Regenerative medicine is a promising therapeutic option for the treatment of type 1 diabetic patients with absolute insulin deficiency. For developing such a therapy, understanding how the pancreas is formed during embryogenesis is a critical issue. Organogenesis is the process of formation of organs through differentiation of cells in the embryonic germ layers, and it totally depends on the functions of a special set of transcription factors that are specific for each organ. Numerous transcription factors are known to be involved in organogenesis of the pancreas. Among these factors, pancreas/duodenum homeobox protein 1 (PDX1) has been considered a key regulatory transcription factors in pancreas development. The importance of PDX1 is evidenced by the findings that Pdx1‐deficient mice show a severe impairment of pancreas development despite the presence of a pancreatic bud, and that deletion of a single nucleotide in the PDX1 coding sequence results in pancreatic agenesis in humans1. Furthermore, in mice, along with PDX1, other transcription factors, such as pancreas transcription factor 1A (PTF1A) and hepatocyte nuclear factor 1B (HNF1B), are essential for formation of the pancreas. Mutations in PTF1A and HNF1B are also associated with pancreatic agenesis or hypoplasia in humans, showing that humans and mice share the fundamental mechanism of pancreas development.
Recently, new molecules have been added to the list of transcription factors that participate in pancreas development. Lango Allen et al.2 examined 27 individuals with pancreatic agenesis, which was defined as neonatal diabetes requiring insulin treatment and exocrine pancreatic insufficiency requiring enzyme replacement therapy. Of these 27, only one individual had a homozygous mutation in the PTF1A splice site, and none of the individuals in the cohort had mutations in PDX1. The exome sequencing technique showed that 15 of the 27 individuals had spontaneous heterozygous loss‐of‐function mutations in the gene encoding the zinc‐finger transcription factor, GATA62. The GATA family of transcription factors comprises six members that share a common deoxyribonucleic acid‐binding motif of two tandem zinc‐finger domains recognizing the consensus sequence A/T‐GATA‐A/G. Among the GATA transcription factors, GATA1, GATA2 and GATA3 are expressed preferentially in hematopoietic cells, and are involved in cell proliferation and differentiation during hematopoiesis. In contrast, GATA4, GATA5 and GATA6 play important roles in specification and differentiation of tissues derived from the mesoderm and endoderm. Furthermore, GATA4 and GATA6 are expressed in the embryonic mouse pancreas. However, as Gata4‐ and Gata6‐null mice are embryonic lethal, the specific roles of these GATA transcription factors in development of the pancreas have not been elucidated.
Recently, the roles of Gata4 and Gata6 in the development of the pancreas were investigated in two different studies. Carrasco et al.3 and Xuan et al.4 both used the Cre/loxP‐based recombination technology to conditionally inactivate Gata4 and Gata6 genes within the pancreas. In both studies, mice carrying a floxed allele of Gata4 or Gata6 were crossbred with transgenic mice expressing Cre recombinase under the control of the Pdx1 promoter to knockout these genes specifically from the pancreas. Unlike in humans, single inactivation of either gene did not affect pancreas formation markedly, suggesting functional redundancy between these transcription factors in mice. However, simultaneous deletion of both Gata4 and Gata6 in the pancreas resulted in pancreatic agenesis, hyperglycemia and early death after birth. Such abnormalities in the double‐mutant mice are a result of the loss of the proliferation ability of the pancreatic progenitors, defects in branching morphogenesis and failure of progenitor cell differentiation. Similar to the results in Pdx1‐deficient mice, the pancreatic bud was formed in the Gata4/Gata6 double‐mutant mice, suggesting that the GATA transcription factors function after pancreas specification.
Notably, in both studies, inactivation of Gata4 and Gata6 was achieved through Pdx1 promoter‐driven expression of Cre recombinase, which ensure that the Gata genes can be removed only after initiation of the pancreatic developmental program. In that case, the roles of GATA transcription factors in the early stages of pancreas formation cannot be elucidated. Xuan et al.4 overcame this problem by using the forkhead box protein a3 (Foxa3)‐Cre mice line, which is generated using the yeast artificial chromosome transgenic technique5. Because Foxa3 is one of the few genes expressed early in the endoderm, but not in other tissues, Gata genes in the endoderm can be deleted before the appearance of the pancreas through crossbreeding Gata4flox/flox; Gata6flox/flox mice with Foxa3‐Cre mice. In the resulting mice, the pancreatic bud was formed despite the absence of the Gata genes in early endoderm. These findings show that the GATA transcription factors are dispensable for pancreas specification and bud outgrowth, but have functions in pancreatic progenitor cells (Figure 1).
Figure 1.
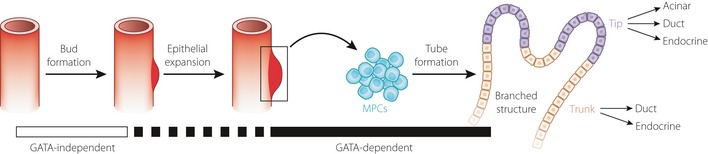
Involvement of GATA transcription factors in pancreas development. In mice, pancreatic budding from the developing gut tube occurs on approximately embryonic day 9.5. Proliferation of epithelial cells results in the formation of microlumen, and then a branched structure with tips and a trunk. Initiation of pancreatic bud formation is a GATA‐independent event. Although it is clear that GATA transcription factors function before the formation of the branched structure, it is not clear whether specification of pancreatic multipotent progenitor cells (MPCs) is GATA‐dependent or not.
During normal pancreas develop‐ment, the pancreatic epithelium forms a branched structure, in which the tips contain carboxypeptidase A1‐positive (CPA1+) multipotent progenitor cells (or acinar‐committed cells) and the trunk contains ductal‐endocrine progenitor cells6. Carrasco et al.3 reported that the pancreatic epithelium of the Gata4/Gata6 double‐mutant mice lacked such branched morphology and showed complete loss of CPA1+ cells. They also observed that these mice did not show acinar morphology and that the number of nurogenin3‐expressing endocrine progenitor cells was decreased3. In addition, the number of PDX1+, PTF1A+ and Sry‐related HMG box‐9‐positve (SOX9+) progenitor cells in the double mutant embryos was reduced partially, but significantly3. Similarly, Xuan et al.4 showed that PDX1 lineage‐labeled cells (i.e., pancreatic progenitor‐derived cells) remained unbranched and undifferentiated in the Gata4/Gata6 double‐mutant mice. Both CPA1+ and neurogenin 3 (NGN3)+ cells were mostly absent in these mice4. However, in contrast to the results of Carrasco et al.3, Xuan et al.4 showed that SOX9+ multipotent progenitor cells were maintained in the Gata4/Gata6 double mutant mice. Therefore, although it is clear that GATA transcription factors function before formation of the lineage‐restricted precursor populations, the specific roles of these factors in specification of pancreatic multipotent progenitor populations remain to be clarified (Figure 1).
Another important inference that can be drawn from the findings of Carrasco et al.3 and Xuan et al.4 is that GATA4 and GATA6 might have distinct roles in pancreas development. In both studies, deletion of only one Gata6 allele and both Gata4 alleles (i.e., retaining one allele of Gata6) markedly reduced pancreatic mass without causing loss of endocrine cells3. Furthermore, the number of pancreatic acinar cells was decreased in these mice. Although the remaining acini were disorganized, these cells expressed exocrine enzymes, such as amylase and elastase, suggesting that acinar cell proliferation, but not differentiation, is sensitive to the level of Gata6 expression. In contrast, mice lacking both Gata6 alleles and retaining one Gata4 allele showed no apparent abnormalities in pancreas formation3. These results suggest that GATA4 and GATA6 do not have identical functions in pancreas formation, despite a significant functional overlap between these factors.
Although the findings from mouse models could provide important information about human diseases, the existence of species‐specific functional differences should be kept in mind. In fact, pancreas formation in humans seems to have greater sensitivity toward loss of GATA factors than that in mice – mutations in only one GATA6 allele are sufficient to prevent normal pancreas development in humans2. Why heterozygous mutations in the GATA6 gene lead to pancreatic agenesis in humans, but not in mice, is unclear at present. Nevertheless, the studies by Carrasco et al.3 and Xuan et al.4 might represent an important step toward understanding the biology of the pancreas and for development of regenerative medicine in type 1 diabetes.
References
- 1.Stoffers DA, Zinkin NT, Stanojevic V, et al Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet 1997; 15: 106–110 [DOI] [PubMed] [Google Scholar]
- 2.Lango Allen H, Flanagan SE, Shaw‐Smith C, et al GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat Genet 2011; 44: 20–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrasco M, Delgado I, Soria B, et al GATA4 and GATA6 control mouse pancreas organogenesis. J Clin Invest 2012; 122: 3504–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xuan S, Borok MJ, Decker KJ, et al Pancreas‐specific deletion of mouse Gata4 and Gata6 causes pancreatic agenesis. J Clin Invest 2012; 122: 3516–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CS, Sund NJ, Behr R, et al Foxa2 is required for the differentiation of pancreatic alpha‐cells. Dev Biol 2005; 278: 484–495 [DOI] [PubMed] [Google Scholar]
- 6.Zhou Q, Law AC, Rajagopal J, et al A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell 2007; 13: 103–114 [DOI] [PubMed] [Google Scholar]


