Abstract
Activity-dependent survival of olfactory sensory neurons (OSNs) may allow animals to tune their olfactory systems to match their odor environment. Activity-dependent genes should play important roles in this process, motivating experiments to identify them. Both unilateral naris occlusion of mice for 6 days and genetic silencing of OSNs decreased S100A5, Lrrc3b, Kirrel2, Slc17a6, Rasgrp4, Pcp4l1, Plcxd3, and Kcnn2 while increasing Kirrel3. Naris occlusion also decreased Eml5, Ptprn, and Nphs1. OSN number was unchanged and stress-response mRNAs were unaffected after 6 days of naris occlusion. This leaves odor stimulation as the most likely cause of differential abundance of these mRNAs, but through a mechanism that is slow or indirect for most because 30–40min of odor stimulation increased only 3 of 11 mRNAs decreased by naris occlusion: S100A5, Lrrc3b, and Kirrel2. Odorant receptor (OR) mRNAs were significantly more variable than the average mRNA, consistent with difficulty in reliably detecting changes in these mRNAs after 6 days of naris occlusion. One OR mRNA, Olfr855, was consistently decreased, however. These results suggest that the latency from the cessation of odor stimulation to effects on activity-dependent OSN survival must be a week or more in juvenile mice.
Key words: microarray, naris occlusion, odorant receptor, smell, transcription
Introduction
Activity-dependent survival of olfactory sensory neurons (OSNs) suggests that mammals might have some capacity to tune their olfactory systems to changes in their odor environments. Differential activity-dependent survival was first revealed in mice with mosaic genetic silencing of OSNs that established competitive situations where inactive OSNs were preferentially lost, leading to a decrease in the frequency of OSNs expressing odorant receptors (ORs) that never experienced odor-stimulated electrical activity (Zhao and Reed 2001). Evidence is accumulating that similar phenomena may occur in response to less drastic conditions, especially changes in an animal’s odor environment (Hudson 1999; Jones et al. 2008; Santoro and Dulac 2012). In mice, at least, the olfactory epithelium appears to be capable of enriching for OSNs that respond to odors encountered, especially odors that become associated with behaviorally important experiences. Although the details of the mechanisms involved are largely unknown, the effect of odor stimulation on OSN survival appears to work, in part, through cAMP signaling and repression of a histone variant that promotes OSN apoptosis (Watt et al. 2004; Sakano 2010; Santoro and Dulac 2012).
Given this fundamental plasticity in the olfactory epithelium, the identities of activity-dependent genes in OSNs are of significant interest (Imai et al. 2006; Serizawa et al. 2006; Imai et al. 2009; Bennett et al. 2010; Williams et al. 2011; Coppola and Waggener 2012; Öztokatli et al. 2012). The initial identification of mRNAs whose abundance depends on odor stimulation can be done by genetically silencing OSNs or by blocking odorant access to the olfactory epithelium. These interventions are highly effective but are not without potential confounding factors (Coppola 2012). First, though naris occlusion is often described as sensory deprivation, it only approximates complete deprivation. Orthonasal stimulation is eliminated, but odors may still access the ipsilateral olfactory epithelium via the nasopharynx (retronasal stimulation) and in rodents, via the septal window at the base of the septum (intranasal stimulation) (Kelemen 1947). However, these alternate routes are insufficient to fully replace lost orthonasal odorant stimulation. Second, the blockage of a naris reduces stress and damage from particulates, pathogens, and chemicals ipsilaterally while simultaneously increasing the amount of these stressors experienced by the open contralateral side because the animal can no longer alternate its breathing between the nares. The importance of this effect is underscored by the ability of filtered air to increase OSN survival and of odorants to cause chemical stress responses in OSNs (Hinds et al. 1984; Sammeta and McClintock 2010). Third, the long survival times typical of both naris occlusion and genetic silencing experiments risk measuring differences due to changes secondary to odor stimulation, such as changes in the cellular composition of the olfactory epithelium or compensatory increases in expression of signaling genes (Farbman et al. 1988; Maruniak et al. 1989; Zhao and Reed 2001; Suh et al. 2006). For example, loss of orthonasal stimulation for more than 10 days after unilateral naris occlusion reduces the number of mature OSNs ipsilaterally (Farbman et al. 1988; Brunjes and Shurling 2003; Suh et al. 2006). Fourth, silencing by genetic manipulation of the mouse germ line, which prevents odorant responses from the onset of formation of the olfactory epithelium, can lead to disorganization of OSN connections to the olfactory bulb (Zhao and Reed 2001; Zou et al. 2004; Col et al. 2007). Given that OSN survival depends on feedback from the olfactory bulb (Schwob et al. 1992), changes in these synaptic connections also might cause changes in mRNA abundance that are independent of the ability of odor stimulation to directly control gene expression in OSNs. Insufficient attention has been given to determining whether activity-dependent changes in OSN mRNAs result directly from differences in odorant stimulation or instead result from loss of mature OSNs, altered feedback from the olfactory bulb, less metabolic activity in OSNs, changes in the lifespan of immature OSNs, or even altered behavior of other cells in the olfactory epithelium (e.g., macrophage recruitment and activation).
To screen for mRNAs likely to be directly sensitive to odorant stimulation, we used unilateral naris occlusion of relatively short duration to detect candidate mRNAs. These candidates were tested for sensitivity to genetic silencing of OSNs, and then for rapid responses to odorant stimulation. We identified 14 genes whose mRNAs were sensitive to naris occlusion, genetic silencing of OSNs, or both. Odorant stimulation was able to rapidly increase the abundance of only 3 of these mRNAs. We also found that ORs are highly variable mRNAs, but confirmed that 1 OR mRNA, Olfr855, was consistently more abundant in the open side’s olfactory epithelium after just 6 days of naris occlusion.
Materials and methods
Mice, naris occlusion, and RNA isolation
C57Bl/6 mice were obtained from Harlan Laboratories. Mice with a targeted deletion of Cnga2 were the kind gift of Dr Peter Mombaerts (Max Planck Institute of Biophysics). All procedures using mice were done in accordance with approved institutional Animal Care and Use protocols and conformed to NIH guidelines. Unilateral naris occlusion was done at age 5 days postnatal (P5) on mice anesthetized by hypothermia. A heated Perfectemp Cautery device (Bovie Medical Corp.) was applied for 1 s to the right external naris, followed by sealing of the cauterized region with cyanoacrylate glue. Verification of sealing was done immediately after the procedure and then again just prior to euthanization at age postnatal day 11 (P11) using a dissecting microscope to watch for bubbling of expired air through a drop of saline applied to the sealed naris. This procedure, which exactly matches previous work on stress-response mRNAs in OSNs, was chosen because it causes no change in the number of OSNs (Sammeta and McClintock 2010). Unlike naris occlusion immediately after birth, it allows both olfactory epithelia to undergo substantial postnatal development while experiencing odor stimulation. Total RNA was prepared separately from the dissected olfactory epithelia of the closed and open sides using Tri Reagent as directed by the manufacturer (Molecular Resource Center).
Odor stimulation
Young adult mice (4 weeks) were placed into inverted conical chambers where a continuous flow (2L/min) of filtered air was pushed down over the mice and through a screen floor to an exhaust directed into a fume hood. After 16h in clean air, valves were switched on to allow diversion of the clean air through a tube containing an odorant mixture or a tube containing vehicle (mineral oil). To minimize the odor background, water, but not food, was available to the mice in these chambers. After 30–40min of odor stimulation, the olfactory epithelia of these mice were collected and homogenized in Tri Reagent in preparation for the isolation of total RNA isolated as described above. This odor stimulation paradigm was chosen to match previous work on odor-sensitive mRNAs (Bennett et al. 2010).
The odorant mixture was a 1:1 mixture of mineral oil with equal volumes of the following odorants, obtained from Sigma-Aldrich unless otherwise noted: R(+)-limonene, coumarin (Santa Cruz Biological), 2-phenethylamine, eugenol, octanal, propionic acid, 2,5-dimethyl pyrazine, menthone (Fluka), (+)-carvone (Fluka), heptaldehyde, acetophenone, farnesol, 3,7-dimethyl-2,6-octadienenitrile, cineole, 1,2-propanediol, isoamyl acetate, and geranyl acetate. The odorants were selected to represent a broad range of chemical structures.
DNA microarray expression profiling
GeneChip Mouse Exon 1.0 arrays, which covers all known and many predicted exons in the mouse genome, were used to measure mRNA abundance in samples from the open and closed sides from 3 mice. Preparation of samples and the initial data reduction were done as described previously by the University of Kentucky Microarray Core facility (McIntyre et al. 2008). Data were analyzed at the transcript cluster level, which combines the signals from the known or predicted exons of each gene into a single measure. The microarray data have been deposited at Gene Expression Omnibus (Accession No. GSE49998). Statistical analysis of the transcript cluster level data was done via paired t-tests. Correction for multiple testing was done using a stepwise procedure at a false discovery rate of 5% (Benjamini and Hochberg 1995; Storey and Tibshirani 2003a,b). To eliminate background, we deleted data from probe sets whose signal intensities in all samples were less than 10% of the global mean signal.
The expression patterns of significantly different mRNAs were evaluated using data from Nickell et al. (2012). These data consist of validated probabilities of expression in individual cell types, or groups of cell types, in the olfactory epithelium. To evaluate the hypothesis that activity-dependent genes should be expressed in mature OSNs, we used the P (in) values for mature OSNs and P (in) values for the other cell type category, which contains all cell types except mature and immature OSNs. P (in) values give inclusive probabilities of expression in a cell type category. The probability of expression specific to a cell type category is a different measure, the P (sp) value.
Quantitative reverse transcription–PCR
Reverse transcription reactions used SuperScript II, random hexamer primers (Life Technologies) and 1 µg of total RNA as we have described previously (Shetty et al. 2005; Yu et al. 2005). PCR primers were designed using Primer Express (Applied Biosystems) and synthesized by Integrated DNA Technologies. Table 1 lists the primer locations. Real-time amplification was performed using an Applied Biosystems 7500 Real-Time PCR system using the Sybr Green Core Reagent Kit. Thermal Cycler conditions were 50 °C for 2min and 95 °C for 10min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1min. Normalization of these data was done using the geometric mean of 4 stable mRNAs (Actb, Gapdh, Hprt1, and Ubc) measured in each cDNA sample (Vandesompele et al. 2002). Student’s t-tests were done to assess significance for unilateral naris occlusion (paired) and Cnga2 mutant mice (unpaired) experiments. Based on expectations from the microarray data, or published data in some cases, these were 1-tailed tests. Correction for multiple testing was done upon potentially significant mRNAs (P < 0.05) using a stepwise procedure at a false discovery rate of 5% (Benjamini and Hochberg 1995; Storey and Tibshirani 2003a,b). The resulting q values for each mRNA are reported.
Table 1.
Primers used for qRT–PCR
| Gene symbol | Gene ID | Nucleotide accession # | Forward primer | Reverse primer |
|---|---|---|---|---|
| Eml5 | 319670 | NM_001081191.1 | 584–603 | 677–658 |
| Gpr158 | 241263 | NM_001004761.1 | 2524–2544 | 2964–2946 |
| Kcnn2 | 140492 | NM_080465.2 | 1497–1517 | 1598–1578 |
| Kirrel2 | 243911 | NM_172898.3 | 895–913 | 970–951 |
| Kirrel3 | 67703 | NM_026324.2 | 1101–1122 | 1180–1159 |
| Lrrc3b | 218763 | 270309120 | 510–530 | 585–565 |
| Mapk10 | 26414 | NM_001081567.1 | 6712–6731 | 6863–6843 |
| Nphs1 | 54631 | NM_019459.2 | 910–929 | 995–972 |
| Olfr199 | 404310 | 231571820 | 129–150 | 217–198 |
| Olfr855 | 258517 | 257196194 | 846–869 | 929–906 |
| Olfr869 | 258550 | 257196224 | 219–240 | 294–271 |
| Omp | 18378 | NM_011010.2 | 344–363 | 394–375 |
| Pcdh10 | 18526 | NM_001098170.1 | 1027–1048 | 1104–1083 |
| Pcp4l1 | 66425 | 157266259 | 1237–1258 | 1312–1290 |
| Plcxd3 | 239318 | NM_177355.3 | 143–163 | 250–230 |
| Ptprn | 19275 | NM_008985.2 | 1785–1805 | 1885–1867 |
| Rasgrp4 | 233046 | NM_145149.3 | 3157–3177 | 3234–3216 |
| S100A5 | 20199 | NM_011312.2 | 197–217 | 278–260 |
| Scl17a6 | 140919 | 188219543 | 822–839 | 898–877 |
| Tubb2a | 22151 | 255958180 | 1256–1276 | 1331–1311 |
Results
Mature OSN mRNAs affected by unilateral naris occlusion
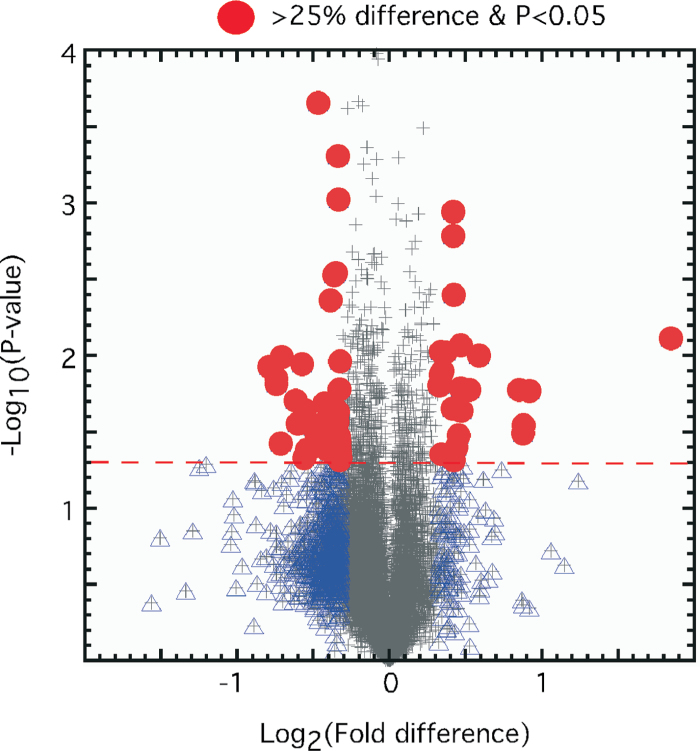
After 6 days of naris occlusion, we detected 491 mRNAs (excluding ORs) that differed in abundance between the open and closed sides (q < 0.05; paired t-tests, n = 3; Supplemental data). Of these, 162 were more abundant on the open side and 329 were more abundant on the closed side. Nearly all of these differences were small in magnitude; in fact, only 36 of these mRNAs differed more than 25% (Figure 1). This set of 36 mRNAs included 4 mRNAs already known to be positively regulated by odor stimulation: S100A5, Pcp4l1, Pcdh10, and Ptprn (Imai et al. 2006; Serizawa et al. 2006; Williams et al. 2011).
Figure 1.
Volcano plot of mRNA abundance in olfactory epithelia contralateral to naris occlusion compared with ipsilateral to naris occlusion. Duration of naris occlusion, 6 days. Triangles: P < 0.05; fold difference < 25%; (+): not significant. This figure is reproduced in color in the online version of the issue.
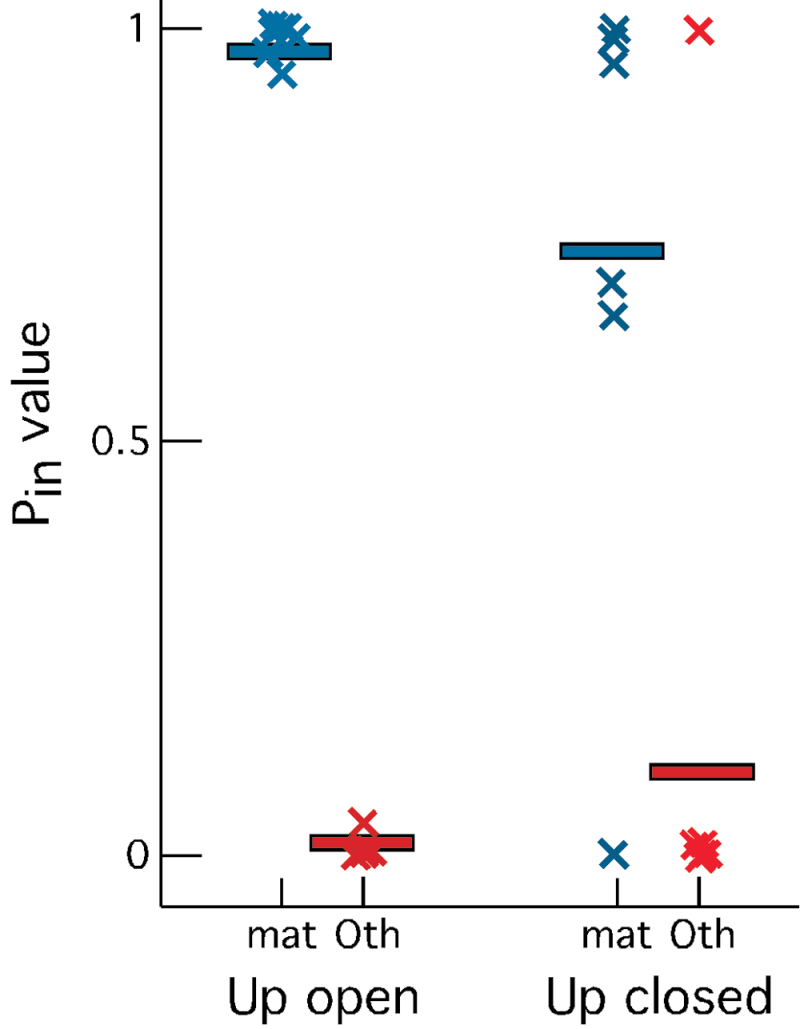
If the loss of orthonasal odor stimulation after naris occlusion is the immediate cause of changes in mRNA abundance in the olfactory epithelium, we expect these changes to arise primarily from genes expressed in mature OSNs. Recent data identifying cell type expression patterns for thousands of mRNAs detected in the olfactory epithelium (Nickell et al. 2012) provided a way to efficiently test this prediction using probabilistic estimates, termed P (in) values, of expression in the 2 relevant cell type categories: mature OSNs and the other cell type category that includes all cell types except OSNs. Transcripts more abundant on the open side than the closed side all had high P (in) mature OSN values and low P (in) other values (Figure 2). Transcripts more abundant on the closed side were also expressed primarily in mature OSNs. These distributions were unlikely to have arisen from random selection of mRNAs. For example, among all genes expressed in the olfactory epithelium, the probability of having mature OSN P (in) = 1 and other P (in) = 0 was 0.06821, but among the affected mRNAs, these extreme values occur 30% of the time. The probability that this occurs by chance is <0.0001. This observation argues that coherent responses of mature OSNs underlie most of the changes in mRNA abundance we observed.
Figure 2.
Differentially abundant mRNAs are nearly all expressed in mature OSNs and have very little expression in samples lacking OSNs. Up open: mRNAs more abundant in the epithelium behind the open naris; up closed: mRNAs more abundant in the epithelium behind the closed naris. mat: mature OSN P (in) values; Oth: other cell type category (non-OSN) P (in) values. Bars: mean P (in) values; X: individual P (in) values. This figure is reproduced in color in the online version of the issue.
Transcripts positively regulated by OSN activity
To focus on genes whose expression might be positively regulated by odor stimulation, we turned our attention to the genes whose mRNAs were more abundant on the open side (Table 2). We first evaluated the possibility that they arose from 2 potential confounding factors, loss of mature OSNs on the closed side and differential stress between the open and closed sides. Differential loss of mature OSNs should preferentially affect mRNAs specific to mature OSNs, but only 4 of these mRNAs are specific to mature OSNs (S100A5, Pcp4l1, Slc17a6, Pcdh10) instead of being expressed in both mature and immature OSNs (Nickell et al. 2012). This is consistent with our previous evidence that naris occlusion at the exact same age and for the same 6-day duration did not alter the number of mature OSNs and contrasts sharply with expression profiling results showing large changes in the abundance of many mature OSN mRNAs when OSN numbers decrease (Shetty et al. 2005; Sammeta and McClintock 2010; Heron et al. 2013). We conclude that the experimental design avoided the potential confound of loss of mature OSNs on the occluded side. Similarly, known stress-response genes in OSNs (Sammeta and McClintock 2010) were not represented among the mRNAs meeting statistical and fold-difference criteria. In fact, only 1 such mRNA, Hspa5 (BiP), exceeded the P value criterion (P = 0.0476), but it had only a 7% difference in abundance between the open and closed sides. Stress response is unlikely to have contributed to the larger changes in abundance we detected in this naris occlusion experiment.
Table 2.
mRNAs higher on the open side after unilateral naris occlusion
| Transcript cluster | Gene name | Gene ID | P value | Fold difference |
|---|---|---|---|---|
| S100a5 | S100 calcium-binding protein A5 | 20199 | 0.0077 | 3.5941 |
| Pcp4l1 | Purkinje cell protein 4-like 1 | 66425 | 0.0170 | 1.8886 |
| EG432649 | Predicted gene 5434 | 432649 | 0.0286 | 1.8409 |
| Eml5 | Echinoderm microtubule-associated protein-like 5 | 319670 | 0.0321 | 1.8364 |
| Rasgrp4 | RAS guanyl-releasing protein 4 | 233046/73833 | 0.0167 | 1.8055 |
| Slc8a1 | Solute carrier family 8 (sodium/calcium exchanger), member 1 | 20541 | 0.0099 | 1.5024 |
| Slc17a6 | Solute carrier family 17 (vesicular glutamate transporter), member 6 | 140919 | 0.0168 | 1.4388 |
| 4930547C10Rik | Riken cDNA 4930547C10 gene | 68274 | 0.0230 | 1.3848 |
| Pcdh10 | Protocadherin 10 | 18526 | 0.0164 | 1.3832 |
| Slc22a13/Slc22a13b-ps | Solute carrier family 22 (organic cation transporter), member 13/pseudone duplicate of Slc22a13 | 109280/102570 | 0.0086 | 1.3823 |
| Plcxd3 | Phosphatidylinositol-specific phospholipase C, X domain containing 3 | 239318 | 0.0332 | 1.3685 |
| EG231885/1700018F24Rik | Predicted gene 4871/Riken cDNA 1700018F24 gene | 231885/69396 | 0.0395 | 1.3564 |
| Nphs1 | Nephrosis 1, nephrin | 54631 | 0.0040 | 1.3382 |
| Cxcr3 | Chemokine (C-X-C motif) receptor 3 | 12766 | 0.0016 | 1.3344 |
| Capza3 | Capping protein (actin filament) muscle Z-line alpha 3 | 12344 | 0.0011 | 1.3344 |
| Ms4a4c/Ms4a4b | Membrane-spanning 4-domains subfamily A, member 4C/4B | 64380/60361 | 0.0221 | 1.3329 |
| Gpr158 | G protein-coupled receptor 158 | 241263 | 0.0095 | 1.2912 |
| Slco1a4/Slco1a5/Slco1a6/ EG625716 | Solute carrier organic anion transporter family, member 1a4/1a5/a6/predicted gene 6614 | 108096/625716/28254/28250 | 0.0126 | 1.2740 |
| Msgn1 | Mesogenin 1 | 56184 | 0.0442 | 1.2666 |
| Klra5/Klra6/ Klra9/Klra19 | Killer cell lectin-like receptor, subfamily A, member 5/member 6/member 9/member 19 | 16640/16637/16636/16639/93971 | 0.0094 | 1.2621 |
| Tlm | T lymphoma oncogene | 21893 | 0.0133 | 1.2619 |
| Ptprn | Protein tyrosine phosphatase receptor type N | 19275 | 0.0156 | 1.2538 |
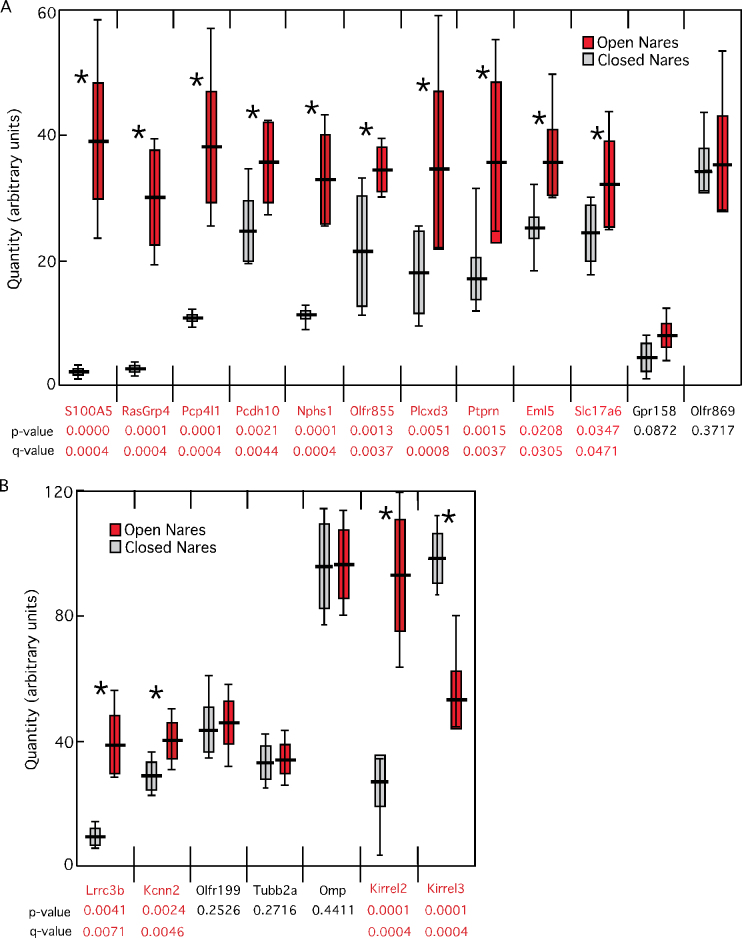
To directly confirm differential abundance, we did quantitative reverse transcription–PCR (qRT–PCR) for 12 of the mRNAs with higher microarray signals on the open side. Ten mRNAs were significantly different between the open and closed sides; only Gpr158 and an OR failed to reach significance (Figure 3A). In addition, we found that Kirrel2 and Lrrc3b mRNAs were more abundant on the open side after naris occlusion (Figure 3B), confirming previous data (Serizawa et al. 2006; Bennett et al. 2010). In contrast, Omp mRNA was unchanged (Figure 3B), additional evidence for stable numbers of mature OSNs at 6 days after naris occlusion. Lastly, we also tested Kcnn2 as a representative of mRNAs that had P <0.05 but did not meet the >25% difference criterion. Kcnn2 proved to be higher on the open side (Figure 3B). These data confirm that our significant difference criteria were valid, and perhaps even conservative, identifiers of mRNAs more abundant on the open side after 6 days of naris occlusion.
Figure 3.
Plots of qRT–PCR data from unilateral naris occlusion samples. (A) Ten mRNAs with increased mRNA abundance on the open side after naris occlusion according to microarray data also had significant differences when tested individually by qRT–PCR. (B) Tests of negative controls (Omp), positive controls (Lrrc3b, Kirrel3), a statistically significant mRNA whose fold difference was smaller than criterion (Kcnn2), and mRNAs that were increased on the closed side in the microarray experiment (Olfr199; Tubb2a). Horizontal line: mean value; solid rectangle: 1 standard deviation; vertical bars: range of data; (*): significant difference. This figure is reproduced in color in the online version of the issue.
Transcripts negatively regulated by OSN activity
The microarray experiment was not similarly effective in identifying mRNAs negatively regulated by OSN activity. The mRNAs predicted as increased on the closed side were mostly ORs, which are discussed below. We detected 14 mRNAs (not including ORs) showing increases on the closed side: BC003266, Cck, 2010005H15Rik, Fbp1, Gucy1b2, Ifitm1, Ndufa7, Ngp, Ptn, Rasl2, Rims3, Stfa3, and Tubb2a. Nearly all are expressed in OSNs (Figure 2), but none were mRNAs known to be activity dependent. We included Tubb2a in our qRT–PCR tests because it had one of the largest differences between the open and closed sides, but this test failed to confirm the microarray result (Figure 3B). In contrast, testing of the same samples readily identified differential abundance of Kirrel3, a mRNA whose abundance was previously shown to be negatively regulated by OSN activity (Serizawa et al. 2006). In the microarray experiment, Kirrel3 showed a 28% decrease but did not reach significance (P = 0.067).
ORs are highly variable mRNAs
In addition to the 36 mRNAs meeting P value and fold-difference criteria mentioned above, 24 ORs also met these same criteria. We treated differences in OR abundance cautiously because the unusual expression pattern of OR genes makes the abundance of their mRNAs in olfactory epithelium samples unusually susceptible to variation. The large numbers of OR genes in mice and the random selection of 1 OR allele by each OSN predicts that OR mRNAs will be more variable than most mRNAs. For example, while 1000 OSNs might express an OR in one mouse, the randomness of OR gene choice argues that a genetically identical sibling from the same cage could easily have 1250 OSNs expressing the same OR. Such random differences in OR gene choice could even occur between the olfactory epithelia on the 2 sides of the nasal cavity in the same animal. This source of variation is lacking in other genes.
Consistent with this reasoning, we found that 16% of the mRNAs with the largest differences in the microarray data were ORs, and 32% of the most variable mRNAs were ORs, both much greater than the representation of ORs among mRNA species detected by the microarray (6%). This finding predicts that OR mRNAs are more likely than the mRNAs of other gene families to be false positives in expression profiling data. Noting that mRNAs with increased abundance on the closed side included 19 OR mRNAs along with the 14 non-OR mRNAs, we became concerned that the mRNAs with the greatest increases on the closed side were false positives. We selected Olfr199, an OR with a large difference (50%) in abundance, for qRT–PCR as a representative of these ORs. We could not confirm a significant difference for this mRNA (Figure 3B).
To assess whether OR mRNAs more abundant on the open side were also false positives, we did qRT–PCR for the 2 that increased the most: Olfr855 and Olfr869. Unlike Olfr199 and Olfr869, Olfr855 was confirmed to be more abundant on the open side after naris occlusion (Figure 3A).
Comparison with genetic silencing
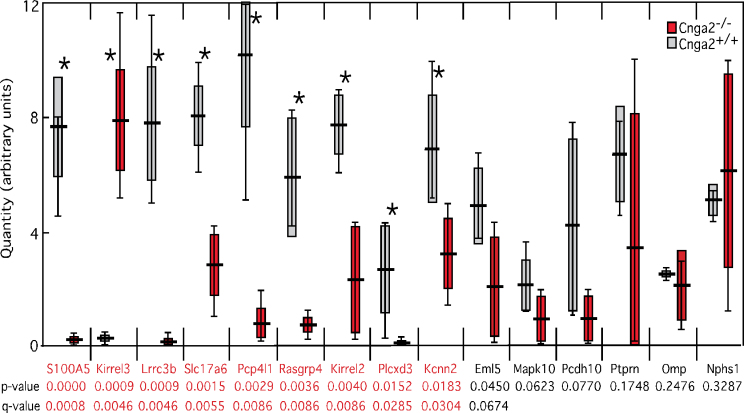
To compare these unilateral naris occlusion data to the effects of genetically silencing all OSNs, we tested 15 mRNAs in olfactory epithelium samples from Cnga2 −/− and Cnga2 +/+ female mice (4 weeks of age) by qRT–PCR (Figure 4). We found significant differences in the abundance of 9 mRNAs: S100A5, Lrrc3b, Rasgrp4, Kirrel2, Kirrel3, Pcp4l1, Kcnn2, Slc17a6, and Plcxd3. The data argue that these mRNAs require not only an increase in cAMP in OSNs but also OR-dependent electrical activity and its downstream signals. The negative control, Omp, was not different and neither were Eml5, Mapk10, Pcdh10, Ptprn, or Nphs1. Transcripts dependent on Cnga2 function are candidates for genes directly and rapidly controlled by odor stimulation.
Figure 4.
Plots of qRT–PCR data comparing olfactory epithelium samples of Cnga2 −/− and Cnga2 +/+ female mice. Horizontal line: mean value; solid rectangle: 1 standard deviation; vertical bars: range of data; (*): significant difference. This figure is reproduced in color in the online version of the issue.
Odor stimulation rapidly affected 3 mRNAs
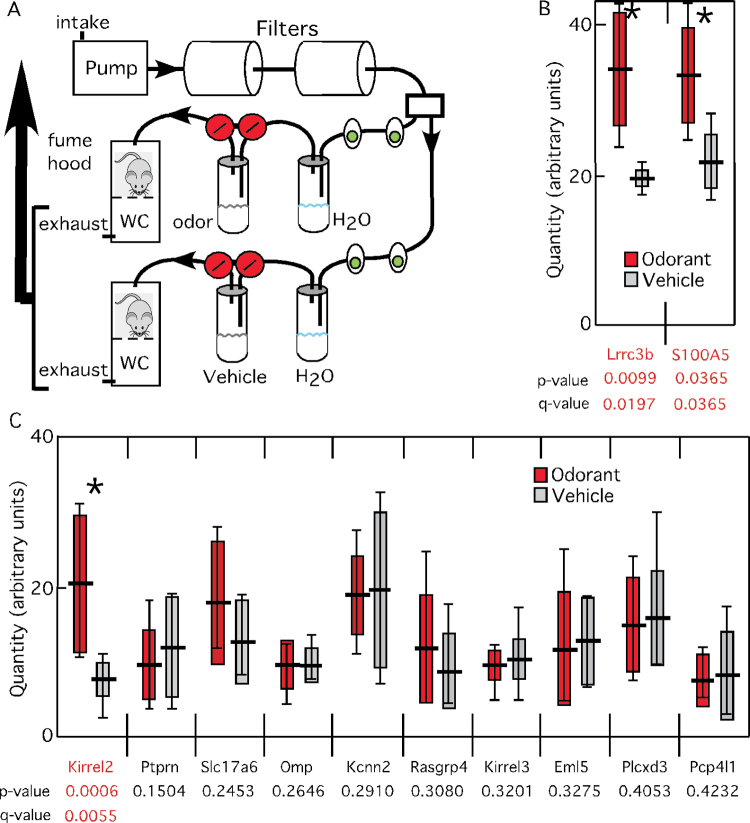
To test whether the transcripts sensitive to both naris occlusion and OR-stimulated electrical activity also respond rapidly to odor stimulation, we first tested a system for controlling the odor environment experienced by mice (Figure 5A). A pilot experiment using 4 mice to test odor-stimulated changes of S100A5 and Lrrc3b confirmed that filtered room air sufficiently reduced the odor background to allow detection of effects of odor stimulation on mRNA abundance (Figure 5B). We then tested 10 additional mRNAs (Figure 5C). Only Kirrel2 showed a significant increase. These data reveal that odor stimulation more strongly and rapidly affects the abundance of S100a5, Lrrc3b, and Kirrel2 mRNAs than other mRNAs sensitive to naris occlusion or genetic silencing of OSNs.
Figure 5.
Odor stimulation rapidly affects 3 mRNAs. (A) Filtered air chambers for controlling odor exposure. Room air is pumped through 2 activated carbon filters, split into 2 streams, hydrated, directed past tubes containing stimuli and then down over individual mice sitting on a wire mesh floor in a conical chamber designed to allow wastes and odors to be carried away to an exhaust stream captured by a fume hood. Capacity is 8 mice: 4 chambers for each of the 2 odor streams. Stimulation with odor or vehicle is done via electronically controlled pinch valves (large circles) that redirect the air stream through a tube containing either odor or vehicle. Small ovals: flow meters. (B) Initial experiment testing S100A5 and Lrrc3b (n = 4). (C) Experiment testing 10 additional mRNAs (n = 8). (B and C) horizontal line: mean value; solid rectangle: 1 standard deviation; vertical bars: range of data; (*): significant difference. This figure is reproduced in color in the online version of the issue.
Discussion
Rapidly responding versus slowly responding activity-dependent OSN mRNAs
Strongly and rapidly odor-responsive OSN mRNAs appear to be relatively few; only 3 of the 11 activity-dependent mRNAs we tested fit this description: S100A5, Lrrc3b, and Kirrel2. The rapidity of their responses argues that they are directly regulated by odor-stimulated electrical activity. In contrast, the other mRNAs affected by the absence of orthonasal stimulation or by genetic silencing of OSNs responded more slowly and usually less strongly. These more slowly responding mRNAs may be representatives of a large number of OSN mRNAs whose abundance is modestly affected by odor stimulation. Our microarray experiment identified 329 mRNAs (excluding ORs) significantly decreased by naris occlusion. We tested 11 of these mRNAs independently and confirmed differential abundance of 10 of them, indicating that the microarray screen successfully identified mRNAs positively regulated by odor stimulation in most instances. However, this experiment did not identify all activity-dependent OSN mRNAs. For example, not only were mRNAs negatively regulated by odor stimulation poorly identified, 3 well-known activity-dependent mRNAs, Lrrc3b, Kirrel2, and Kirrel3, were not among the significant mRNAs (Serizawa et al. 2006; Bennett et al. 2010). Lrrc3b and Kirrel3 mRNAs had relatively large average differences between open and closed sides of 53% and −28%, respectively, but had P values of 0.063 and 0.067, respectively. The Kirrel2 transcript cluster did not have signal above background and could not be evaluated in the microarray experiment. While not a complete set of activity-dependent OSN mRNAs, our data are consistent with previous expression profiling data from longer durations of naris occlusion (Coppola and Waggener 2012; Santoro and Dulac 2012) because they continue to indicate that hundreds of OSN mRNAs may be modestly sensitive to odor stimulation.
The slowly responding mRNAs, albeit less obviously linked to odor stimulation, were still likely to be responding to changes in odor stimulation. We were able to exclude the 2 potentially confounding factors of greatest concern. First, activity-dependent effects occurred in the absence of any change in the number of OSNs. Second, the detection of several mRNAs (S100A5, Lrrc3b, Kirrel2, Rasgrp4, Pcp4l, Plcxd3, Kcnn2, and Slc17a6) that responded to both naris occlusion, where exposure to ambient stressors differs greatly between the nares, and genetic silencing where these stressors do not differ between the nares, argues that stress or damage cannot explain our results. We conclude that S100A5, Lrrc3b, Kirrel2, Rasgrp4, Pcp4l, Plcxd3, Kcnn2, and Slc17a6 are activity-dependent mRNAs sensitive to odor stimulation. The activity-dependent mRNAs that were not affected by genetic silencing of OSNs—Eml5, Nphs1, Ptprn, and Pcdh10—may also be regulated by odor stimulation, albeit through mechanisms not requiring the cyclic nucleotide-gated cation channel.
How odor stimulation contributes to regulation of 2 sets of activity-dependent mRNAs with different temporal profiles is as yet unclear. One possible explanation is that slowly responding mRNAs require integration of multiple factors, only one of which is odor stimulation, and that this is a slow process. Alternatively, odor stimulation might regulate these mRNAs indirectly. For example, activity-dependent feedback from the olfactory bulb could regulate them, thereby accounting for the slow response (Schwob et al. 1992). These hypotheses, which are not mutually exclusive, are worthy targets for future experiments.
Activity-dependent mRNAs encoding axonal and calcium-binding proteins
Of the activity-dependent mRNAs we identified and confirmed, 6 were mRNAs whose activity dependence was previously unknown: Eml5, Kcnn2, Nphs1, Plcxd3, Rasgrp4, and Slc17a6. Nphs1 belongs to an immunoglobulin family of cell adhesion proteins and helps form the glomerular filtration barrier in the kidney (Ristola and Lehtonen 2014). What it might be doing in OSNs is difficult to predict. Plcxd3 encodes a protein about which relatively little is known. It has catalytic activity against phosphatidylinositol, is expressed abundantly in neural tissues, and when overexpressed in cultured cell lines, locates to cytoplasmic organelles (Gellatly et al. 2012). In contrast, the functions of the proteins encoded by the other newly identified activity-dependent mRNAs add further support to evidence that odor stimulation tends to regulate mRNAs encoding proteins with roles in axonal or synaptic function and maintenance and in calcium regulation of as yet undefined biochemical events (Imai et al. 2009; Bennett et al. 2010). S100A5, Kirrel2, Kirrel3, Pcdh10, Slc17a6, Ptprn, and Eml5 proteins are found in either axons or secretory vesicles (Schäfer et al. 2000; O’Connor et al. 2004; Serizawa et al. 2006; Uemura et al. 2007; Takeyama et al. 2009; Suckale and Solimena 2010; Allen et al. 2011; Williams et al. 2011). Kirrel2 and Kirrel3 are important for the fasciculation and targeting of OSN axons. Slc17a6 encodes Vglut2, a vesicular glutamate transporter characteristic of the synaptic vesicles of glutamatergic neurons such as OSNs. Pcdh10 regulates the growth of developing striatal axons. Ptprn is a receptor protein tyrosine phosphatase known to be involved in axon guidance, for example, in developing retinal ganglion cells (Ensslen-Craig and Brady-Kalnay 2005). S100A5 is a calcium- and zinc-binding protein of unknown function that is abundant in OSN axons. Eml5, which belongs to a family of proteins that regulate microtubule dynamics, is found in axons in several regions of the brain. These data are consistent with several other known activity-dependent mRNAs that encode axonal or synaptic proteins (Imai et al. 2006; Kaneko-Goto et al. 2008; Imai et al. 2009). Another commonality among the activity-dependent OSN mRNAs is the ability of several of their encoded proteins to bind calcium directly or to interact with calcium-binding proteins. To this set of proteins, we can now add Rasgrp4 and Kcnn2. Kcnn2 is a component of the SK2 calcium-activated potassium channel. Rasgrp4 is a calcium- and diacylglycerol-stimulated guanine nucleotide-releasing factor for Ras family monomeric G-proteins and has also been implicated in activation of phosphatidylinositol-3-kinase γ (Li et al. 2003; Suire et al. 2012).
Activity-dependent gene expression and the latency to OSN loss
By making inactive OSNs more susceptible to apoptosis (Watt et al. 2004; Sakano 2010; Santoro and Dulac 2012), an animal may be setting the stage for tuning its OSN population to better match its odor environment. If so, then balancing the period of survival of unstimulated OSNs with the likelihood of reencountering an odorant agonist would be critical. Activity-dependent genes could act as a molecular memory of odor stimulation history and help set the survival period. At 6 days after naris occlusion, we saw evidence of a broad pattern of small changes in OSN mRNA abundance, but only one piece of evidence that changes in tuning might have begun; this being the differential abundance of Olfr855. By 3–4 weeks of naris occlusion, however, the relative abundances of numerous OR mRNAs have changed due to differential OSN survival (Coppola and Waggener 2012; Santoro and Dulac 2012; Zhao et al. 2013). Consistent with changes in the number of OSNs only after more than a week of naris occlusion (Farbman et al. 1988; Brunjes and Shurling 2003; Suh et al. 2006), our data also indicate that the latency period for OSN survival after the cessation of odor stimulation is about a week.
Supplementary material
Supplementary material can be found at http://www.chemse.oxfordjournals.org/
Funding
This work was supported by grants from the National Institute on Deafness and Other Communication Disorders [R01 DC002736 to T.S.M. and F32 DC011427 to P.M.H.]; the National Institute of General Medical Sciences [5P20GM103436 to A.J.S.]; and the National Center for Advancing Translational Sciences [UL1TR000117 to A.J.S.].
Supplementary Material
Acknowledgements
We thank W. Titlow for technical support and Dr D. Coppola for comments on a draft of the manuscript.
References
- Allen D, Bond CT, Lujan R, Ballesteros-Merino C, Lin MT, Wang K, Klett N, Watanabe M, Shigemoto R, Stackman RW, Jr, et al. 2011. The SK2-long isoform directs synaptic localization and function of SK2-containing channels. Nature Neurosci. 14:744–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 57:289–300 [Google Scholar]
- Bennett MK, Kulaga HM, Reed RR. 2010. Odor-evoked gene regulation and visualization in olfactory receptor neurons. Mol Cell Neurosci. 43:353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunjes P, Shurling DC. 2003. Cell death in the nasal septum of normal and naris-occluded rats. Brain Res Dev Brain Res. 146:25–28 [DOI] [PubMed] [Google Scholar]
- Col JA, Matsuo T, Storm DR, Rodriguez I. 2007. Adenylyl cyclase-dependent axonal targeting in the olfactory system. Development. 134:2481–2489 [DOI] [PubMed] [Google Scholar]
- Coppola DM. 2012. Studies of olfactory system neural plasticity: the contribution of the unilateral naris occlusion technique. Neural Plasticity. 2012:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola DM, Waggener CT. 2012. The effects of unilateral naris occlusion on gene expression profiles in mouse olfactory mucosa. J Mol Neurosci. 47:604–618 [DOI] [PubMed] [Google Scholar]
- Ensslen-Craig SE, Brady-Kalnay SM. 2005. PTP mu expression and catalytic activity are required for PTP mu-mediated neurite outgrowth and repulsion. Mol Cell Neurosci. 28:177–188 [DOI] [PubMed] [Google Scholar]
- Farbman AI, Brunjes PC, Rentfro L, Michas J, Ritz S. 1988. The effect of unilateral naris occlusion on cell dynamics in the developing rat olfactory epithelium. J Neurosci. 8:3290–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellatly SA, Kalujnaia S, Cramb G. 2012. Cloning, tissue distribution and sub-cellular localisation of phospholipase C X-domain containing protein (PLCXD) isoforms. Biochem Biophys Res Commun. 424:651–656 [DOI] [PubMed] [Google Scholar]
- Heron PM, Stromberg AJ, Breheny P, McClintock TS. 2013. Molecular events in the cell types of the olfactory epithelium during adult neurogenesis. Mol Brain. 6:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds JW, Hinds PL, McNelly NA. 1984. An autoradiographic study of the mouse olfactory epithelium: evidence for long-lived receptors. Anat Rec. 210:375–383 [DOI] [PubMed] [Google Scholar]
- Hudson R. 1999. From molecule to mind: the role of experience in shaping olfactory function. J Comp Physiol A. 185:297–304 [DOI] [PubMed] [Google Scholar]
- Imai T, Suzuki M, Sakano H. 2006. Odorant receptor-derived cAMP signals direct axonal targeting. Science. 314:657–661 [DOI] [PubMed] [Google Scholar]
- Imai T, Yamazaki T, Kobayakawa R, Kobayakawa K, Abe T, Suzuki M, Sakano H. 2009. Pre-target axon sorting establishes the neural map topography. Science. 325:585–590 [DOI] [PubMed] [Google Scholar]
- Jones SV, Choi DC, Davis M, Ressler KJ. 2008. Learning-dependent structural plasticity in the adult olfactory pathway. J Neurosci. 28:13106–13111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko-Goto T, Yoshihara S, Miyazaki H, Yoshihara Y. 2008. BIG-2 mediates olfactory axon convergence to target glomeruli. Neuron. 57:834–846 [DOI] [PubMed] [Google Scholar]
- Kelemen G. 1947. The junction of the nasal cavity and the pharyngeal tube in the rat. Arch Otolaryngol. 45:159–168 [DOI] [PubMed] [Google Scholar]
- Li L, Yang Y, Stevens RL. 2003. RasGRP4 regulates the expression of prostaglandin D2 in human and rat mast cell lines. J Biol Chem. 278:4725–4729 [DOI] [PubMed] [Google Scholar]
- Maruniak JA, Lin PJ, Henegar JR. 1989. Effects of unilateral naris closure on the olfactory epithelia of adult mice. Brain Res. 490:212–218 [DOI] [PubMed] [Google Scholar]
- McIntyre JC, Bose SC, Stromberg AJ, McClintock TS. 2008. Emx2 stimulates odorant receptor gene expression. Chem Senses. 33:825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell MD, Breheny P, Stromberg AJ, McClintock TS. 2012. Genomics of mature and immature olfactory sensory neurons. J Comp Neurol. 520:2608–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor V, Houtman SH, De Zeeuw CI, Bliss TV, French PJ. 2004. Eml5, a novel WD40 domain protein expressed in rat brain. Gene. 336:127–137 [DOI] [PubMed] [Google Scholar]
- Öztokatli H, Hörnberg M, Berghard A, Bohm S. 2012. Retinoic acid receptor and CNGA2 channel signaling are part of a regulatory feedback loop controlling axonal convergence and survival of olfactory sensory neurons. FASEB J. 26:617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristola M, Lehtonen S. 2014. Functions of the podocyte proteins nephrin and Neph3 and the transcriptional regulation of their genes. Clin Sci (Lond). 126:315–328 [DOI] [PubMed] [Google Scholar]
- Sakano H. 2010. Neural map formation in the mouse olfactory system. Neuron. 67:530–542 [DOI] [PubMed] [Google Scholar]
- Sammeta N, McClintock TS. 2010. Chemical stress induces the unfolded protein response in olfactory sensory neurons. J Comp Neurol. 518:1825–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Dulac C. 2012. The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. Elife. 1:e00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer BW, Fritschy JM, Murmann P, Troxler H, Durussel I, Heizmann CW, Cox JA. 2000. Brain S100A5 is a novel calcium-, zinc-, and copper ion-binding protein of the EF-hand superfamily. J Biol Chem. 275:30623–30630 [DOI] [PubMed] [Google Scholar]
- Schwob JE, Szumowski KE, Stasky AA. 1992. Olfactory sensory neurons are trophically dependent on the olfactory bulb for their prolonged survival. J Neurosci. 12:3896–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Takeuchi H, Yamagishi Y, Suzuki M, Sakano H. 2006. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell. 127:1057–1069 [DOI] [PubMed] [Google Scholar]
- Shetty RS, Bose SC, Nickell MD, McIntyre JC, Hardin DH, Harris AM, McClintock TS. 2005. Transcriptional changes during neuronal death and replacement in the olfactory epithelium. Mol Cell Neurosci. 30:90–107 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003a. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 100:9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003b. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol. 224:149–157 [DOI] [PubMed] [Google Scholar]
- Suckale J, Solimena M. 2010. The insulin secretory granule as a signaling hub. Trends Endocrinol Metab. 21:599–609 [DOI] [PubMed] [Google Scholar]
- Suh KS, Kim SY, Bae YC, Ronnett GV, Moon C. 2006. Effects of unilateral naris occlusion on the olfactory epithelium of adult mice. Neuroreport. 17:1139–1142 [DOI] [PubMed] [Google Scholar]
- Suire S, Lecureuil C, Anderson KE, Damoulakis G, Niewczas I, Davidson K, Guillou H, Pan D, Jonathan C, Phillip TH, et al. 2012. GPCR activation of Ras and PI3Kc in neutrophils depends on PLCb2/b3 and the RasGEF RasGRP4. EMBO J. 31:3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeyama N, Ano Y, Wu G, Kubota N, Saeki K, Sakudo A, Momotani E, Sugiura K, Yukawa M, Onodera T. 2009. Localization of insulinoma associated protein 2, IA-2 in mouse neuroendocrine tissues using two novel monoclonal antibodies. Life Sci. 84:678–687 [DOI] [PubMed] [Google Scholar]
- Uemura M, Nakao S, Suzuki ST, Takeichi M, Hirano S. 2007. OL-Protocadherin is essential for growth of striatal axons and thalamocortical projections. Nat Neurosci. 10:1151–1159 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt WC, Sakano H, Lee ZY, Reusch JE, Trinh K, Storm DR. 2004. Odorant stimulation enhances survival of olfactory sensory neurons via MAPK and CREB. Neuron. 41:955–967 [DOI] [PubMed] [Google Scholar]
- Williams EO, Sickles HM, Dooley AL, Palumbos S, Bisogni AJ, Lin DM. 2011. Delta Protocadherin 10 is Regulated by Activity in the Mouse Main Olfactory System. Front Neural Circuits. 5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TT, McIntyre JC, Bose SC, Hardin D, Owen MC, McClintock TS. 2005. Differentially expressed transcripts from phenotypically identified olfactory sensory neurons. J Comp Neurol. 483:251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Reed RR. 2001. X inactivation of the OCNC1 channel gene reveals a role for activity-dependent competition in the olfactory system. Cell. 104:651–660 [DOI] [PubMed] [Google Scholar]
- Zhao S, Tian H, Ma L, Yuan Y, Yu CR, Ma M. 2013. Activity-dependent modulation of odorant receptor gene expression in the mouse olfactory epithelium. PLoS One. 8:e69862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou DJ, Feinstein P, Rivers AL, Mathews GA, Kim A, Greer CA, Mombaerts P, Firestein S. 2004. Postnatal refinement of peripheral olfactory projections. Science. 304:1976–1979 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.