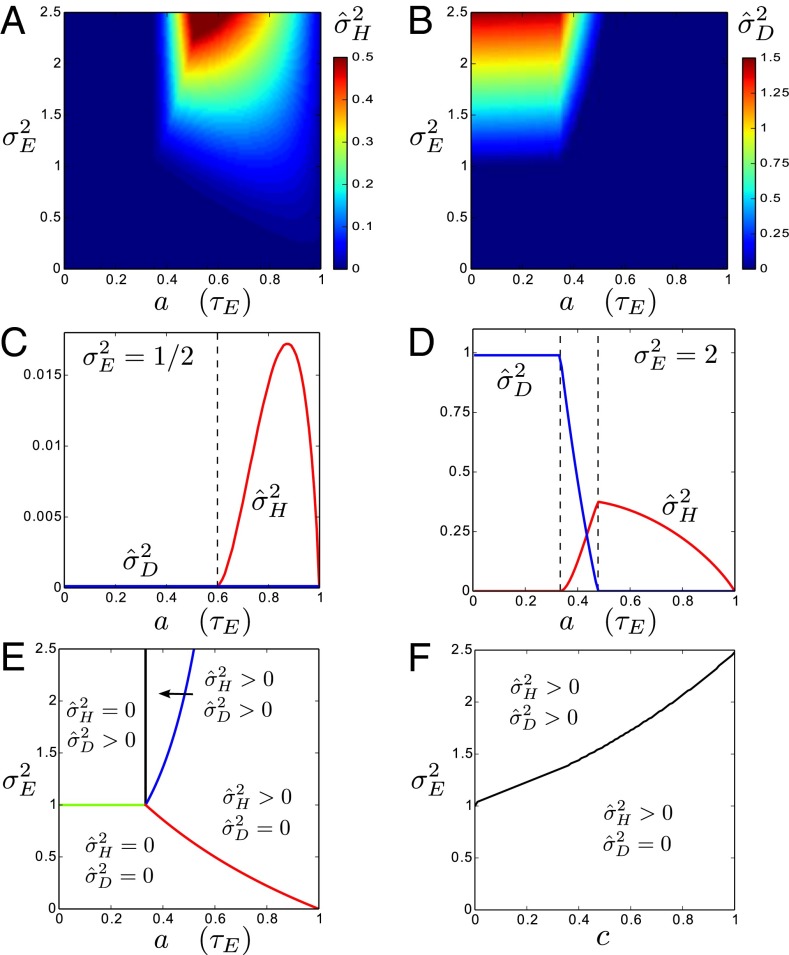
that optimizes the growth rate Λ for given environmental parameters 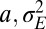 . The results of this optimization are shown in Fig. 2A: whereas larger environmental fluctuations are conducive to larger mutations rates as may be expected, the dependence into the temporal correlation is less intuitive, with a nonmonotonic dependence on a for
. The results of this optimization are shown in Fig. 2A: whereas larger environmental fluctuations are conducive to larger mutations rates as may be expected, the dependence into the temporal correlation is less intuitive, with a nonmonotonic dependence on a for 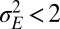 . A transition line, given by
. A transition line, given by 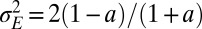 and represented in Fig. 2B, separates environmental situations where variations tend to be suppressed,
and represented in Fig. 2B, separates environmental situations where variations tend to be suppressed, 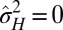 , and environmental situations where they are favored,
, and environmental situations where they are favored, 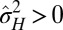 .
.
Fig. 2.
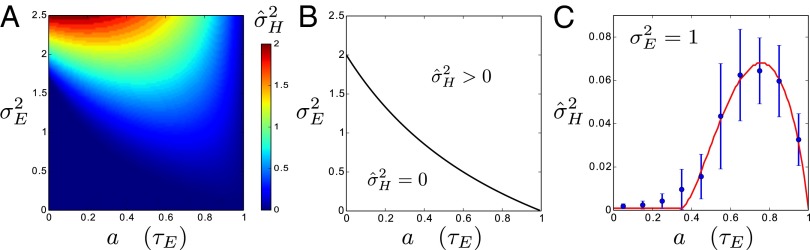
How much variations to introduce? (A) For the simple model described by Eq. 18 and depicted in Fig. 1A, optimal value of the degree 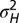 of genotypic variations as a function of
of genotypic variations as a function of 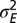 , the amplitude of the environmental fluctuations and a, representing the characteristic timescale
, the amplitude of the environmental fluctuations and a, representing the characteristic timescale 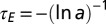 of their relaxation. (B) “Phase diagram” for the nonzero optimal values of
of their relaxation. (B) “Phase diagram” for the nonzero optimal values of 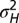 as a function of
as a function of 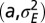 . (C) In red, optimal values of
. (C) In red, optimal values of 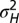 as a function of a for
as a function of a for 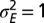 . In blue, results of numerical simulations of the dynamics of a population in which
. In blue, results of numerical simulations of the dynamics of a population in which 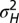 varies between parent and offspring as
varies between parent and offspring as 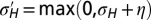 with
with 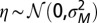 ; the mean and SD of
; the mean and SD of 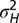 are computed over
are computed over 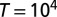 generations of a population of size
generations of a population of size 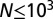 , with
, with 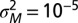 (see also SI Appendix, Fig. S1, for other values of
(see also SI Appendix, Fig. S1, for other values of 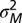 ).
).
The approach of optimizing the growth rate Λ with respect to one of its parameters, here 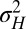 , is predicated on the assumption that a population for which this parameter mutates at a slow rate will eventually evolve toward the value of this parameter optimizing Λ. Fig. 2C shows that this assumption is indeed verified in numerical simulations of the population dynamics where
, is predicated on the assumption that a population for which this parameter mutates at a slow rate will eventually evolve toward the value of this parameter optimizing Λ. Fig. 2C shows that this assumption is indeed verified in numerical simulations of the population dynamics where 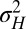 is taken to be part of the genotype and transmitted as
is taken to be part of the genotype and transmitted as 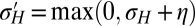 with
with 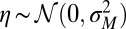 (the max is here to ensure that
(the max is here to ensure that 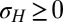 ). We performed these simulations by imposing a maximum total population size (SI Appendix), and the results of Fig. 2C therefore also demonstrate that the asymptotic growth rate Λ can describe well the behavior of finite populations. Finally, the same simulations repeated with non-Gaussian distributions for the selection, the mutations or the environmental fluctuations lead to similar results (SI Appendix, Fig. S1), indicating that our conclusions are not crucially dependent on the Gaussian assumptions that we make for computational convenience.
). We performed these simulations by imposing a maximum total population size (SI Appendix), and the results of Fig. 2C therefore also demonstrate that the asymptotic growth rate Λ can describe well the behavior of finite populations. Finally, the same simulations repeated with non-Gaussian distributions for the selection, the mutations or the environmental fluctuations lead to similar results (SI Appendix, Fig. S1), indicating that our conclusions are not crucially dependent on the Gaussian assumptions that we make for computational convenience.
Generalizations
The previous formulae solve the simple model schematically represented in Fig. 1A. A generalization of this model, depicted in Fig. 1B, explicitly distinguishes between the inherited genotype γ and the phenotype ϕ of an individual. The phenotype arises from the inherited genotype through a process of “development,” represented by a stochastic kernel D, which can show some dependence on the external environment, i.e., developmental plasticity (arrow P in Fig. 1B). The generalized model also incorporates the possibility of acquiring information from the environment and directly modifying the heritability kernel H (arrow L). The latter can also be influenced internally by a feedback from the phenotype to the transmitted genotype (arrow F). This more general model, several limits of which we analyze in the following sections, can also be solved analytically.
More precisely, the general model is characterized by the following:
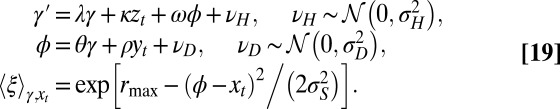 |
The first equation specifies how the transmitted genotype 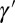 depends on the inherited genotype γ, on some external information
depends on the inherited genotype γ, on some external information 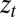 coming from the environment and on the current phenotype ϕ. The second equation defines how this current phenotype depends on the inherited genotype γ and on some possibly available external information
coming from the environment and on the current phenotype ϕ. The second equation defines how this current phenotype depends on the inherited genotype γ and on some possibly available external information 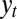 . The third equation, finally, gives the expected number of offsprings for individuals with phenotype ϕ in environment
. The third equation, finally, gives the expected number of offsprings for individuals with phenotype ϕ in environment 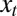 .
.
The description of the model is completed by the equations governing the dynamics of the environment. As before, it is supposed to be fixed (quenched) independently of the dynamics of the population:
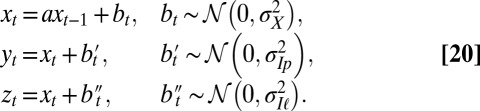 |
The signals 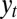 and
and 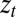 are thus assumed to derive from
are thus assumed to derive from 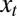 via additive white Gaussian noise channels, one of the simplest models of signal transmission.
via additive white Gaussian noise channels, one of the simplest models of signal transmission.
Although involving more parameters, the derivation of the solution for this general model follows the same principles as for the simple model presented previously (SI Appendix). We take advantage of an analytical formula for its growth rate Λ to analyze three particular variants of the model, which address the three specific questions raised in the introduction. In each case, we characterize the most adaptive scheme for generating and transmitting variations by considering the value of the parameters that optimize Λ—the expected outcome of an evolutionary dynamics where these parameters evolve on a timescale longer than the characteristics timescale 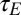 of the environment. [It can be shown that Λ is concave with respect to its parameters (32), implying an absence of local maxima into which the evolutionary dynamics could otherwise be trapped.]
of the environment. [It can be shown that Λ is concave with respect to its parameters (32), implying an absence of local maxima into which the evolutionary dynamics could otherwise be trapped.]
Where to Introduce Variation?
The introduction of new variations is a requirement for sustained evolution: a population into which no new variations are introduced will eventually become monomorphic even in absence of selection, simply as a consequence of “random drift.” However, if the characters of the individuals are so new as to be uncorrelated with those of their parents, inheritance is negated, and adaptation by natural selection impossible. Therefore, an appropriate “degree” of new variations must lie between these two extremes. This logical conclusion raises a first class of questions, which we started to address with the model of Fig. 2: What is this intermediate degree of variations? What sets its value? In addition, if an optimal degree exists, can it be selected for? Several biological observations hint at a positive answer to this last point: a complex molecular machinery has for instance evolved to ensure a faithful replication of DNA, and thus to “set” its mutation rate; moreover, not all genes are treated equally: in bacteria, for example, the positioning of genes on the leading or lagging strand of the chromosome, where they are subject to different mutation rates, is correlated to the nature of the selective pressure that they experience (33).
A second class of questions follows from noticing that new variations may not only vary in degree but also in “nature.” In particular, new variations may be phenotypic, thus affecting survival and reproduction but not being transmitted to the next generation, or/and genotypic, thus being transmitted to the next generation but not directly affecting survival and reproduction. How different are these two (nonexclusive) types of variation from an evolutionary standpoint? Is one type of variations more advantageous than the other? Or, does each have its own optimal degree? Here again, several observations support the biological relevance of these questions. Both types of variations are indeed found simultaneously in every living organism. An example of universally shared genotypic variations is provided by DNA mutations, whereas the so-called nongenetic individuality of bacteria (34) is an example of phenotypic variations. The latter, although often assimilated to noise, may in fact confer a selective advantage to the organisms (35).
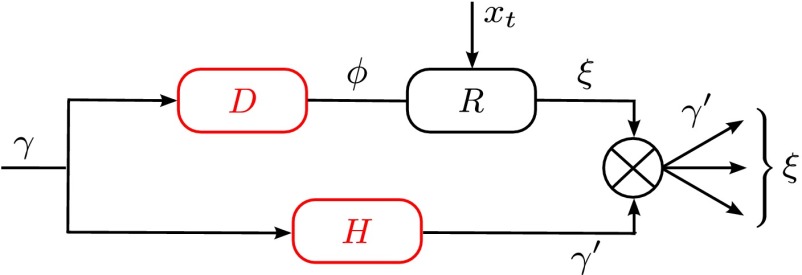
Addressing questions about the degree and nature of new variations requires a model that is both quantitative and rich enough to allow for nontrivial selective pressures. Our model meets these criteria; in fact, it is even sufficient to consider its simplified version, in which all three mechanisms depicted by red arrows in Fig. 1B, namely plasticity, feedback, and Lamarckian effects, are absent. This limiting case, represented in Fig. 3, is described by the following equations:
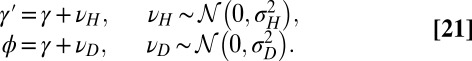 |
In this particular instance of the model (obtained from the general model by taking 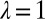 ,
, 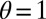 ,
, 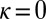 ,
, 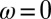 ,
, 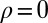 ), new variations are generated independently of the state of the environment and are introduced at two levels: at the genotypic level, through the random variable
), new variations are generated independently of the state of the environment and are introduced at two levels: at the genotypic level, through the random variable 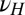 with variance
with variance 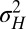 , and at the phenotypic level, through the random variable
, and at the phenotypic level, through the random variable 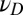 with variance
with variance 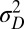 . The two possible types of new variations are thus tunable at various degrees. By studying how the long-term growth Λ may be optimized with respect to
. The two possible types of new variations are thus tunable at various degrees. By studying how the long-term growth Λ may be optimized with respect to 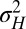 and
and 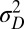 , we can thus analyze quantitatively the adaptive value of these two sources of variation as a function of the characteristics of the environment, its temporal correlation a and its stochasticity
, we can thus analyze quantitatively the adaptive value of these two sources of variation as a function of the characteristics of the environment, its temporal correlation a and its stochasticity 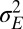 .
.
Fig. 3.
Where to introduce variation? This question is addressed within a model where the two sources of noise, the developmental kernel 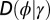 and heredity kernel
and heredity kernel 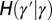 , are jointly optimized to yield the largest long-term growth rate. The optimization is performed over the two parameters
, are jointly optimized to yield the largest long-term growth rate. The optimization is performed over the two parameters 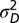 and
and 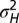 , which define
, which define 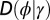 and
and 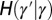 by the relations
by the relations 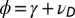 and
and 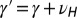 , with
, with 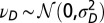 and
and 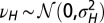 .
.
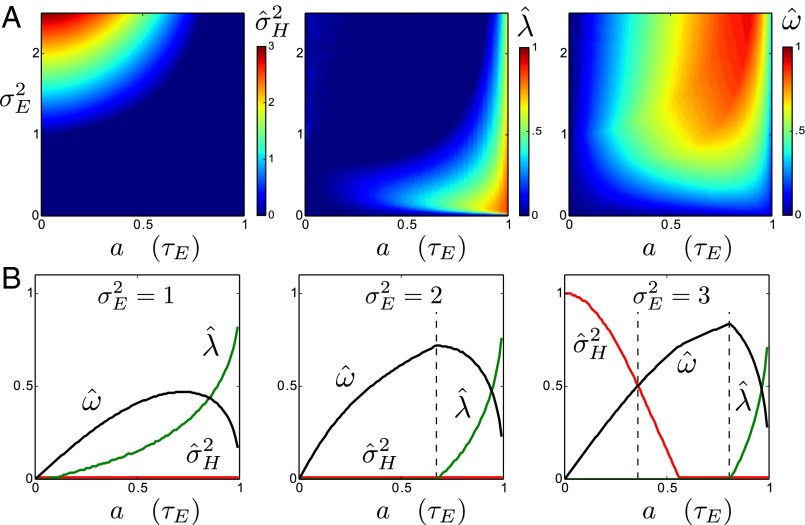
Specifically, we analyze here the values (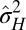 ,
, 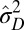 ) of the variables
) of the variables 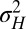 and
and 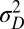 that optimize the long-term growth Λ, for given values of
that optimize the long-term growth Λ, for given values of 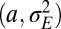 (fixing without loss of generality
(fixing without loss of generality 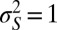 ); this corresponds to the expected outcome of a competition between populations characterized by different values of
); this corresponds to the expected outcome of a competition between populations characterized by different values of 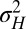 and
and 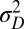 , or, equivalently, to the expected outcome of an evolutionary dynamics where
, or, equivalently, to the expected outcome of an evolutionary dynamics where 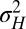 and
and 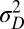 are themselves slowly varying. The results, presented in Fig. 4 A and B, show that the nature of the most adaptive variations indeed depends on the statistics of the environment. For instance, phenotypic noise
are themselves slowly varying. The results, presented in Fig. 4 A and B, show that the nature of the most adaptive variations indeed depends on the statistics of the environment. For instance, phenotypic noise 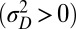 is preferred over genotypic noise
is preferred over genotypic noise 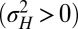 for weakly correlated environments (low a). The optimization can be performed analytically to derive a phase diagram with distinct phases, defined by the presence or absence of phenotypic or genotypic stochasticity in the optimal solution. The different boundaries, shown in Fig. 4E, are given by the following:
for weakly correlated environments (low a). The optimization can be performed analytically to derive a phase diagram with distinct phases, defined by the presence or absence of phenotypic or genotypic stochasticity in the optimal solution. The different boundaries, shown in Fig. 4E, are given by the following:
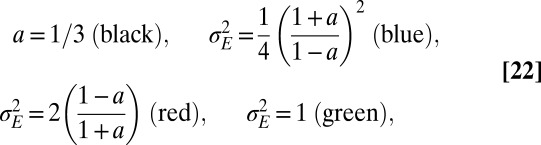 |
with the central point at 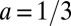 and
and 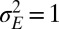 . When
. When 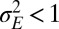 , as in Fig. 4C, we thus encounter two phases as a is varied; when
, as in Fig. 4C, we thus encounter two phases as a is varied; when 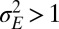 , as in Fig. 4D, three phases are traversed.
, as in Fig. 4D, three phases are traversed.
Fig. 4.
Where to introduce variation? (A) For the model described by Eq. 21 and depicted in Fig. 3, optimal values of the degree 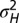 of genotypic variations as a function of the environmental variables
of genotypic variations as a function of the environmental variables 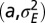 when optimizing jointly over
when optimizing jointly over 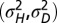 . (B) Optimal values of the degree
. (B) Optimal values of the degree 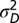 of phenotypic variations as a function of
of phenotypic variations as a function of 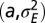 when optimizing jointly over
when optimizing jointly over 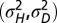 . (C) Optimal values of
. (C) Optimal values of 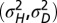 as a function of a for
as a function of a for 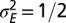 . (D) Optimal values of
. (D) Optimal values of 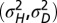 as a function of a for
as a function of a for 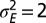 . (E) Phase diagram for the optimal nonzero values of
. (E) Phase diagram for the optimal nonzero values of 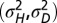 as a function of
as a function of 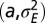 . (F) Same phase diagram for an environment undergoing directed changes,
. (F) Same phase diagram for an environment undergoing directed changes, 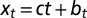 with
with 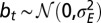 , in which case the environmental parameters are
, in which case the environmental parameters are 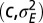 .
.
A first conclusion from these results is that a population can be adapted to a fluctuating environment (here have optimal 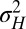 and
and 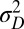 ) even though its individuals are not undergoing any change: this is the case for the regions of the phase diagram where
) even though its individuals are not undergoing any change: this is the case for the regions of the phase diagram where 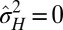 , corresponding to a population with a homogeneous and constant genotype. In this sense, adaptation of a population to a fluctuating environment does not require variations in the attributes of the individuals. However, an absence of evolution does not necessarily imply an absence of diversity. When
, corresponding to a population with a homogeneous and constant genotype. In this sense, adaptation of a population to a fluctuating environment does not require variations in the attributes of the individuals. However, an absence of evolution does not necessarily imply an absence of diversity. When 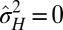 but
but 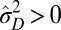 , the population is phenotypically diverse, although rather than transmitted from one generation to the next, the same diversity is reproduced at each generation. Moreover, a population may also be adapted to a fluctuating environment without showing any diversity: for small environment fluctuations, we find indeed that
, the population is phenotypically diverse, although rather than transmitted from one generation to the next, the same diversity is reproduced at each generation. Moreover, a population may also be adapted to a fluctuating environment without showing any diversity: for small environment fluctuations, we find indeed that 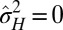 and
and 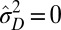 (Fig. 4E); in this case, natural selection favors the suppression of any variation. The diversity of the population, either genotypic or phenotypic, can be more precisely quantified within the model: we thus have
(Fig. 4E); in this case, natural selection favors the suppression of any variation. The diversity of the population, either genotypic or phenotypic, can be more precisely quantified within the model: we thus have 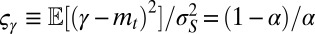 , for the genotypic diversity, and,
, for the genotypic diversity, and, 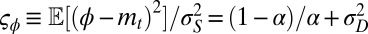 for the phenotypic diversity.
for the phenotypic diversity.
Although new variations are beneficial when the fluctuations of the environment are large enough, our model predicts that natural selection should favor their introduction at different levels, depending on the statistical structure of these fluctuations. As indicated in Fig. 4E, phenotypic variations are suppressed 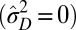 when the environmental stochasticity is small enough or the environmental correlation large enough: this may be interpreted as a selection for “canalization,” i.e., reduction of the phenotypic diversity of genotypically identical individuals, a phenomenon indeed observed in biological organisms (13). Genotypic variations are suppressed
when the environmental stochasticity is small enough or the environmental correlation large enough: this may be interpreted as a selection for “canalization,” i.e., reduction of the phenotypic diversity of genotypically identical individuals, a phenomenon indeed observed in biological organisms (13). Genotypic variations are suppressed 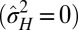 , however, when the environment is not strongly correlated (a small). This may be rationalized by noticing that nontrivial inheritance is relevant only when successive generations share correlated selective pressures.
, however, when the environment is not strongly correlated (a small). This may be rationalized by noticing that nontrivial inheritance is relevant only when successive generations share correlated selective pressures.
The study can be extended to an environment undergoing directed changes, 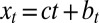 with
with 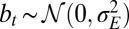 (SI Appendix). In this case, we find that nonzero genotypic variations
(SI Appendix). In this case, we find that nonzero genotypic variations 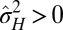 are always needed to keep up with the environmental changes, but a transition between phenotypic canalization
are always needed to keep up with the environmental changes, but a transition between phenotypic canalization 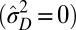 and phenotypic plasticity
and phenotypic plasticity 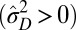 is still observed as the environmental fluctuations
is still observed as the environmental fluctuations 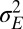 increase, or as the speed c of the environmental changes decreases, as summarized in Fig. 4F.
increase, or as the speed c of the environmental changes decreases, as summarized in Fig. 4F.
Living organisms do not harbor a single trait, but many, each potentially subject to a selective pressure with a different statistical structure. For instance, in bacteria, the strength of selection may be very different between central metabolism and mechanisms of resistance to antibiotics. Our model suggests that this diversity of selective pressures may be responsible for the evolution of the diversity of ways in which new traits are generated and transmitted. However, our model is obviously extremely schematic and does not account for a number of features that affect the evolution of mechanisms of inheritance. In particular, it does not consider the cost of these mechanisms, which may strongly limit their actual diversity: suppressing any variation by error corrections, checkpoints, canalization, etc., may be prohibitively expensive, and evolving a different system of inheritance for every trait simply impossible (any mechanism for introducing variations in a trait defines a new trait into which variations may be introduced).
When to Separate Phenotype and Transmitted Genotype?
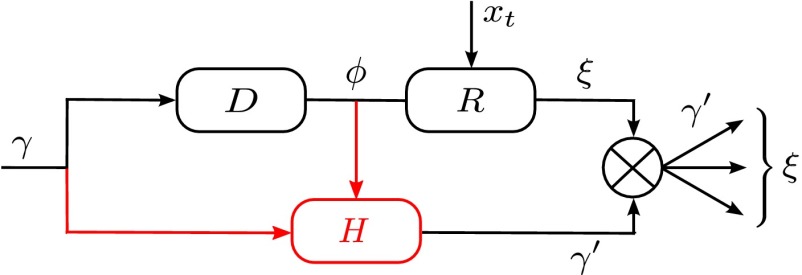
The previous version of the model assumes an independent germ line, with a transmitted genotype 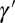 that is not influenced by the phenotype ϕ. Such a separation between a germplasm γ and a “soma” ϕ is central to the view that new traits are exclusively generated by random mutations in the gametes, independently of any event occurring during the lifetime of the individual. This separation, however, cannot be taken for granted, and is in fact absent in many if not most living organisms, including notably plants (36). However, mammals do seem to possess specific mechanisms to enforce a separation; for instance, murine primordial germ cells undergo resetting and erasing of maternal and paternal imprints, genome-wide DNA methylation, extensive histone modifications, and inactive X-chromosome reactivation (18). As a very first step in trying to understand the origin of such mechanisms, it is instructive to abstract from the many constraints that may limit the evolution of systems of inheritance, and look for the way in which genotypic and phenotypic features should ideally be combined to ensure a maximal growth rate of the population; if a “Weismann’s barrier” segregating a germ line from the soma is never found in such conditions, this implies that its origin must reside elsewhere. We follow here this approach by examining within model the selective value of a feedback from phenotype to transmitted genotype.
that is not influenced by the phenotype ϕ. Such a separation between a germplasm γ and a “soma” ϕ is central to the view that new traits are exclusively generated by random mutations in the gametes, independently of any event occurring during the lifetime of the individual. This separation, however, cannot be taken for granted, and is in fact absent in many if not most living organisms, including notably plants (36). However, mammals do seem to possess specific mechanisms to enforce a separation; for instance, murine primordial germ cells undergo resetting and erasing of maternal and paternal imprints, genome-wide DNA methylation, extensive histone modifications, and inactive X-chromosome reactivation (18). As a very first step in trying to understand the origin of such mechanisms, it is instructive to abstract from the many constraints that may limit the evolution of systems of inheritance, and look for the way in which genotypic and phenotypic features should ideally be combined to ensure a maximal growth rate of the population; if a “Weismann’s barrier” segregating a germ line from the soma is never found in such conditions, this implies that its origin must reside elsewhere. We follow here this approach by examining within model the selective value of a feedback from phenotype to transmitted genotype.
Three factors potentially contribute to the genotype 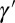 transmitted to the offsprings: the genotype γ inherited from the parent, the phenotype ϕ of the individual, and random variations
transmitted to the offsprings: the genotype γ inherited from the parent, the phenotype ϕ of the individual, and random variations 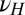 . To analyze their relative adaptive value, we study the model depicted by Fig. 5, which allows for different combinations of these three elements:
. To analyze their relative adaptive value, we study the model depicted by Fig. 5, which allows for different combinations of these three elements:
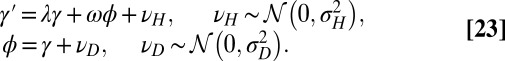 |
Each factor is controlled by a parameter, λ for γ, ω for ϕ, and 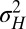 for
for 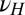 . We thus consider optimizing the long-term growth rate Λ over
. We thus consider optimizing the long-term growth rate Λ over 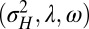 , for various values of
, for various values of 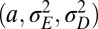 , using the expression for Λ of the general model, with
, using the expression for Λ of the general model, with 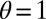 ,
, 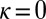 ,
, 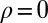 [we consider
[we consider 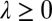 and
and 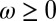 ; see also SI Appendix, Fig. S2, for an alternative analysis where the optimization is performed over discrete values,
; see also SI Appendix, Fig. S2, for an alternative analysis where the optimization is performed over discrete values, 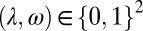 ].
].
Fig. 5.
When to separate phenotype and transmitted genotype? This question is addressed within a model where the heredity kernel 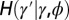 is optimized. The optimization is made over the three parameters λ, ω,
is optimized. The optimization is made over the three parameters λ, ω, 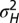 , which define
, which define 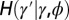 by the relation
by the relation 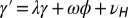 with
with 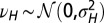 .
.
The result of a numerical optimization of Λ are presented in Fig. 6. They show that a feedback from phenotype to transmitted genotype is prevented 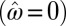 in two limits: the limit of uncorrelated environments,
in two limits: the limit of uncorrelated environments, 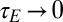
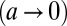 , and the limit of deterministic environments
, and the limit of deterministic environments 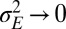 . In the first limit,
. In the first limit, 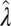 vanishes as well; hence the absence of feedback does not imply an isolated germ line, but simply an absence of nontrivial heredity (for small a but large
vanishes as well; hence the absence of feedback does not imply an isolated germ line, but simply an absence of nontrivial heredity (for small a but large 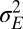 , the solution
, the solution 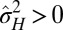 ,
, 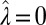 ,
, 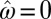 indicates that only noise is transmitted to the offsprings). The system of inheritance most reminiscent to Weismann’s scenario is obtained in the limit of constant environments
indicates that only noise is transmitted to the offsprings). The system of inheritance most reminiscent to Weismann’s scenario is obtained in the limit of constant environments 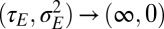
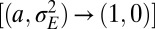 , suggesting that a separation of germplasm from soma is beneficial only for those aspects of the phenotype subject to nonfluctuating selective pressures (“housekeeping” genes, for instance).
, suggesting that a separation of germplasm from soma is beneficial only for those aspects of the phenotype subject to nonfluctuating selective pressures (“housekeeping” genes, for instance).
Fig. 6.
When to separate phenotype and transmitted genotype? (A) For the model described by Eq. 23 and depicted in Fig. 5, the values of 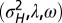 that jointly optimize Λ are represented as a function of the environmental parameters
that jointly optimize Λ are represented as a function of the environmental parameters 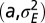 for a fixed developmental noise
for a fixed developmental noise 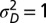 . (B) Same results presented as a function of a for three fixed values of
. (B) Same results presented as a function of a for three fixed values of 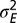 .
.
Where to Acquire Information?
In the two previous models, the role of the environment is confined to selection, and any new variation is introduced independently of the environmental state. Examples, however, abound of living organisms generating new traits that are correlated with the environment. It is thus well recognized that the current phenotype of an individual is affected not only by the genotype received from its parents but also by the environment in which it develops; such a variation, which may be adaptive, is generally referred to as “plasticity.” As already mentioned in the introduction, the question of whether environmentally induced variations can be transmitted to the progeny was, and still remains, a subject of hot debates. This possibility was central to Lamarck’s theory of evolution, and is often referred to as “Lamarckism.” Evolution by natural selection does not require it, but it does not exclude it. Several examples of environmentally induced traits have indeed been observed. Proving that such traits confer a selective advantage is generally delicate, but a particularly striking example is provided, for instance, by the bacterial immune system called CRISPR (37); this system relies on the insertion of phage-specific sequences into bacterial genomes and has been shown to protect against phage infection the bacteria that inherit them from their parent. This example implies a specific mechanism for incorporating and exploiting the environmental “signal” (here, the presence of a particular strain of phage). The constraints to which the evolution of such mechanisms are subject are determining but potentially nongeneric, and, in any case, difficult to model. Here, we consider a thought experiment (or Gedankenexperiment), where we assume a mechanism for incorporating external signals, but question the way in which it is optimally “plugged in.” This approach allows us, without discussing the mechanisms themselves, to compare the Darwinian and Lamarckian “modalities,” and test the conjecture that each of them is tuned to a different type of selective pressure (38).
We thus compare two models, where the same information 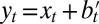 with
with 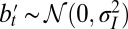 is available, but where it is either processed at the phenotypic level (model P for “plasticity,” for which
is available, but where it is either processed at the phenotypic level (model P for “plasticity,” for which 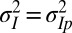 ; Fig. 7A), or at the genotypic level (model L for “Lamarckism,” for which
; Fig. 7A), or at the genotypic level (model L for “Lamarckism,” for which 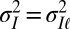 ; Fig. 7B). Formally, model P is described by the following:
; Fig. 7B). Formally, model P is described by the following:
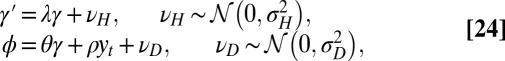 |
thus corresponding to the general model with 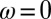 and
and 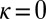 , whereas model L is described by the following:
, whereas model L is described by the following:
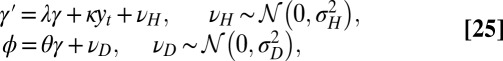 |
thus corresponding to the general model with 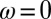 and
and 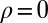 . We consider optimizing the long-term growth rate Λ over the parameters that control the contributions of each factor:
. We consider optimizing the long-term growth rate Λ over the parameters that control the contributions of each factor: 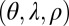 for model P, and
for model P, and 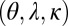 for model L (considering here again only positive values of these parameters).
for model L (considering here again only positive values of these parameters).
Fig. 7.
Where to acquire information? This question is addressed by comparing two models in presence of the same information 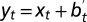 with
with 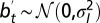 . (A) Model P, described by Eq. 24, where the information
. (A) Model P, described by Eq. 24, where the information 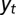 is incorporated to the phenotype and where the growth rate Λ is optimized with respect to the three parameters θ, λ, ρ, which define
is incorporated to the phenotype and where the growth rate Λ is optimized with respect to the three parameters θ, λ, ρ, which define 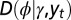 and
and 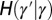 by the relations
by the relations 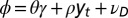 and
and 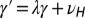 with
with 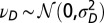 and
and 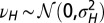 , and fixed
, and fixed 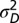 ,
, 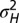 . (B) Model L, described by Eq. 25, where the information
. (B) Model L, described by Eq. 25, where the information 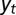 is incorporated to the transmitted genotype and where the growth rate Λ is optimized with respect to the three parameters θ, λ, κ, which define
is incorporated to the transmitted genotype and where the growth rate Λ is optimized with respect to the three parameters θ, λ, κ, which define 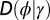 and
and 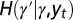 by the relations
by the relations 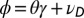 and
and 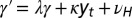 with
with 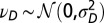 and
and 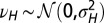 , and fixed
, and fixed 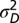 ,
, 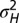 .
.
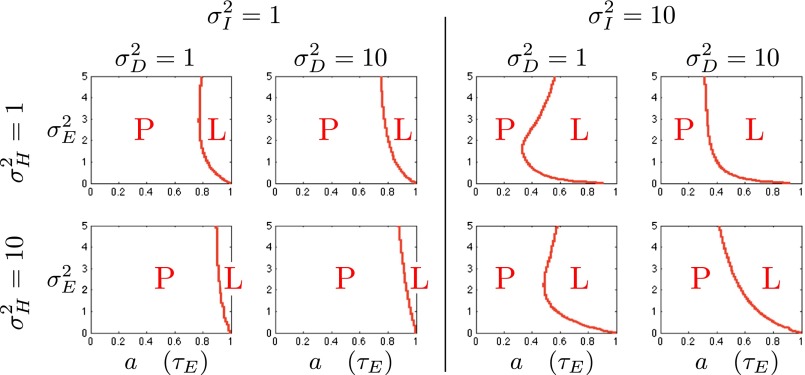
The results of a comparison between the two models are shown in Fig. 8, where, as a function of 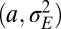 and for different values of the fixed parameters
and for different values of the fixed parameters 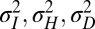 , we present which of the two models, P or L, yields the highest growth rate. The main controlling parameter appears to be the correlation a (or equivalently
, we present which of the two models, P or L, yields the highest growth rate. The main controlling parameter appears to be the correlation a (or equivalently 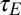 ) of the environmental fluctuations, with the Lamarckian modality systematically becoming more favorable when this correlation is large, in line with the intuition that transmitting acquired information is beneficial when the selective pressure experienced by the offspring is sufficiently similar to that experienced by the parents. Note that this simple conclusion conceals in fact a much richer diversity of strategies, revealed by considering the values of the parameters optimizing the two models (SI Appendix, Fig. S3).
) of the environmental fluctuations, with the Lamarckian modality systematically becoming more favorable when this correlation is large, in line with the intuition that transmitting acquired information is beneficial when the selective pressure experienced by the offspring is sufficiently similar to that experienced by the parents. Note that this simple conclusion conceals in fact a much richer diversity of strategies, revealed by considering the values of the parameters optimizing the two models (SI Appendix, Fig. S3).
Fig. 8.
Where to acquire information? Boundary between 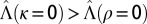 , when acquiring information at the phenotypic level is more beneficial, indicated by “P,” and
, when acquiring information at the phenotypic level is more beneficial, indicated by “P,” and 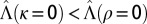 , when acquiring information at the genotypic level is more beneficial, indicated by “L,” for all eight combinations of
, when acquiring information at the genotypic level is more beneficial, indicated by “L,” for all eight combinations of 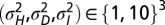 .
. 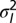 represents
represents 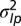 for model P and
for model P and 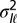 for model L.
for model L.
Connections
Of the many studies that share part of our intents or methods, two lines of work stand out: (i) a different approach, based on Price’s equation, has been proposed with the same goal of uniting the various forms of inheritance within a common mathematical framework (39, 40); (ii) a different problem, pertaining to control in engineering, has been solved using a closely related mathematical framework (24). We discuss here the relations between our model and these two lines of work.
Link to Price Equation.
Price equation is a general formula, applicable to any model of population dynamics, which uses a covariance formalism to express the change in the mean value of a trait between successive generations (41). Being a mathematical identity, it necessarily holds true. Its virtue is to provide a decomposition of evolutionary change that can illuminate its origins; initially derived for models of cooperation, it has contributed to clarify the nature of group/kin selection (42). Applied in many other contexts, it has also been used as a general mathematical framework for studying the different possible modes of inheritance (39, 40). As for any other model of population dynamics, a Price equation can be written for our model (SI Appendix).
Price equation is, however, limited to a short-term description of the dynamics: as it considers only the mean values of traits, it cannot be iterated to describe changes in subsequent generations; this indeed requires the full distribution 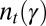 . As illustrated in the previous sections, systems of inheritance may, however, have qualitatively different implications in environments with different statistical structures. Only a long-term analysis of the dynamics of a population can thus fully reveal their evolutionary properties. In our approach, this is achieved by coupling Price equation, the recursion over the mean
. As illustrated in the previous sections, systems of inheritance may, however, have qualitatively different implications in environments with different statistical structures. Only a long-term analysis of the dynamics of a population can thus fully reveal their evolutionary properties. In our approach, this is achieved by coupling Price equation, the recursion over the mean 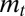 of the trait, Eq. 9, with a recursion over the variance
of the trait, Eq. 9, with a recursion over the variance 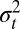 of the trait, Eq. 10, and by considering their asymptotic properties, which consist of a fixed point for
of the trait, Eq. 10, and by considering their asymptotic properties, which consist of a fixed point for 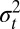 and a stationary distribution for
and a stationary distribution for 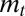 . Within the Gaussian assumptions that define our model, these two quantities are sufficient to fully capture the population dynamics. More importantly, our formalism involves a central quantity not present in covariance formalisms, the Lyapunov exponent Λ. Because this quantity decides the eventual fate of two competing populations, it allows us to associate with each scheme of inheritance an adaptive value, and thus to derive the conditions under which a given scheme confers a selective advantage over the others.
. Within the Gaussian assumptions that define our model, these two quantities are sufficient to fully capture the population dynamics. More importantly, our formalism involves a central quantity not present in covariance formalisms, the Lyapunov exponent Λ. Because this quantity decides the eventual fate of two competing populations, it allows us to associate with each scheme of inheritance an adaptive value, and thus to derive the conditions under which a given scheme confers a selective advantage over the others.
Link to the Kalman Filter.
At the core of adaptation is the problem of anticipating the next state of the environment. From this perspective, different systems of inheritance can be considered as different ways to share with the current generation the “knowledge” accumulated, through natural selection, by the previous generations. In a stochastically fluctuating environment, no system of inheritance can, however, perform better than direct sensing of the present environment by the individual that experiences it. Sensors, when not altogether absent, are generically imperfect. The individual with such an imperfect access to its current environment thus faces a dilemma: it has to arbitrate between two valuable but unreliable sources of information, the germplasm γ inherited from the parent, and the cues 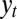 or
or 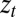 that it gained from its own perception. Interestingly, the very same dilemma is encountered in various problems of engineering, such as for instance the automatic guidance of aircrafts, where decisions must also be made based on two potentially conflicting sources: the past states of the system, and the signals from the sensors. A classical algorithm for solving this problem is the Kalman filter (24). Maybe not surprisingly, it involves the same essential mathematical ingredients that make our model solvable: linearity and Gaussianity. We present here a limit case of our model where the two approaches formally coincide, thus revealing that the scope of the concepts of inheritance extends beyond the study of biological organisms.
that it gained from its own perception. Interestingly, the very same dilemma is encountered in various problems of engineering, such as for instance the automatic guidance of aircrafts, where decisions must also be made based on two potentially conflicting sources: the past states of the system, and the signals from the sensors. A classical algorithm for solving this problem is the Kalman filter (24). Maybe not surprisingly, it involves the same essential mathematical ingredients that make our model solvable: linearity and Gaussianity. We present here a limit case of our model where the two approaches formally coincide, thus revealing that the scope of the concepts of inheritance extends beyond the study of biological organisms.
Control in engineering typically involves a single system, rather than a population of diverse individuals. We thus obtain a formal analogy with the Kalman filter only when the population is perfectly homogeneous, and described by a single “state” γ. This corresponds in our model to a limit where no developmental noise is present, 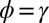 (i.e.,
(i.e., 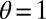 ,
, 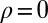 ,
, 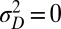 ), and where the environment is perfectly selective,
), and where the environment is perfectly selective, 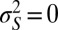 , so that each and every individual has a common
, so that each and every individual has a common 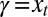 . In this limit case, the heredity kernel
. In this limit case, the heredity kernel 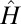 that optimizes Λ can be computed exactly for any stationary Markovian environmental process (not necessarily Gaussian).
that optimizes Λ can be computed exactly for any stationary Markovian environmental process (not necessarily Gaussian).
First, assuming that no external information is available, with a model described by 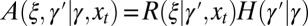 where
where 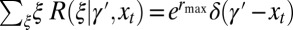 (note the slight difference with Eq. 6), and denoting by
(note the slight difference with Eq. 6), and denoting by 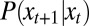 the stochastic kernel defining the environmental process, we have the following:
the stochastic kernel defining the environmental process, we have the following:
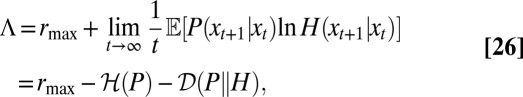 |
where 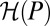 denotes the entropy rate of the process P, and
denotes the entropy rate of the process P, and 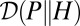 the rate of relative entropy of P with respect to H (23). [They are defined by
the rate of relative entropy of P with respect to H (23). [They are defined by 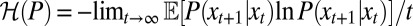 and
and 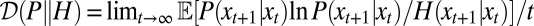 (43).] The later verifies
(43).] The later verifies 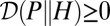 , with
, with 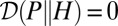 if and only if
if and only if 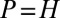 (43). The growth rate Λ is therefore minimal for
(43). The growth rate Λ is therefore minimal for 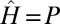 , an instance of the so-called proportional betting strategy (44), which consists in matching the stochasticity of the environment. In particular, when taking a Gaussian environment with
, an instance of the so-called proportional betting strategy (44), which consists in matching the stochasticity of the environment. In particular, when taking a Gaussian environment with 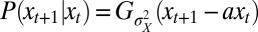 , we obtain
, we obtain 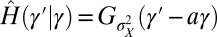 , corresponding to
, corresponding to 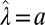 and
and 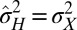 [Gσ2 (x) denotes a Gaussian function with zero mean and variance
[Gσ2 (x) denotes a Gaussian function with zero mean and variance 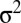 ].
].
Assuming now that some information 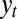 is available, which is derived from
is available, which is derived from 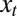 as
as 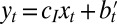 with
with 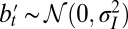 , we can extend this result to a model with
, we can extend this result to a model with 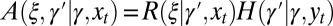 . (We previously assumed
. (We previously assumed 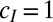 ; κ and
; κ and 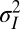 can indeed always be rescaled to be in this case. We introduce here
can indeed always be rescaled to be in this case. We introduce here 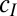 only to make the correspondence with the Kalman filter where this rescaling is generally not assumed.) This model, which corresponds to a continuous version of the model studied in ref. 23, was previously analyzed in ref. 45. In the limit where
only to make the correspondence with the Kalman filter where this rescaling is generally not assumed.) This model, which corresponds to a continuous version of the model studied in ref. 23, was previously analyzed in ref. 45. In the limit where 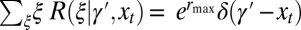 , the growth rate Λ is optimized by following a Bayesian strategy:
, the growth rate Λ is optimized by following a Bayesian strategy:
where 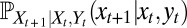 denotes the conditional probability of having the environmental random variable
denotes the conditional probability of having the environmental random variable 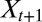 taking the value
taking the value 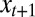 at time
at time 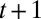 given that it took the value
given that it took the value 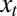 at time t, and that the population observed
at time t, and that the population observed 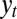 at this time. With a Gaussian environment and Gaussian noisy channel,
at this time. With a Gaussian environment and Gaussian noisy channel, 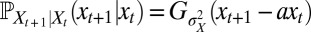 and
and 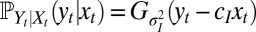 , this yields for
, this yields for 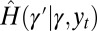 :
:
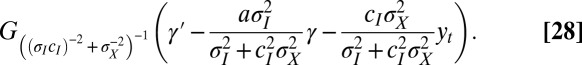 |
In this case, it is thus proved that the optimal form of the heritability kernel H is Gaussian. The two coefficients 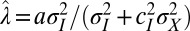 and
and 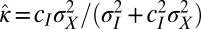 correspond exactly to the gains for the Kalman filter (24); in this context, they prescribe how the previous state γ of the system and the newly acquired information
correspond exactly to the gains for the Kalman filter (24); in this context, they prescribe how the previous state γ of the system and the newly acquired information  must be linearly combined to optimally define the subsequent state
must be linearly combined to optimally define the subsequent state 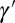 of the system.
of the system.
The formal correspondence with the solution to the Kalman filter holds only in the limit of infinite selectivity 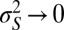 , where the population is perfectly homogeneous at every time. This limit, where the optimal scheme for processing information follows the Bayesian principles, is also the limit in which the value of the information
, where the population is perfectly homogeneous at every time. This limit, where the optimal scheme for processing information follows the Bayesian principles, is also the limit in which the value of the information 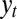 can be quantified by the usual concepts of information theory (23). We may thus view our model as a generalization of the problem of stochastic control encountered in engineering by incorporating biological features that are absent in this context, notably a diverse and growing population, and a distinction between genotype and phenotype. Reciprocally, the Kalman filter has been extended along several lines since its original formulation (46), and the mathematical formalisms thus developed may suggest ways along which generalizations of our model could be analyzed.
can be quantified by the usual concepts of information theory (23). We may thus view our model as a generalization of the problem of stochastic control encountered in engineering by incorporating biological features that are absent in this context, notably a diverse and growing population, and a distinction between genotype and phenotype. Reciprocally, the Kalman filter has been extended along several lines since its original formulation (46), and the mathematical formalisms thus developed may suggest ways along which generalizations of our model could be analyzed.
Conclusion
Classical models of population genetics take the mechanisms for generating and transmitting new traits as given. Several previous studies have extended these models to analyze how the mechanisms of inheritance may themselves evolve, starting from works on the evolution of mutation rates (27) and including, among several other examples, studies of maternal effects (47), nongenetic inheritance (48), plasticity and memory (49), and relationship quantitative trait loci (50). Here, we proposed a simple model to compare the adaptive value of different schemes for generating and transmitting variations in populations. Its analysis indicates that different modalities of inheritance are favored depending on the statistical structure of the fluctuations of the environment. For an organism with various traits, each potentially subject to a different selective pressure, this analysis suggests that multiple inheritance systems operating in parallel may be selected for, consistently with observations.
Our model is schematic but captures a key feature of evolutionary dynamics: information can be transmitted between generations along different canals, and not only does the topology of these canals affect the dynamics, but this topology can be itself subject to selection. Certainly, the model does not encompass the full diversity of possible modes of inheritance, but it can still be extended along several lines while retaining its analytical tractability. For instance, rather than combining the inherited and the acquired information into a single attribute, it could include two channels of transmission, one for the germ line and another for somatic elements, as for instance in ref. 47; because Gaussian formulae extend to multidimensional variables, the model remains indeed solvable when considering the generation and transmission of multiple traits. It can similarly be extended to account for multiple timescales, for instance by introducing a temporal delay between the developmental stage and the time of reproduction (formally 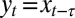 , for
, for 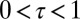 ). Different environmental processes can also be analyzed, which do not need to be Gaussian for the model to be solvable; e.g., adding a linearly varying component to the environment (as in Fig. 4F) provides a framework for studying the implications of different modes of adaptations to the survival of a population facing a directed change of selective pressure (51).
). Different environmental processes can also be analyzed, which do not need to be Gaussian for the model to be solvable; e.g., adding a linearly varying component to the environment (as in Fig. 4F) provides a framework for studying the implications of different modes of adaptations to the survival of a population facing a directed change of selective pressure (51).
The model is abstracted from material implementations and, in particular, does not refer to the genetic or nongenetic nature of what is transmitted. It cannot, therefore, account for the constraints and costs that the evolution of any specific mechanism for generating and transmitting variations must face. The model can certainly be extended to include such costs, both constitutive (attached to the mechanisms) or inductive (stemming from their use), but only at the price of introducing new, ad hoc parameters. This limitation for example prevents us from making a meaningful comparison of the relative benefit of “selection” versus “instruction,” because selection results automatically from the interaction with the environment, whereas a specific mechanism is needed to acquire and incorporate environmental signals. More generally, the model does not account for the fact that a reliable hereditary mechanism must precede the evolution of a Lamarckian mechanism, if this mechanism is to be faithfully transmitted.
Despite these limitations, we hope that our approach may be of value for providing theoretical limits to the evolution of systems of inheritance, in the spirit of the theoretical limits that Shannon derived for the communication of signals over noisy channels, after similarly abstracting from practical costs and constraints (17). As shown in our previous work (23), the similarity between the two problems extends beyond the mere analogy: the fundamental quantities of information theory are recovered as a limit of our model. We exposed here, in the same limit, another formal analogy, with the solution to the Kalman filter used in stochastic control (24). The model presented in this paper thus provides a versatile, analytically tractable framework for clarifying and unifying common issues and concepts in population genetics, information theory, and stochastic control, which may contribute to stimulate further crossbreeding between these disciplines.
Supplementary Material
Acknowledgments
We thank B. Chazelle, B. Houchmandzadeh, D. Huse, D. Jordan, and S. Kuehn for comments. This work was supported by Agence Nationale de la Recherche, Grant CoevolInterProt (to O.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.B.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323901111/-/DCSupplemental.
References
- 1.Darwin C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- 2.Darwin C. The Variation of Animals and Plants under Domestication. London: John Murray; 1868. [Google Scholar]
- 3.Weismann A. The Germ-Plasm. A Theory of Heredity. New York: Charles Scribner’s Sons; 1893. [Google Scholar]
- 4.Romanes GJ. Darwin, and After Darwin. II. Post-Darwinian Questions: Heredity and Utility. Chicago: The Open Court Publishing Company; 1895. [Google Scholar]
- 5.Huxley J. Evolution: The Modern Synthesis. London: Allen and Unwin; 1942. [Google Scholar]
- 6.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lederberg J. Genes and antibodies. Science. 1959;129(3364):1649–1653. doi: 10.1126/science.129.3364.1649. [DOI] [PubMed] [Google Scholar]
- 8.Monod J. From enzymatic adaptation to allosteric transitions. Science. 1966;154(3748):475–483. doi: 10.1126/science.154.3748.475. [DOI] [PubMed] [Google Scholar]
- 9.Crick F. Central dogma of molecular biology. Nature. 1970;227(5258):561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 10.Baldwin J. A new factor in evolution. Am Nat. 1896;30:441–451. [Google Scholar]
- 11.Morgan CL. On modification and variation. Science. 1896;4(99):733–740. doi: 10.1126/science.4.99.733. [DOI] [PubMed] [Google Scholar]
- 12.Osborn HF. Ontogenic and phylogenic variation. Science. 1896;4(100):786–789. doi: 10.1126/science.4.100.786. [DOI] [PubMed] [Google Scholar]
- 13.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 14.Soyfer VN. Lysenko and the Tragedy of Soviet Science. New Brunswick, NJ: Rutgers Univ Press; 1994. [Google Scholar]
- 15.Bonduriansky R. Rethinking heredity, again. Trends Ecol Evol. 2012;27(6):330–336. doi: 10.1016/j.tree.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Jablonka E, Lamb ML. Evolution in Four Dimensions: Genetic, Epigenetic, Behavioral, and Symbolic Variation in the History of Life. Cambridge, MA: The MIT Press; 2005. [Google Scholar]
- 17.Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27:379. [Google Scholar]
- 18.Sabour D, Schöler HR. Reprogramming and the mammalian germline: The Weismann barrier revisited. Curr Opin Cell Biol. 2012;24(6):716–723. doi: 10.1016/j.ceb.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 19.West-Eberhard MJ. Developmental Plasticity and Evolution. New York: Oxford Univ Press; 2003. [Google Scholar]
- 20.Jablonka E, Lamb MJ, Avital E. “Lamarckian” mechanisms in darwinian evolution. Trends Ecol Evol. 1998;13(5):206–210. doi: 10.1016/S0169-5347(98)01344-5. [DOI] [PubMed] [Google Scholar]
- 21.Fisher RA. The Genetical Theory of Natural Selection. Oxford: Clarendon; 1930. [Google Scholar]
- 22.Maybeck PS. Stochastic Models, Estimation and Control. New York: Academic; 1979. [Google Scholar]
- 23.Rivoire O, Leibler S. The value of information for populations in varying environments. J Stat Phys. 2011;142:1–43. [Google Scholar]
- 24.Kalman R. A new approach to linear filtering and prediction problems. J Basic Eng. 1960;82:35–45. [Google Scholar]
- 25.Tuljapurkar S. Population Dynamics in Variable Environments. Berlin: Springer; 1990. [Google Scholar]
- 26.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 27.Kimura M. On the evolutionary adjustment of spontaneous mutation rates. Genet Res. 1967;9:23–34. [Google Scholar]
- 28.Leigh EG., Jr The evolution of mutation rates. Genetics. 1973;73(Suppl 73):1–18. [PubMed] [Google Scholar]
- 29.Gillespie JH. Mutation modification in a random environment. Evolution. 1981;35:468–476. doi: 10.1111/j.1558-5646.1981.tb04910.x. [DOI] [PubMed] [Google Scholar]
- 30.Ishii K, Matsuda H, Iwasa Y, Sasaki A. Evolutionarily stable mutation rate in a periodically changing environment. Genetics. 1989;121(1):163–174. doi: 10.1093/genetics/121.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. The evolution of mutation rates: Separating causes from consequences. Bioessays. 2000;22(12):1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 32.Cohen JE. Convexity properties of products of random nonnegative matrices. Proc Natl Acad Sci USA. 1980;77(7):3749–3752. doi: 10.1073/pnas.77.7.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H. Accelerated gene evolution through replication-transcription conflicts. Nature. 2013;495(7442):512–515. doi: 10.1038/nature11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spudich JL, Koshland DE., Jr Non-genetic individuality: Chance in the single cell. Nature. 1976;262(5568):467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- 35.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467(7312):167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitham TG, Slobodchikoff CN. Evolution by individuals, plant-herbivore interactions, and mosaics of genetic variability: The adaptive significance of somatic mutations in plants. Oecologia. 1981;49:287–292. doi: 10.1007/BF00347587. [DOI] [PubMed] [Google Scholar]
- 37.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 38.Koonin EV, Wolf YI. Is evolution Darwinian or/and Lamarckian? Biol Direct. 2009;4:42. doi: 10.1186/1745-6150-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helanterä H, Uller T. The Price equation and extended inheritance. Philos Theor Biol. 2010;2:e101. [Google Scholar]
- 40.Day T, Bonduriansky R. A unified approach to the evolutionary consequences of genetic and nongenetic inheritance. Am Nat. 2011;178(2):E18–E36. doi: 10.1086/660911. [DOI] [PubMed] [Google Scholar]
- 41.Price GR. Selection and covariance. Nature. 1970;227(5257):520–521. doi: 10.1038/227520a0. [DOI] [PubMed] [Google Scholar]
- 42.Price GR. Extension of covariance selection mathematics. Ann Hum Genet. 1972;35(4):485–490. doi: 10.1111/j.1469-1809.1957.tb01874.x. [DOI] [PubMed] [Google Scholar]
- 43.Cover TM, Thomas JA. Elements of Information Theory. New York: Wiley; 1991. [Google Scholar]
- 44.Kelly JL. A new interpretation of information rate. Information Theory. 1956;2:185–189. [Google Scholar]
- 45.Haccou P, Iwasa Y. Optimal mixed strategies in stochastic environments. Theor Popul Biol. 1995;47:212–243. [Google Scholar]
- 46.Simon D. Optimal State Estimation: Kalman, H Infinity, and Nonlinear Approaches. Hoboken, NJ: Wiley-Interscience; 2006. [Google Scholar]
- 47.Kirkpatrick M, Lande R. The evolution of maternal characters. Evolution. 1989;43:485–503. doi: 10.1111/j.1558-5646.1989.tb04247.x. [DOI] [PubMed] [Google Scholar]
- 48.Lachmann M, Jablonka E. The inheritance of phenotypes: An adaptation to fluctuating environments. J Theor Biol. 1996;181(1):1–9. doi: 10.1006/jtbi.1996.0109. [DOI] [PubMed] [Google Scholar]
- 49.Pal C. Plasticity, memory and the adaptive landscape of the genotype. Proc Biol Sci. 1998;265:1319. [Google Scholar]
- 50.Pavlicev M, Cheverud JM, Wagner GP. Evolution of adaptive phenotypic variation patterns by direct selection for evolvability. Proc Biol Sci. 2011;278(1713):1903–1912. doi: 10.1098/rspb.2010.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chevin L-M, Lande R, Mace GM. Adaptation, plasticity, and extinction in a changing environment: Towards a predictive theory. PLoS Biol. 2010;8(4):e1000357. doi: 10.1371/journal.pbio.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.