Significance
We have discovered a critical role for ten-eleven translocation 3-mediated hydroxylation of 5-methycytosine in the adult prefrontal cortex in mediating rapid behavioral adaptation. 5-hydroxymethylcytosine (5-hmC) is highly dynamic in response to fear extinction training, and rather than simply reflecting a functional intermediary of active DNA demethylation, the learning-induced intergenic accumulation of 5-hmC creates an epigenetic state that promotes experience-dependent gene expression and behavioral adaptation.
Abstract
5-hydroxymethylcytosine (5-hmC) is a novel DNA modification that is highly enriched in the adult brain and dynamically regulated by neural activity. 5-hmC accumulates across the lifespan; however, the functional relevance of this change in 5-hmC and whether it is necessary for behavioral adaptation have not been fully elucidated. Moreover, although the ten-eleven translocation (Tet) family of enzymes is known to be essential for converting methylated DNA to 5-hmC, the role of individual Tet proteins in the adult cortex remains unclear. Using 5-hmC capture together with high-throughput DNA sequencing on individual mice, we show that fear extinction, an important form of reversal learning, leads to a dramatic genome-wide redistribution of 5-hmC within the infralimbic prefrontal cortex. Moreover, extinction learning-induced Tet3-mediated accumulation of 5-hmC is associated with the establishment of epigenetic states that promote gene expression and rapid behavioral adaptation.
Epigenetic mechanisms are critically involved in the regulation of gene expression underlying learning and memory (1). Dynamic variation in the accumulation of a particular epigenetic mark, 5-methycytosine (5-mC), has emerged as a key factor in experience-dependent plasticity and the formation of fear-related memory (2). However, 5-mC is not the only covalent modification of DNA in eukaryotes, as methylated cytosine guanine (CpG) dinucleotides can be successively oxidized and converted to 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC), and 5-carboxylcytosine by the Tet family of DNA dioxygenases (3, 4). Although little is known about the functional relevance of 5-fC and 5-carboxylcytosine (5, 6), an understanding of 5-hmC is starting to emerge. 5-hmC is highly enriched in the adult brain (7), dynamically regulated by neural activity (8), and accumulates across the lifespan (9). This epigenetic mark is critically involved in neuronal differentiation and in the reprogramming of pluripotent stem cells (10), and rather than being an intermediate state of active DNA demethylation, 5-hmC can be either dynamic or stable (8, 10). Unlike its repressive cousin, 5-mC, which is primarily found along CpG-rich gene promoters, 5-hmC is enriched within gene bodies and at intron–exon boundaries of synaptic plasticity-related genes, as well as within distal cis-regulatory elements, which together point to an important role for 5-hmC in coordinating transcriptional activity (11–13). Thus, it is evident that the relationship between this particular covalent modification of DNA and gene expression is far more complex than currently realized.
The inhibition of learned fear is an evolutionarily conserved behavioral adaptation that is essential for survival. This learning process, known as extinction, involves rapid reversal of previously learned contingencies, which depend on gene expression and protein synthesis. Impairments in the neural mechanisms that promote this beneficial response to threat can lead to the development of posttraumatic stress disorder and phobia (14). Fear extinction has long been recognized as an invaluable tool for investigating the neural mechanisms of fear-related learning and memory (15). Using this experimental paradigm, the important contribution of the medial prefrontal cortex toward fear extinction has been demonstrated (16, 17). For example, lesions or infusions of protein synthesis inhibitors into the infralimbic prefrontal cortex (ILPFC) dramatically impair fear extinction learning (18, 19).
We, and others, have recently identified epigenetic regulatory mechanisms in the ILPFC, including histone modifications and small noncoding RNAs that are selectively involved in the extinction of conditioned fear (20–26). Recent evidence indicates that Tet1 promotes active DNA demethylation in the adult hippocampus (8) and that the accumulation of 5-hmC and associated effects on gene expression are involved in adult neurogenesis, spatial learning, and the extinction of contextual fear (27–29). In contrast, Tet3 is highly expressed in the adult cortex (30), although its function with respect to fear extinction memory has yet to be determined. We therefore set out to explore the role of Tet3-mediated hydroxylation within the ILPFC and to elucidate whether it is involved in the rapid behavioral adaptation supporting the extinction of conditioned fear.
Results
Tet3, but Not Tet1, Is Activity-Dependent in Primary Cortical Neurons and Necessary for Rapid Behavioral Adaptation.
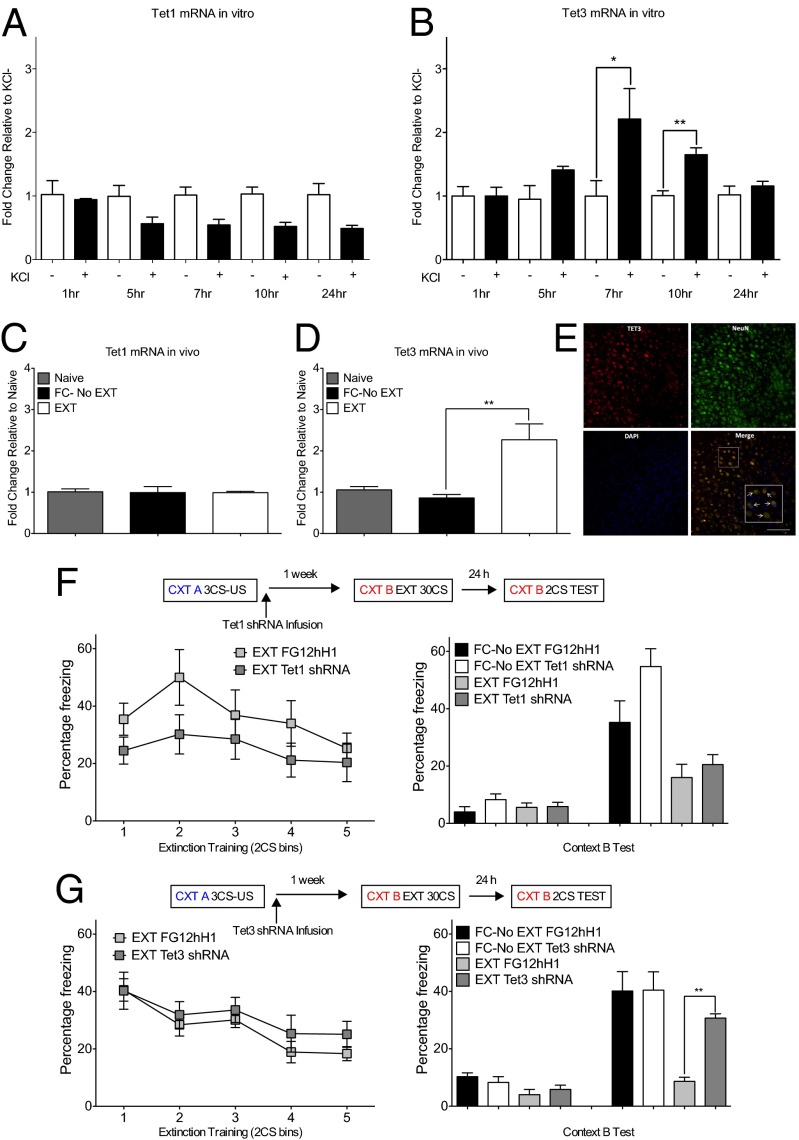
To elucidate the underlying mechanism by which experience-dependent hydroxylation of 5-mC occurs in the prefrontal cortex, we first examined the activity-dependent expression of Tet1 and Tet3 in primary cortical neurons in vitro and within the ILPFC after fear extinction learning in vivo. In primary cortical neurons, in contrast to previous reports using hippocampal neurons (8, 27), Tet1 did not respond in an activity-dependent manner, whereas Tet3 exhibited a time-dependent increase in mRNA expression (Fig. 1 A and B). Moreover, although there was no effect on Tet1, extinction training led to a significant increase in Tet3 mRNA levels within cortical neurons (Fig. 1 C–E). These findings indicate that Tet3 is selectively activated within the adult neocortex in an experience-dependent manner. We next generated Tet1 or Tet3 knockdown lentiviral plasmids, using previously published protocols (25), and functionally validated them in N2A neuroblastoma cells (SI Appendix, Fig. S1 A and B). Importantly, our Tet3 shRNA reduces global 5-hmC in vitro (SI Appendix, Fig. S1C) and is functional in vivo (SI Appendix, Fig. S1 D and E). Knockdown of either Tet1 or Tet3 via intra-ILPFC shRNA infusion had no significant effect on within-session performance during the first 10 conditioned stimulus exposures during extinction training. Although there was also no significant effect of either Tet1 or Tet3 shRNA on the ability to express fear in mice that were fear conditioned and exposed to a novel context without extinction training (FC-No EXT), mice trained in the presence of Tet3 shRNA showed a significant impairment in fear extinction memory (Fig. 1 F and G). Moreover, NMDA receptor antagonist-mediated inhibition of fear extinction learning by systemic Dizocilpine (MK-801) treatment before training blocked the extinction-learning-induced increase in Tet3 mRNA expression in the ILPFC (SI Appendix, Fig. S2). Together, these data demonstrate a critical role for Tet3 within the ILPFC in the regulation of this important behavioral adaptation.
Fig. 1.
Tet3-mediated hydroxylation of 5-mC is required for rapid behavioral adaptation. (A) Under KCl-induced depolarization conditions, there was an overall reduction in Tet1 mRNA (n = 3 per group; F9,29 = 3.78; P < 0.01). (B) In contrast, there was a significant increase in Tet3 mRNA expression 7 and 10 h poststimulation (n = 3 per group; F9,29 = 3.78; P < 0.01). (C) Although there was no effect of behavioral training on Tet1 mRNA within the ILPFC when measured 2 h after training, (D) there was a selective increase in Tet3 mRNA expression (n = 4 per group; F2,11 = 10.79; P < 0.01; Tukey’s post hoc analysis FC-No EXT vs. EXT, *P < 0.05). (E) Tet3 is highly expressed within the ILPFC. (F) There was no significant effect of Tet1 shRNA on within-session extinction across (Left) and no effect of Tet1 shRNA on fear extinction memory at test 24 h after training (Right). (G) Although there was no significant effect of Tet3 knockdown on within-session extinction (Left), there was a significant impairment in fear extinction memory when tested 24 h after training (Right, n = 7 per group; F3,26 = 8.44; P < 0.01; Tukey’s post hoc analysis EXT FG12hH1 vs. EXT Tet3 shRNA, *P < 0.05). Error bars represent standard error of the mean.
Genome-Wide Patterns of 5-hmC Are Dramatically Redistributed in Response to Extinction Learning.
To gain further insight into the putative role of 5-hmC in regulating gene expression associated with fear extinction, we developed a protocol that capitalizes on the addition of “DNA barcodes” to individual samples before 5-hmC capture, followed by deep sequencing. Although there are single-base resolution approaches for profiling 5-hmC across the genome (31, 32), we adopted a cost-effective approach that enables the mapping of 5-hmC within the ILPFC derived from individual mice, without the need to pool samples (SI Appendix, Materials and Methods). This 5-hmC antibody-based method is comparable to chemical-based labeling with respect to the enrichment of both clustered and sparsely distributed 5-hmC marks in genomic DNA derived from samples in vivo (8), which makes it particularly suitable for detecting the regional distribution of 5-hmC across the genome in the adult cortex. With this approach in hand, a genome-wide map of 5-hmC was generated from 8 biological replicates per treatment group, with an average of 60 million high-quality mapped reads per sample (SI Appendix, Table S1).
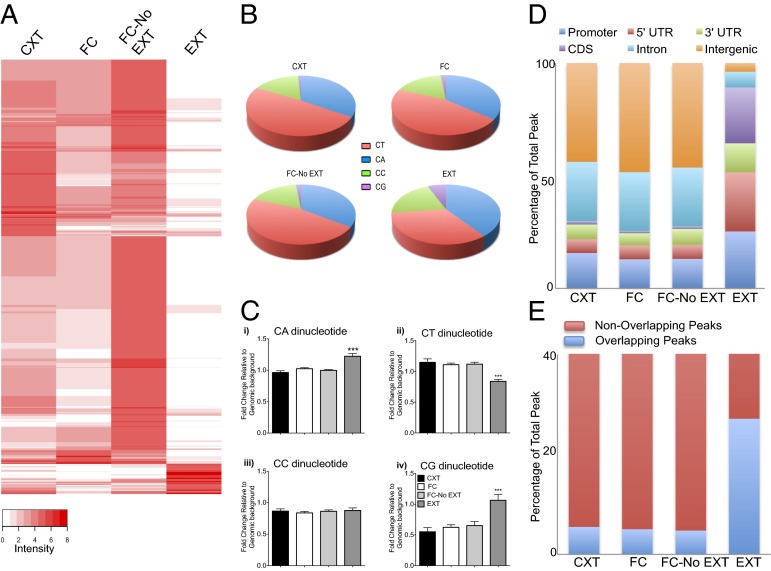
A comparison between ILPFC samples derived from mice trained to fear an auditory cue and extinction-trained mice revealed dramatic experience-dependent genome-wide differences in the accumulation of 5-hmC in response to learning (Fig. 2A). In all significant peaks where 5-hmC accumulated after extinction training, 35–40% of nucleotides spanning a 100-bp region directly under the summit of the peak appeared to contain either CA or CT dinucleotide repeats (Fig. 2B). Furthermore, very few peaks were enriched for CG dinucelotides, suggesting that in the adult ILPFC, 5-hmC may not prefer CpG-rich regions, which are typically found surrounding transcription start site (TSS) in proximal gene promoters (Fig. 2C). Although speculative at this stage, and requiring further validation using single base resolution approaches, these findings bear resemblance to previous observations of 5-hmC within proximal gene promoters during embryonic development (33). However, our data also raise the possibility that in response to experience, 5-hmC may accumulate in nonpromoter regions containing repetitive elements, a finding that accords with the emerging consensus that DNA methylation is more widely distributed across the genome than previously thought (8, 34, 35). We favor the latter interpretation because the pattern of 5-hmC within CA- or CT-rich regions was not equally distributed across the four treatment groups. Moreover, there was a dramatic shift in the presence of 5-hmC, with a significant reduction in intronic and intergenic regions. This was accompanied by an accumulation of 5-hmC within distal gene promoters, 5′-UTR (untranslated region), 3′UTR, and coding DNA sequence (CDS) (Fig. 2D and SI Appendix, Table S2). Using the ENCODE database as a reference, we also observed a significant overlap between 5-hmC and DNaseI-hypersensitive regions across the genome, which expanded from 5–27% of total peaks called after extinction training (Fig. 2E and SI Appendix, Table S3). As DNaseI-hypersensitive regions are typically present within noncoding regulatory regions of the genome (36), an experience-dependent shift in 5-hmC within these regions may reflect the activation or suppression of distal enhancers.
Fig. 2.
Experience-dependent redistribution of 5-hmC within the ILPFC. (A) Representative heat map of genome-wide 5-hmC enrichment after behavioral training, derived from 8 biological replicates per treatment group. Note the cluster of candidate genes, which showed enrichment specifically after fear extinction learning. (B) In all significant peaks in which 5-hmC accumulated after extinction training, 35–40% of nucleotides spanning a 100-bp region directly under the summit of the peak contained either CA or CT dinucleotide repeats (n = 8 per group; CA F3, 31 = 18.02; CT F3, 31 = 14.23; CG F3, 31 = 10.38; Tukey’s post hoc analysis for all EXT vs. context (CXT), FC, FC-No EXT, ***P < 0.001). (C) Relative to all other groups, a unique pattern of 5-hmC distribution with an increase in CG-rich regions occurred in response to fear extinction learning. (D) There was an extinction training-induced decrease in 5-hmC peaks detected within intronic and intergenic regions, which was accompanied by increased 5-hmC at gene promoters, 5′-UTR, 3′-UTR, and within CDS. (E) After extinction training, there was a significant overlap between 5-hmC and DNaseI-hypersensitive regions across the genome, which expand from 5–27% of total peaks called. CXT, context only, 24 h; FC, fear conditioned, 24 h; FC-No EXT, fear conditioned and context B exposed without extinction, 2 h; EXT, extinction, 2 h. Error bars represent standard error of the mean.
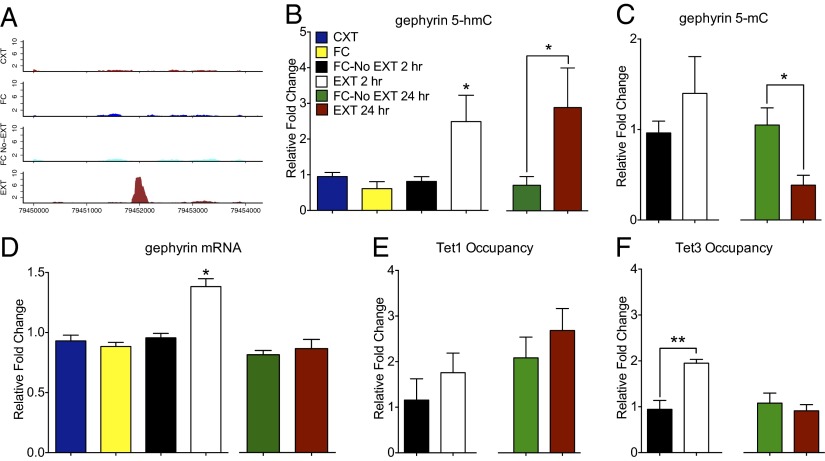
A gene ontology analysis was performed on 233 genes associated with 5-hmC peaks that were selectively induced by extinction training, revealing that 16% belong to a network associated with synaptic signaling (SI Appendix, Table S4). As described in SI Appendix, Table 5, 13 candidate genes were selected for validation on the basis of their role in neural plasticity; these were subsequently confirmed in a an independent biological cohort by 5-hmC capture followed by quantitative PCR (SI Appendix, Fig. S3). Of particular interest was gephyrin, which anchors GABA receptors to the postsynaptic membrane and is directly involved in fear extinction (37, 38). We observed an accumulation of 5-hmC within an intron of the gephyrin gene, which was accompanied by a concomitant decrease in 5-mC 24 h postextinction training (Fig. 3 A–C and SI Appendix, Fig. S4). Importantly, these effects were selective for this locus, as 5-hmC did not accumulate within the gene body upstream or downstream of this intron (SI Appendix, Fig. S5). Gephyrin mRNA expression increased transiently 2 h after extinction training and returned to baseline 24 h later (Fig. 3D). Furthermore, there was also a transient increase in Tet3 occupancy surrounding the gephyrin gene (Fig. 1 E and F). DNA methylation can be persistently altered after learning (5), and it has been proposed that discordance between DNA methylation status and gene expression might reflect a form of genomic metaplasticity that serves to promote gene expression on subsequent stimulation (39).
Fig. 3.
Fear extinction learning increased the accumulation of 5-hmC and Tet3 surrounding the extinction-related gene, gephyrin. (A) Extinction training led to a persistent increase in 5-hmC within an intron of the gene encoding gephyrin. Shown is the normalized depth of coverage for this peak and its nearby region in each of the conditions. (B) A significant enrichment of 5-hmC within the intronic region of gephyrin occurred after fear extinction training [n = 3–4 per group, 2 h; F3,14 = 5.29 (P < 0.01); Tukey’s post hoc analysis FC-No EXT 2 h vs. EXT 2 h (*P < 0.05), 24 h; t6 = 2.24 (P < 0.05). (C) This effect was accompanied by a reduction in 5-mC 24 h after extinction training (n = 4 per group, 24 h, t6 = 3.03; P < 0.05. (D) Fear extinction led to a transient increase in gephyrin mRNA expression (n = 4–5 per group; F3,19 = 4.08; P < 0.05; Tukey’s post hoc analysis FC-No EXT vs. EXT, *P < 0.05). (E) There was a trend toward an increase in Tet1 occupancy within the intronic region of the gephyrin gene 24 h after extinction training. (F) Fear extinction learning leads to a transient increase in Tet3 occupancy (n = 3–5 per group; F3,15 = 17.58; P < 0.001; Tukey’s post hoc analysis FC-No EXT 2 h vs. EXT 2 h, **P < 0.01).
Fear Extinction Leads to a Tet3-Mediated Accumulation of 5-hmC, Which Is Associated with a Permissive Epigenetic State.
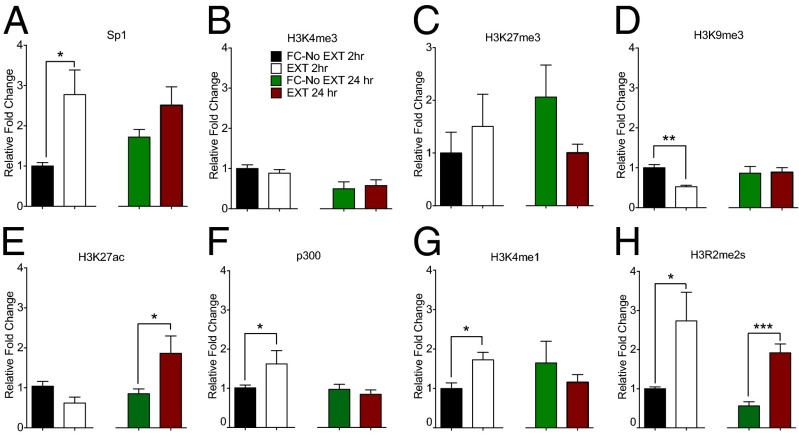
Emerging evidence suggests a role for 5-hmC outside gene promoters in the regulation of gene expression, potentially through its effects on alternative splicing or through an interaction with the DNA methyl binding protein, MECP2 (11, 12). However, an understanding of how 5-hmC influences this process within the context of learning and memory has not been achieved. It has recently been suggested that 5-hmC can regulate gene expression by recruiting “readers” to regions across the genome in which this epigenetic mark accumulates (40). On the basis of our genome-wide 5-hmC mapping data, we reasoned that the presence of 5-hmC within intronic and intergenic regions influences the local chromatin landscape, thereby providing a link between this epigenetic modification and experience-dependent regulation of gene expression. To explore this possibility, we interrogated the chromatin environment surrounding the accumulation of 5-hmC within the gephyrin gene, revealing increased binding of specificity protein 1 (Sp1) (Fig. 4A). This transcription factor contributes to gene regulation by protecting active genomic loci from DNA remethylation (41). There was no effect on the occupancy of H3K4me3 or H3K27me3 (Fig. 4 B and C), indicating that the development of a bivalent chromatin state (42) at this locus is not associated with the experience-dependent accumulation of 5-hmC. However, in contrast, we observed a transient reduction in the heterochromatin-related histone mark H3K9me3 (Fig. 4D), which was associated with a delayed increase in H3K27ac and a transient increase in p300 and H3K4me1 occupancy (Fig. 4 E–G). These chromatin modifications are associated with regulatory elements across the genome and promote gene expression (43, 44). Finally, there was a significant increase in the occupancy of symmetric dimethyl H3 arginine 2 (H3R2me2s) within the gephyrin intronic locus (Fig. 4H), which paralleled the persistent accumulation of 5-hmC. This novel histone mark, which is crucial for the maintenance of a euchromatic or “primed” state, is preferentially associated with sites of recombination and recently activated genes (45, 46). Importantly, these effects were specific to extinction training and did not occur in mice that had been fear conditioned, followed by a single reactivation trial, therefore arguing against the possibility that such epigenetic modifications are nonspecifically induced by the retrieval or reconsolidation of the original fear memory (SI Appendix, Fig. S6).
Fig. 4.
Fear extinction learning is associated with an altered chromatin landscape. Fear extinction led to (A) a transient increase in the occupancy of Sp1 (t7 = 2.54; P < 0.01) with no effect on bivalent chromatin marks H3K4me3 (B) or H3K27me3 (C), (D) a transient reduction in the heterochromatin mark H3K9me3 (t7 = 3.12; P < 0.05), (E) a delayed increase in H3K27ac (n= 4–5 per group; t7 = 2.013; P < 0.05), a transient increase in presence of the enhancer-related elements p300 (f; t7 = 2.74; P < 0.05) and H3K4me1 (G) (t6 = 0.85; P < 0.05), and a persistent increase in accumulation of the euchromatic mark, H3R2me2s (H) [(2 h, t6 = 2.21 (P < 0.05); 24 h, t6 = 5.46 (P < 0.001)] within the ILPFC. Error bars represent standard error of the mean.
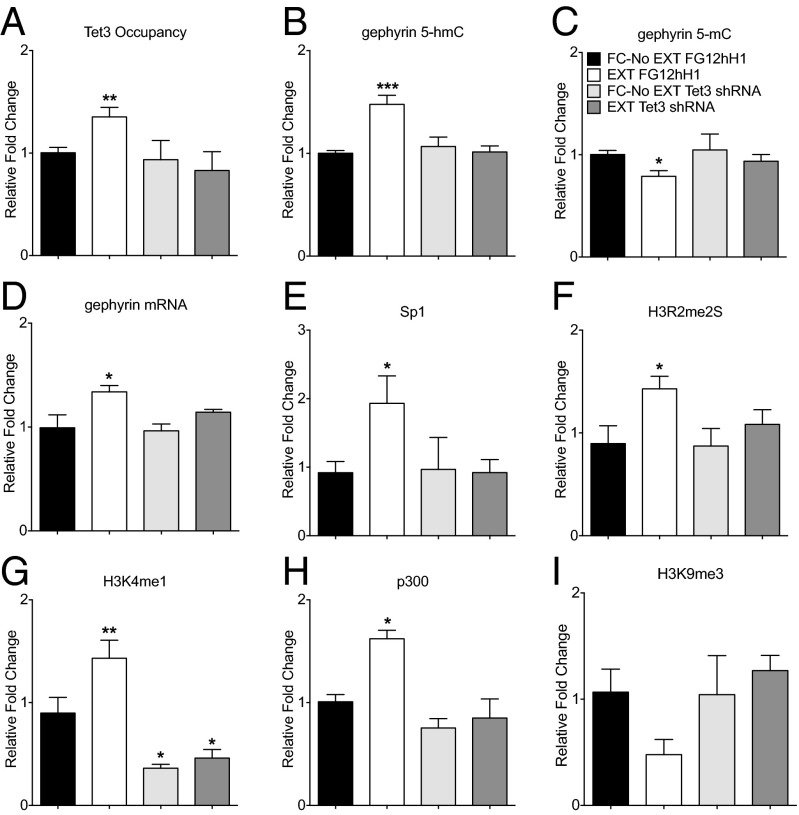
Together, the data suggest that fear extinction learning leads to a redistribution of 5-hmC within the ILPFC, which subsequently promotes a transcriptionally active chromatin state, possibly by driving the selective accumulation of 5-hmC within noncoding DNA regulatory regions of the genome. The generality of these findings was tested by interrogating a 5-hmC peak found within a distal promoter region of the gene encoding Lin7a, where the pattern of chromatin modification was quite different, suggesting that the accumulation of 5-hmC induced by extinction training may have functionally distinct consequences depending on the sequence context in which it occurs (SI Appendix, Figs. S7 and S8). Finally, the effect of extinction learning on Tet3 occupancy at the gephyrin locus, as well as the dynamic changes in the accumulation of 5-hmC and 5-mC, gephyrin mRNA, and associated effects on the chromatin landscape were completely blocked in the presence of Tet3 shRNA (Fig. 5 A–I). Thus, Tet3 activity within the ILPFC is necessary for the learning-dependent accumulation of 5-hmC and related chromatin modifications, which underpins rapid behavioral adaptation.
Fig. 5.
Tet3 is required for extinction training-induced accumulation of 5-hmC and associated effects on the chromatin landscape. (A) Tet3 occupancy at gephyrin locus was reduced in the presence of Tet3 shRNA (n = 4/group; F3,12 = 5.27; P < 0.05; Dunnett’s post hoc test FC-No EXT FG12hH1 vs. EXT FG12hH1, **P < 0.01). (B) Tet3 knockdown blocked the accumulation of 5-hmC after fear extinction training (n = 4; F3,12 = 11.63; P < 0.001; Dunnett’s post hoc test FC-No EXT FG12hH1 vs. EXT FG12hH1, ***P < 0.001). (C) There was no effect on 5-mC after extinction training in the presence of Tet3 shRNA (n = 4; F3,12 = 4.26; P < 0.05; Dunnett’s post hoc test FC-No EXT FG12hH1 vs. EXT FG12hH1, *P < 0.05). (D) There was no significant increase in gephyrin mRNA expression after extinction training in the presence of Tet3 shRNA (n = 4–6 per group; F3,19 = 6.364; P < 0.01; Dunnett’s FC-No EXT FG12hH1 vs. EXT FG12hH1, *P < 0.05). Knockdown Tet3 mRNA blocked the effect of extinction training on Sp1 (E) (n = 4/group; F3,12 = 3.76; P < 0.05; Dunnett’s post hoc test FC-No EXT FG12hH1 vs. EXT FG12hH1, *P < 0.05), H3R2me2S (F) (n = 3–4/group; F3,12 = 3.52; P < 0.05; Dunnett’s post hoc test FC-No EXT FG12hH1 vs. EXT FG12hH1, *P < 0.05), H3K4me1 (G) (n = 4/group; F3,12 = 23.56; P < 0.0001; Dunnett’s post hoc test FC-No EXT FG12hH1 vs. EXT FG12hH1, **P < 0.01, vs. FC-No EXT Tet3 shRNA, *P < 0.05), and p300 occupancy at the gephyrin locus (H) (n = 4/group; F3,12 = 11.88; P < 0.001; Dunnett’s post hoc test FC-No EXT FG12hH1 vs. EXT FG12hH1, *P < 0.05), (I) There was no significant effect of Tet3 shRNA on H3K9me3. Error bars represent standard error of the mean.
Discussion
This study has generated 3 novel findings: Tet3 expression is activity-dependent in primary cortical neurons and, within the adult prefrontal cortex, is necessary for rapid behavioral adaptation; the genome-wide pattern of 5-hmC in the ILPFC is dramatically redistributed in response to fear extinction learning; and fear extinction leads to a Tet3-mediated accumulation of 5-hmC, which is associated with a permissive epigenetic state.
The observation that Tet3, but not Tet1, expression is activity-dependent and necessary for rapid behavioral adaptation (Fig. 1) appears, at first glance, to be at odds with the recent discovery that Tet1 is involved in neuronal-activity-regulated gene expression and extinction (29). However, the general interpretation of this work is limited by the use of developmental Tet1 knockout mice, rather than spatiotemporally restricting the ablation of Tet1 to the adult prefrontal cortex. Tet1-mediated accumulation of 5-hmC is critically involved in neuronal differentiation and in the reprogramming of pluripotent stem cells (33), thus suggesting a distinct role for Tet1 during early brain development. Tet3, in contrast, is the most highly expressed isoform in the adult cortex (30), and the highest level of 5-hmC in the adult brain is found in cortical neurons (7), where it continues to accumulate across the lifespan (9). Together, these data suggest a potential developmental stage- and/or region-specific function for each Tet isoform and its influence on the regulation of 5-hmC, which would be difficult to disentangle from the use of mice that have had Tet1 knockout throughout early development. Moreover, Rudenko and colleagues (29) primarily focused on hippocampal-dependent cognitive tasks, including the extinction of spatial memory and the extinction of contextual fear, whereas our analysis was specifically targeted toward understanding the role of Tet3 in the adult brain and in a cognitive task that is highly dependent on the ILPFC. Notwithstanding these differences, the evidence is clear that, within the ILPFC, Tet3 expression is activity-dependent and plays a key role in the formation of fear extinction memory.
A key question is whether neocortical accumulation of 5-hmC has a role in regulating fear extinction that is distinct from its role during early embryonic development. In contrast to 5-mC, which in general is considered to be a repressive mark associated with CpGs in proximal promoters, although this perception is rapidly changing (35, 47), the pervasive distribution of 5-hmC throughout the genome suggests it is involved in regulating transcriptional activation. Indeed, previous work has shown that 5-hmC is enriched within highly transcribed gene bodies and at intron–exon boundaries of synaptic plasticity-related genes, as well as within distal cis-regulatory elements (6, 9, 11–13, 31–33, 48–50). Our observation of a dramatic redistribution of 5-hmC in response to fear extinction training, with subsequent effects on chromatin modifications and gene expression, lends support to this idea and further suggests that, at least in the case of fear extinction, the accumulation of 5-hmC is associated with a transcriptionally permissive chromatin environment, although the pattern differs depending on the genomic locus (Fig. 4 and SI Appendix, Fig. S7). With regard to the gephyrin locus, we observed a persistent increase in the presence of the euchromatic mark H3R2me2s, which is dependent on Tet3-mediated accumulation of 5-hmC (Fig. 5). In addition, the extinction training-induced increase in 5-hmC was also present 24 h after extinction training, whereas the expression of gephyrin mRNA returned to baseline at the same time (Fig. 3). This suggests a potentially time-dependent relationship between experience-dependent DNA modifications, epigenetic states, and gene expression that occur in response to extinction learning. We recently proposed that experience-dependent variations in DNA methylation represent a form of metaplasticity that serves to prime the genome to respond to later events by regulating transcriptional capacity, rather than by mediating enduring changes in gene expression (39). Given the recent observation that 5-fC is involved in long-lasting epigenetic priming and preferentially occurs at regulatory elements, including poised enhancers (6), it is conceivable that 5-hmC will be replaced by 5-fC to promote a metaplastic or “primed” epigenetic state, which will then be reflected by greater induction of gene expression in response to further extinction training.
Although the precise mechanism by which extinction training leads to a Tet3-mediated redistribution of nonpromoter 5-hmC and select chromatin modifications is not yet known, an understanding of the unique characteristics of Tet3 may shed light on this process. Together with a catalytic C-terminal domain, which provides hydroxylase activity, Tet1 and Tet3 also harbor a CxxC binding motif (CXXC) zinc finger-binding domain that promotes high-affinity binding at genomic loci containing CpGs. However, in contrast to Tet1, the Tet3 CXXC binding domain recognizes unmodified C followed by A, T, C, or G, with a preference for nonpromoter CpGs (51). Moreover, Tet3 also contains the novel-binding domain PRK12323, the function of which has not been determined (52). Tet3 is also endowed with a function that is independent of its enzymatic activity: Tet3 is a direct binding partner with O-linked N-acetylglucosamine transferase and colocalizes with this transferase on chromatin at active promoters enriched for H3K4me3, which is critically involved in transcriptional activation (53, 54). However, at least with respect to the gephyrin locus after extinction training, we observed no change in H3K4me3 occupancy (Fig. 4), and we found that that Tet3 is catalytically active by virtue of its effect on the experience-dependent accumulation of 5-hmC and its related effect on the chromatin landscape (Fig. 5). These data therefore point to a critical role for Tet3-mediated hydroxylation of 5-mC within the ILPFC in rapid behavioral adaptation.
The relationship between different DNA base modifications, gene expression, and the chromatin landscape may also vary depending on the genomic locus in which the modifications occur in response to different learning conditions. For example, the intragenic accumulation of 5-hmC induced by extinction training may be associated with Tet3, whereas the accumulation of 5-hmC within distal regulatory elements and proximal promoters may be associated with the recruitment of Tet1 after fear learning. Indeed, Tet1 accumulation in promoter regions after hippocampal-dependent contextual fear conditioning has previously been demonstrated (28). The accumulation of these DNA base modifications may have different effects depending on whether they occur at single bases within key binding motifs, which would suggest a role in regulating transcription factor activity, and potentially alternative splicing, or if they are more broadly distributed, the pattern of accumulation may be indicative of a role in the regulation of DNA structure or chromatin looping (39). Our findings on the critical role of Tet3-mediated redistribution of 5-hmC in the adult brain provide the foundation for future studies on this interesting mode of epigenetic regulation and highlight the importance of examining the full repertoire of DNA base modifications across brain regions to elucidate how epigenetic states are established in response to different forms of learning.
Materials and Methods
Details of DNA/RNA extraction, qRT-PCR, 5-hmC and 5-mC capture, DNA sequencing, lentiviral construct design, immunohistochemistry, chromatin immunoprecipitation, primary cortical neuron culture, and 5-hmC dot blot are included in the SI Appendix, Materials and Methods. Details of protocols for subjects, cannulation surgery, and behavioral methods. A description of bioinformatics and sequencing analyses are also described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Ms Rowan Tweedale for helpful editing of the manuscript. The authors acknowledge grant support from the National Health and Medical Research Council of Australia (APP1033127-TWB), the Australian Research Council (DP1096148-TWB), and the National Institutes of Health (GM68804-YZ). X.L. is supported by postgraduate scholarships from the University of Queensland and the ANZ Trustees Queensland for medical research.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318906111/-/DCSupplemental.
References
- 1.Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70(5):813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller CA, et al. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13(6):664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen L, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153(3):692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song C-X, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153(3):678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo JU, Su Y, Zhong C, Ming G-L, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szulwach KEK, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn MA, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3(2):291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khare T, et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat Struct Mol Biol. 2012;19(10):1037–1043. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336(6083):934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 14.Orsini CA, Maren S. Neural and cellular mechanisms of fear and extinction memory formation. Neurosci Biobehav Rev. 2012;36(7):1773–1802. doi: 10.1016/j.neubiorev.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis M. Pharmacological and anatomical analysis of fear conditioning using the fear-potentiated startle paradigm. Behav Neurosci. 1986;100(6):814–824. doi: 10.1037//0735-7044.100.6.814. [DOI] [PubMed] [Google Scholar]
- 16.Milad MR, Quirk GJ. Fear extinction as a model for translational neuroscience: Ten years of progress. Annu Rev Psychol. 2012;63:129–151. doi: 10.1146/annurev.psych.121208.131631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons RG, Ressler KJ. Implications of memory modulation for post-traumatic stress and fear disorders. Nat Neurosci. 2013;16(2):146–153. doi: 10.1038/nn.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laurent V, Westbrook RF. Inactivation of the infralimbic but not the prelimbic cortex impairs consolidation and retrieval of fear extinction. Learn Mem. 2009;16(9):520–529. doi: 10.1101/lm.1474609. [DOI] [PubMed] [Google Scholar]
- 19.Sierra-Mercado D, Padilla-Coreano N, Quirk GJ. Dissociable roles of prelimbic and infralimbic cortices, ventral hippocampus, and basolateral amygdala in the expression and extinction of conditioned fear. Neuropsychopharmacology. 2011;36(2):529–538. doi: 10.1038/npp.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bahari-Javan S, et al. HDAC1 regulates fear extinction in mice. J Neurosci. 2012;32(15):5062–5073. doi: 10.1523/JNEUROSCI.0079-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei W, et al. p300/CBP-associated factor selectively regulates the extinction of conditioned fear. J Neurosci. 2012;32(35):11930–11941. doi: 10.1523/JNEUROSCI.0178-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bredy TW, et al. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14(4):268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bredy TW, Barad M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learn Mem. 2008;15(1):39–45. doi: 10.1101/lm.801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marek R, et al. Paradoxical enhancement of fear extinction memory and synaptic plasticity by inhibition of the histone acetyltransferase p300. J Neurosci. 2011;31(20):7486–7491. doi: 10.1523/JNEUROSCI.0133-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Q, et al. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat Neurosci. 2011;14(9):1115–1117. doi: 10.1038/nn.2891. [DOI] [PubMed] [Google Scholar]
- 26.Stafford JM, Lattal KM. Is an epigenetic switch the key to persistent extinction? Neurobiol Learn Mem. 2011;96(1):35–40. doi: 10.1016/j.nlm.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang RR, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13(2):237–245. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaas GA, et al. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79(6):1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudenko A, et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79(6):1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38(19):e181–e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25(7):679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie W, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148(4):816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernstein BE, et al. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chhatwal JP, Myers KM, Ressler KJ, Davis M. Regulation of gephyrin and GABAA receptor binding within the amygdala after fear acquisition and extinction. J Neurosci. 2005;25(2):502–506. doi: 10.1523/JNEUROSCI.3301-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin H-C, Mao S-C, Gean P-W. Block of γ-aminobutyric acid-A receptor insertion in the amygdala impairs extinction of conditioned fear. Biol Psychiatry. 2009;66(7):665–673. doi: 10.1016/j.biopsych.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Baker-Andresen D, Ratnu VS, Bredy TW. Dynamic DNA methylation: A prime candidate for genomic metaplasticity and behavioral adaptation. Trends Neurosci. 2013;36(1):3–13. doi: 10.1016/j.tins.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152(5):1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Boumber YA, et al. An Sp1/Sp3 binding polymorphism confers methylation protection. PLoS Genet. 2008;4(8):e1000162. doi: 10.1371/journal.pgen.1000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 43.Xiao S, et al. Comparative epigenomic annotation of regulatory DNA. Cell. 2012;149(6):1381–1392. doi: 10.1016/j.cell.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231):854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Migliori V, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19(2):136–144. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 46.Yuan C-C, et al. Histone H3R2 symmetric dimethylation and histone H3K4 trimethylation are tightly correlated in eukaryotic genomes. Cell Rep. 2012;1(2):83–90. doi: 10.1016/j.celrep.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473(7347):389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 49.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12(6):R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Y, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151(6):1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139(11):1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ito R, et al. TET3-OGT interaction increases the stability and the presence of OGT in chromatin. Genes Cells. 2014;19(1):52–65. doi: 10.1111/gtc. [DOI] [PubMed] [Google Scholar]
- 54.Deplus R, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32(5):645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.