Abstract
Aim: The present study was conducted to define the relationship between the anti-aging effect of ubiquinol-10 supplementation and mitochondrial activation in senescence-accelerated mouse prone 1 (SAMP1) mice. Results: Here, we report that dietary supplementation with ubiquinol-10 prevents age-related decreases in the expression of sirtuin gene family members, which results in the activation of peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), a major factor that controls mitochondrial biogenesis and respiration, as well as superoxide dismutase 2 (SOD2) and isocitrate dehydrogenase 2 (IDH2), which are major mitochondrial antioxidant enzymes. Ubiquinol-10 supplementation can also increase mitochondrial complex I activity and decrease levels of oxidative stress markers, including protein carbonyls, apurinic/apyrimidinic sites, malondialdehydes, and increase the reduced glutathione/oxidized glutathione ratio. Furthermore, ubiquinol-10 may activate Sirt1 and PGC-1α by increasing cyclic adenosine monophosphate (cAMP) levels that, in turn, activate cAMP response element-binding protein (CREB) and AMP-activated protein kinase (AMPK). Innovation and Conclusion: These results show that ubiquinol-10 may enhance mitochondrial activity by increasing levels of SIRT1, PGC-1α, and SIRT3 that slow the rate of age-related hearing loss and protect against the progression of aging and symptoms of age-related diseases. Antioxid. Redox Signal. 20, 2606–2620
Introduction
Coenzyme Q10 (CoQ10) is a well-known essential electron transporter in complexes I (NADH–ubiquinone oxidoreductase, EC 1.6.5.3), II (succinate–ubiquinone oxidoreductase, EC 1.3.5.1), and III (ubiquinone–cytochrome c oxidoreductase, EC 1.10.2.2) of the mitochondrial respiratory chain (RC) (24, 26). Ubiquinone-10 (oxidized form of CoQ10) is enzymatically reduced to ubiquinol-10 (the reduced form of CoQ10, 2,3-dimethoxy-5-methyl-6-decaprenyl-1,4-benzoquinol) (32, 67), which acts as a fat-soluble antioxidant that effectively protects lipid membranes and lipoproteins from oxidative damage and has beneficial effects in preventing DNA damage (53). In many tissues, ubiquinol-10 biosynthesis and levels decrease with age, suggesting that ubiquinol-10 may have possible anti-aging effects (1). Furthermore, nuclear DNA (nDNA) damage in rat blood lymphocytes, mitochondrial DNA (mtDNA) damage in rat heart, and mitochondria deletion and age-related molecular changes in rat brain mitochondria were suppressed by ubiquinone-10 (39, 45, 46). These findings may support the possibility that ubiquinol-10 has anti-aging effects, because ubiquinone-10 is readily reduced to ubiquinol-10 after dietary uptake (33, 63) and a ratio of ubiquinol-10 to total CoQ10 of more than 85% is maintained in the plasma (40, 70). Dietary supplementation with ubiquinol-10 produces beneficial effects for several age-related diseases, such as cardiovascular disease, diabetes, age-related hearing loss (AHL), and Parkinson's disease (50, 61, 62, 68). Our recent study identified many ubiquinol-10-sensitive genes that are regulated by peroxisome proliferator-activated receptor-alpha (PPARα) in the liver (54).
Innovation.
In this study, we observed the decelerated age-related accumulation of oxidative damage in ubiquinol-10-supplemented mice with accelerated senescence. Our major finding is that ubiquinol-10 induces a pathway that activates SIRT1, SIRT3, peroxisome proliferator-activated receptor γ coactivator 1α, and mitochondrial function. Activation of this pathway modulates oxidative damage in the liver and inner ear by enhancing the activity of mitochondrial antioxidant defense systems, which results in significantly delayed senescence and age-related hearing loss. These findings suggest that pharmaceutical treatments which include ubiquinol-10 supplementation to activate sirtuin pathways could prevent mitochondrial decay associated with aging and decelerate age-related diseases by increasing oxidative stress resistance.
Senescence-accelerated mouse (SAM) strains provide a unique model system for studying the aging process in higher organisms. SAM strains include the accelerated senescence-prone senescence-accelerated mouse prone (SAMP) strains that have markedly shorter life spans and exhibit early signs of aging (64). SAMP1, a strain in the SAMP series, exhibits accelerated progression of many age-associated degenerative diseases such as senile amyloidosis, impaired immune response, hyperinflation of the lungs, and AHL (31, 65). AHL is a universal feature of mammalian aging and is the most common sensory disorder in the elderly (62). SAMP1 mice show age-related hearing impairment, which is closely related to chronological age, and also manifestations of accelerated senescence (51, 52). These SAMP strains also showed mitochondrial dysfunction and a higher oxidative stress status (11). Therefore, as a model of presbycusis, SAMP1 mice should prove useful. Here we examined whether dietary supplementation with ubiquinol-10 could exert beneficial effects against accelerated senescence and age-related diseases in SAMP1 mice and elucidate the mechanism of the anti-aging effect of ubiquinol-10.
Aging and age-related diseases in higher organisms is a complex process that is likely controlled by a combination of many different genetic, environmental, nutritional, and pathologic factors. While the specific role of oxidative stress in aging and development of age-related diseases is an area of active investigation, the exact mechanisms that may define this complex relationship are unclear (69). A better understanding of this relationship may help identify useful biomarkers of oxidative stress, which can be objectively measured as indicators of normal and pathologic processes that result in age-related diseases. Cell aging is thought to be due to cumulative cellular and genomic damage that results in permanent cell-cycle arrest, apoptosis, or senescence (49). A major source of cellular damage is reactive oxygen species (ROS), which are mainly generated at complexes I and III of the cellular RC (2). ROS generation was estimated to represent 0.1%–2% of total oxygen consumption (15, 35), and an early study suggested that its generation rate is thought to increase with age due to the accumulation of damaged mitochondria in a vicious cycle in which ROS-induced mitochondrial impairment results in increased ROS production, which, in turn, leads to further mitochondrial damage in aging (10, 14). However, recent research revealed that mitochondria ROS generation does not need to increase with age to cause aging (3). Nutrient intake and numerous genetic mutations affect the rate of aging with a concomitant alteration of mitochondrial metabolism and ROS accumulation, suggesting that mitochondrial homeostasis can be regulated during the aging process. However, recent research revealed that besides the harmful effects of ROS, ROS play an important physiological role as “redox messengers” in intracellular signaling and regulation by modulating fundamental cell death and cell survival processes, including apoptosis and autophagy (28). In addition, since many exceptions and contradictory findings to the ROS accumulation theory have been reported recently (6, 18), further research to understand the role of oxidative damage and mitochondria dynamics in the aging process is needed.
Metabolic pathways are co-ordinated through reversible acetylation of metabolic enzymes in response to nutrient availability and calorie intake (59). The sirtuin family has emerged as key regulators of the nutrient-sensitive metabolic regulatory circuit. Sir2 plays a direct role in the anti-aging effects of calorie restriction (CR) in yeast (21), while overexpression of SIRT1, the mammalian Sir2 homolog, was reported to protect mice from age-related phenotypes such as type 2 diabetes (2), cancer (19), and Alzheimer's disease (13). However, there are inconsistent results as to whether SIRT1 activity extends the lifespan of Caenorhabditis elegans (19). SIRT3 regulates the global acetylation landscape of mitochondrial proteins, and SIRT3-initiated metabolic adaptations enhance mitochondrial management of ROS. SIRT3 increases the activity of antioxidants such as superoxide dismutase 2 (SOD2) and reduced glutathione (GSH), and promotes ROS scavenging (44, 66). A recent study on AHL in mice clearly showed that ROS production could diminish the effects of sirtuins (62). Mice sustain cumulative oxidative damage to hair cells and spiral ganglia neurons of the inner ear cochlea, resulting in hearing loss in aged mice. CR induced SIRT3 protein expression in mice, so it is likely that this sirtuin mediates ROS management, mitochondrial integrity, and sensory function, at least in those neurons which govern hearing.
Our recent study indicated that ubiquinol-10 up-regulated genes are downstream of PPARα (54). PPARα is found mainly in oxidative tissues such as cardiac muscle, skeletal muscle, and liver, and its activation is known to be regulated by peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) (12), which controls mitochondrial biogenesis and respiration in tissues. Thus, we hypothesized that an important mediator of the metabolic effects of ubiquinol-10 may be PGC-1α activation. Since SIRT1 is known to deacetylate and activate PGC-1α, we further hypothesized that ubiquinol-10 may increase the activity of sirtuin families and PGC-1α in mice in a way similar to CR, and, in turn, reduce the oxidative damage that can promote AHL.
In this report, we show that ubiquinol-10 can prevent age-related oxidative stress and activate mitochondrial function by inducing sirtuin and Pgc-1α gene expression in SAMP1 mice, which protects against the symptoms of age-related diseases.
Results
Ubiquinol-10 reduced oxidative damage in SAMP1 mice
To investigate whether ubiquinol-10 plays a role in senescence, we conducted a study using SAMP1 mice. We supplemented the diets of young, middle, and old SAMP1 mice with ubiquinol-10, starting at 1, 7, and 13 months of age, respectively. Mice supplemented with ubiquinol-10 from a young age showed no significant changes in body weight, but a significant decrease was observed in mice that were supplemented with ubiquinol-10 beginning in middle or old age as compared with mice fed the control diet (Supplementary Fig. S1A, B; Supplementary Data are available online at www.liebertpub.com/ars). Similar to the results from our previous study (74), in terms of the degree of senescence, a significant deceleration was observed for mice fed ubiquinol-10 from a young age as compared with mice that received the control diet (Supplementary Fig. S1C, D). A significant decrease in food intake was observed only in mice started with ubiquinol-10 during middle age (7–9 months) (Supplementary Fig. S1E). The life spans of mice fed either a control diet or one supplemented with ubiquinol-10 from a young age, during middle and old age were similar to control animals (Supplementary Fig. S1F).
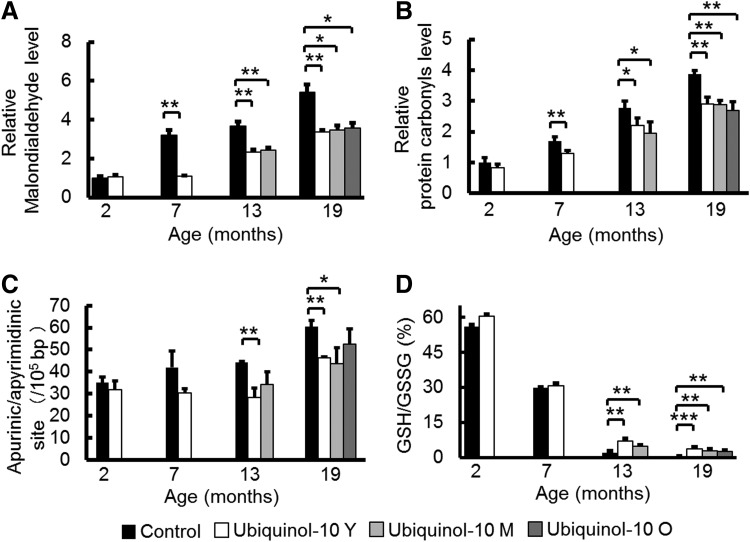
Since we reported that ubiquinol-10 showed its most distinct effects on gene expression in the liver, to elucidate the mechanism of decelerated aging effects, we first measured oxidative damage to lipids, protein, and DNA in SAMP1 mouse livers (54). We evaluated malondialdehyde levels as a measure of oxidative damage to lipids in the liver of control and ubiquinol-10 diet mice at 2, 7, 13, and 19 months and observed age-related increases in lipid damage in control diet mice that were suppressed by ubiquinol-10 supplementation (Fig. 1A). The same result was found for oxidative damage to proteins (e.g., protein carbonyls) (Fig. 1B) and DNA as assessed by apurinic/apyrimidinic (AP) sites (Fig. 1C). Glutathione acts as the major antioxidant in cells, with NADPH-dependent glutathione reductase regenerating GSH from oxidized glutathione (GSSG). We measured the ratio of GSH:GSSG in the livers of control and ubiquinol-10 diet SAMP1 mice at 2, 7, 13, and 19 months of age and found that age-related decreases in the ratios of GSH:GSSG could be suppressed in mice supplemented with ubiquinol-10 from a young age, as well as during middle and old age (Fig. 1D). These results showed that ubiquinol-10 reduces oxidative damage to lipids, proteins, and DNA in SAMP1 mice and that ubiquinol-10 may promote a more reductive environment for liver mitochondria.
FIG. 1.
Ubiquinol-10 decelerated the increase in oxidative stress associated with aging in SAMP1 mice. (A, B) Western blot analysis of oxidative damage to lipids (malondialdehyde) (A) and proteins (protein carbonyls) (B) in livers from 2-, 7-, 13-, and 19-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet beginning in young, middle, or old age (n=3–5). (C) Oxidative damage to DNA (AP sites) was measured in livers from 2-, 7-, 13-, and 19-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet beginning during young, middle, or old age (n=3–5). (D) Ratios of liver GSH:GSSG were measured (n=3–5). *p<0.05; **p<0.01; ***p<0.001. AP, apurinic/apyrimidinic; GSH, reduced glutathione; GSSG, oxidized glutathione; SAMP1, senescence-accelerated mouse prone 1.
To confirm the accumulation of CoQ in the liver, homogenate, cytosol, and mitochondrial concentrations of total coenzyme Q9 (CoQ9) (dominant CoQ form in mice) and total CoQ10 (the sum of the concentrations of the reduced and oxidized forms, respectively) were determined (23). Ubiquinol-10 supplementation in SAMP1 mice produced different changes in the liver concentrations of CoQ10 and CoQ9 from 7 months of age. Supplementation with ubiquinol-10 for 7 months significantly elevated homogenate liver, cytosol, and mitochondrial concentrations of CoQ10. However, there were no significant increases in the CoQ9 concentrations in SAMP1 mice supplemented with ubiquinol-10 (Supplementary Fig. S2).
Ubiquinol-10 suppressed age-related decreases in the expression of genes and proteins related to mitochondrial function
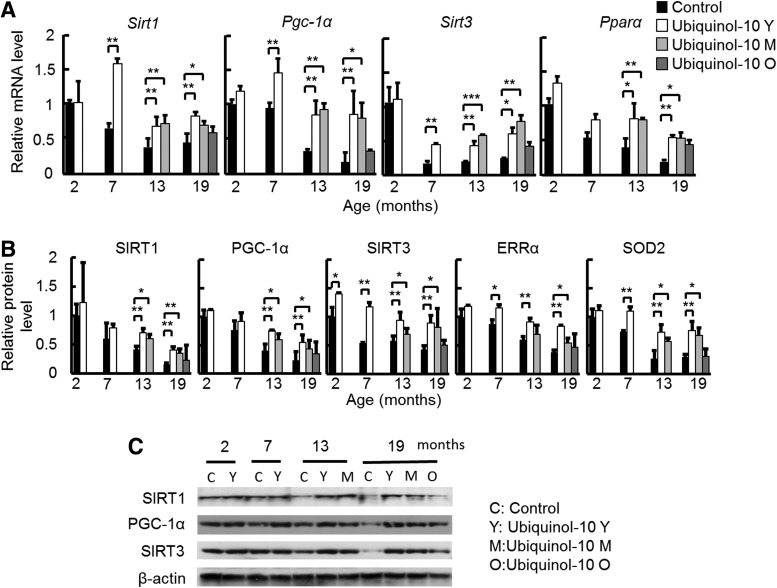
CoQ10 is an electron carrier from enzyme complex I and complex II to complex III in mitochondria, while ubiquinol-10 may enhance the antioxidant activity of mitochondria. Therefore, we hypothesize that ubiquinol-10 supplementation might increase mitochondrial activity. To test this possibility, we extracted total RNA and proteins from the livers of 2-, 7-, 13-, and 19-month-old mice and examined age-related changes in sirtuin family members and Pgc-1α gene and protein expression levels by real-time polymerase chain reaction (PCR) and western blot, respectively. We found an age-related decrease in the expression of Sirt1, Sirt3, and Pgc-1α mRNA, and ubiquinol-10 supplementation in mice from a young, middle, and old age suppressed these age-related decreases (Fig. 2A). To confirm the results from our previous study (54), we measured Pparα mRNA expression and found similar increases in expression. In addition, western blot analyses using antibodies to SIRT1, PGC-1α, SIRT3, estrogen-related receptor alpha (ERRα), and SOD2 showed that the levels of these mitochondria-related proteins also decreased with age, and these decreases were significantly reduced for young and middle age groups after ubiquinol-10 supplementation (Fig. 2B, C). Thus, these data provide strong biochemical evidence that ubiquinol-10 suppresses age-related decreases in the expression of genes and proteins related to mitochondrial functions.
FIG. 2.
Ubiquinol-10 decelerated the age-related decline in the expression of genes and proteins related to mitochondrial function. (A) Real-time PCR analyses of Sirt1, Pgc-1α, Sirt3, and Pparα gene expression levels in livers from 2-, 7-, 13-, and 19-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet (n=3–5). (B) Western blot analyses of SIRT1, PGC-1α, SIRT3, ERRα, and SOD2 protein expression levels in livers from 2-, 7-, 13-, and 19-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet (n=3–5). (C) A typical primary blot of SIRT1, SIRT3, and PGC-1α in liver samples is shown. *p<0.05; **p<0.01; ***p<0.001. ERRα, estrogen-related receptor alpha; PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1α; SOD2, superoxide dismutase 2; PCR, polymerase chain reaction.
Supplementation of ubiquinol-10 decelerates senescence and suppresses AHL progression
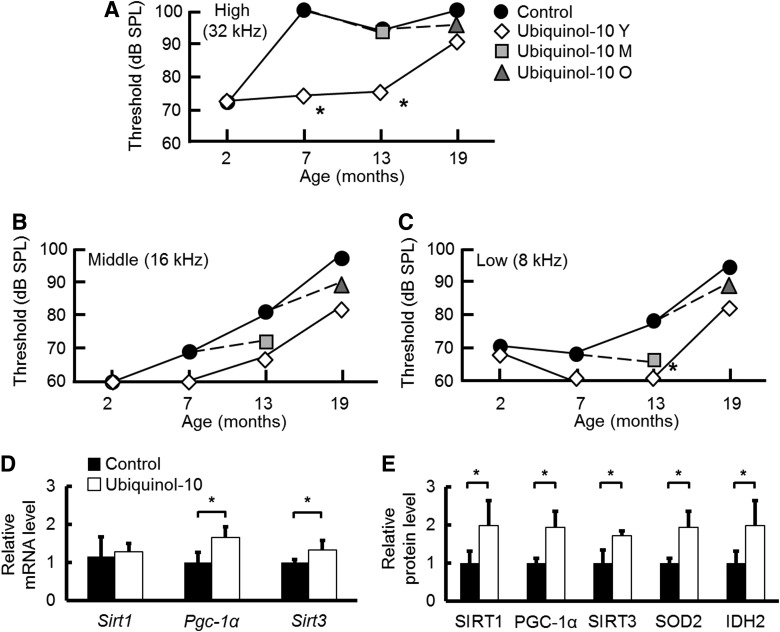
SAMP1 mice exhibit accelerated progression of many age-associated degenerative diseases, including AHL. To investigate whether ubiquinol-10 plays a role in AHL, we analyzed the auditory brainstem response (ABR) in 2-, 7-, 13-, and 19-month-old mice. For control mice, we observed an age-related increase in the ABR hearing thresholds at high (32 kHz), middle (16 kHz), and low (8 kHz) frequencies (Fig. 3A–C), while mice supplemented with ubiquinol-10 from a young age had hearing thresholds at high frequencies that were significantly lower at 7 and 13 months (Fig. 3A). For middle (16 kHz) and low (8 kHz) frequencies, age-associated hearing losses were suppressed by ubiquinol-10 supplementation (Fig. 3B, C). These results demonstrate that ubiquinol-10 plays an essential role in AHL in SAMP1 mice.
FIG. 3.
Ubiquinol-10 delayed the progression of AHL in SAMP1 mice. (A–C) ABR hearing thresholds were measured at high (32 kHz) (A), middle (16 kHz) (B), and low (8 kHz) (C) frequencies at 2, 7, 13, and 19 months of age for SAMP1 mice fed control (●) or ubiquinol-10-supplemented diets started from young (⋄), middle age (gray square) and old age (gray triangle) (n=2–5). (D) Real-time PCR analysis of Sirt1, Pgc-1α, and Sirt3 mRNA expression levels in the cochleae from 7-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet (n=5). *p<0.05. (E) Western blot analysis of SIRT1, PGC-1α, SIRT3, SOD2, and IDH2 protein levels in cochleae from 10-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet (n=5). *p<0.05. ABR, auditory brainstem response; AHL, age-related hearing loss; IDH2, isocitrate dehydrogenase 2.
We confirmed that aging resulted in increased ABR hearing thresholds at high (32 kHz) frequencies in 7-month-old SAMP1 mice fed a control diet (Fig. 3A), indicating that these mice displayed hearing loss. Ubiquinol-10 suppressed the exacerbation of AHL in mice fed a supplemented diet. Since ubiquinol-10 was shown to regulate mitochondrial metabolic activity in the liver, we hypothesize that ubiquinol-10 may similarly regulate mitochondrial metabolic activity in the cochleae. By determining the expression of mitochondria-related genes and proteins in cochleae, we could show that, in agreement with the ABR test and liver results, Sirt3 and Pgc-1α mRNA and protein expression was significantly increased after ubiquinol-10 supplementation (Fig. 3D, E). These results provide strong biochemical evidence that ubiquinol-10 regulates mitochondrial metabolic activity by increasing SIRT1, PGC-1α, and SIRT3 levels in the cochleae of SAMP1 mice.
Ubiquinol-10 decelerated the decline in mitochondrial function associated with aging
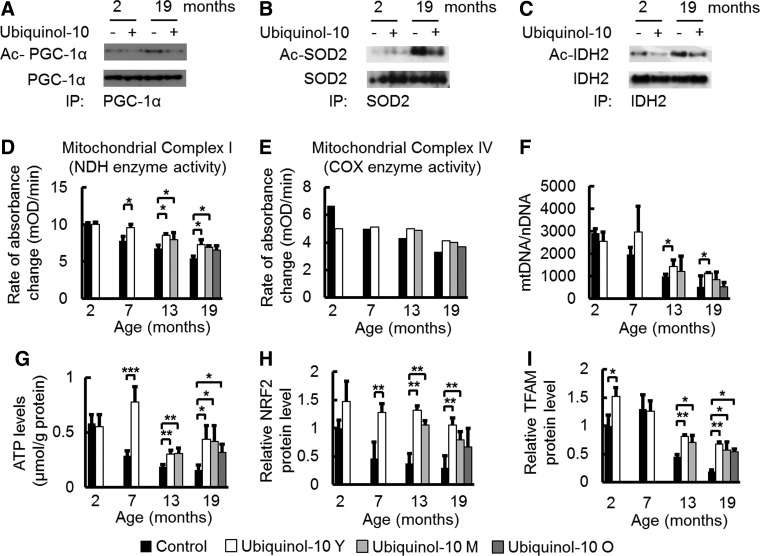
PGC-1α is known to be an important mediator of the metabolic effects of SIRT1. SIRT1 deacetylates and activates PGC-1α (48), while SIRT3 deacetylates and activates isocitrate dehydrogenase 2 (IDH2) (20) and SOD2 (44, 66). We observed that ubiquinol-10 supplementation suppressed the age-related decrease in Sirt1 and Sirt3 levels in the liver (Fig. 2B). Based on these findings, we examined the acetylation levels of PGC-1α, SOD2, and IDH2. PGC-1α, SOD2, and IDH2 were immunoprecipitated with their respective antibodies and acetylation levels of PGC-1α, SOD2, and IDH2 were detected with an anti-acetyl lysine antibody. We found that acetylation levels of PGC-1α, SOD2, and IDH2 increased in old mice (19 months), and ubiquinol-10 supplementation suppressed this increase (Fig. 4A–C). Thus, our findings provide evidence that ubiquinol-10 activates PGC-1α, SOD2, and IDH2 after SIRT1-dependent deacetylation and activation of SIRT3.
FIG. 4.
Ubiquinol-10 decelerated the age-related decline in mitochondrial function. (A–C) Immunoblot analysis of the acetylation stages of PGC-1α, SOD2, and IDH2 measured in livers from 2- and 19-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet (mixed protein samples from three mice) started during young age. (D, E) Mitochondria complex I (NDH) and IV (COX) enzyme activities were measured in livers from 2-, 7-, 13-, and 19-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet [complex I: n=3; complex IV: mixed protein samples (n=3)]. (F) Analysis of mtDNA copy number per nDNA in 2-, 7-, 13-, and 19-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet (n=3–5). (G) ATP level analysis in livers from 2-, 7-, 13-, and 19-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet (n=3–5). (H, I) Western blot analysis of NRF2 and TFAM protein levels in livers from 2-, 7-, 13-, and 19-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet (n=3–5). *p<0.05, **p<0.01, ***p<0.001. COX, cytochrome c oxidase; mtDNA, mitochondrial DNA; NDH, NADH dehydrogenase; NRF2, nuclear factor erythroid 2-related factor 2; TFAM, mitochondrial transcription factor A; nDNA, nuclear DNA.
The primary function of mitochondria is production of ATP via oxidative phosphorylation, which is conducted by the four RC complexes (complexes I–IV) and ATP synthase (complex V), all of which are located in the inner mitochondrial membrane. To provide evidence that ubiquinol-10 increases mitochondria function in SAMP1 mice, we determined the effect of ubiquinol-10 on the activity of complexes I and IV (cytochrome-c oxidase, EC 1.9.3.1) in the liver, as the activities of these complexes were found to be decreased with age (36). We found that both complex I and IV activity decreased with age, and compared with control animals, decreased complex I activity was significantly suppressed in mice supplemented with ubiquinol-10 during young and middle age (Fig. 4D, E). In several studies, an increase in mtDNA damage and mutations resulted in decreased amounts of mitochondria. As such, we investigated the effect of ubiquinol-10 on mtDNA copy number during aging. Ubiquinol-10 supplementation significantly suppressed a decrease in mtDNA copy number when ubiquinol-10 supplementation started at a young age (Fig. 4F). In addition, age-related decreases in liver ATP levels in control diet mice were suppressed by ubiquinol-10 supplementation started from young, middle, and old age (Fig. 4G). We observed an effect of ubiquinol-10 on PGC-1α downstream targets and mitochondrial biogenesis-related proteins such as nuclear factor erythroid 2-related factor 2 (NRF2) and mitochondrial transcription factor A (TFAM). Western blot analyses showed that the levels of these proteins also decreased with age, but these decreases were significantly reduced for young and middle age groups after ubiquinol-10 supplementation (Fig. 4H, I). Thus, ubiquinol-10 promoted deacetylation of PGC-1α, SOD2, and IDH2 via SIRT1 and SIRT3 and increased the function and amount of mitochondria via activation of PGC-1α.
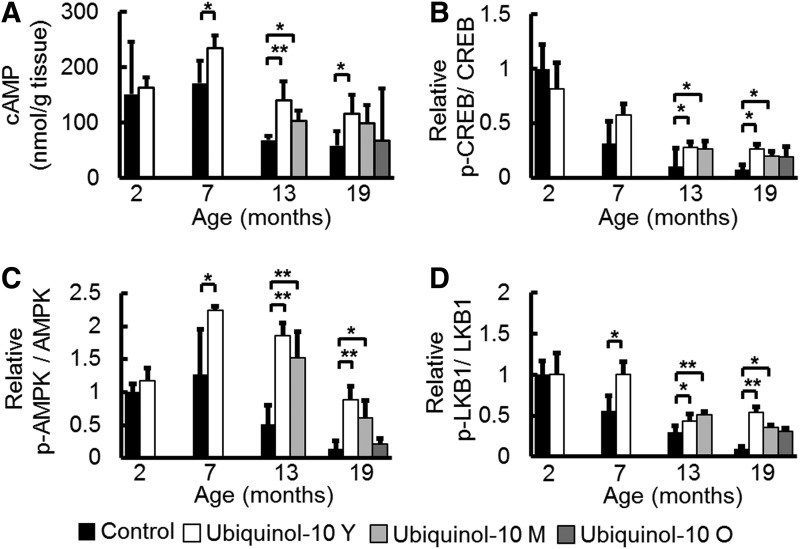
Ubiquinol-10 suppressed age-related decreases in cAMP levels and the expression of phosphorylated CREB, AMPK, and LKB1
While the mechanism by which ubiquinol-10 activates SIRT1 and PGC-1α remains unclear, a recent study revealed that increased cyclic adenosine monophosphate (cAMP) levels mediated the metabolic effects of PGC-1α on mitochondrial biogenesis and function by increasing PGC-1α expression (43). We found that cAMP levels decreased with aging, and compared with control animals, ubiquinol-10 supplementation from young and middle age significantly suppressed this decrease (Fig. 5A). The cAMP response element-binding protein (CREB) is a transcription factor that is induced by a variety of growth factors and inflammatory signals and subsequently mediates the transcription of genes containing a cAMP-responsive element. A previous study showed that CREB binds to a target sequence in the PGC-1α promoter, thereby enhancing its expression in response to cAMP. We found that age-related decreases in the ratio of phosphorylated-CREB/CREB were suppressed by ubiquinol-10 supplementation (Fig. 5B). AMP-activated protein kinase (AMPK) is a trimeric complex that senses nutrient deprivation by responding to changes in the AMP/ATP (9) and ADP/ATP (73) ratios. AMPK, which is emerging as a key regulator of whole-body metabolism, was shown to increase NAD+ levels and activate SIRT1 and PGC-1α (8). We determined that phosphorylated AMPK levels were reduced by aging and that these reductions could be slowed by ubiquinol-10 (Fig. 5C). We also found that ubiquinol-10 supplementation suppressed age-related decreases in phosphorylated serine/threonine kinase B1 (LKB1), which is the critical upstream kinase required for AMPK phosphorylation (Fig. 5D). These data provide biochemical evidence that ubiquinol-10 regulates mitochondrial metabolic activity by increasing the activity of the cAMP-AMPK-Sirt1-PGC-1α pathway.
FIG. 5.
Ubiquinol-10 suppressed age-related decreases in cAMP levels and the expression of phosphorylated CREB, AMPK, and LKB1. (A) ELISA analysis of cAMP levels in livers from 2-, 7-, 13-, and 19-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet (n=3–5). (B) Western blot analysis of CREB and phosphorylated CREB, (C) AMPK and phosphorylated AMPK, (D) LKB1 and phosphorylated LKB1 expression levels in livers from 2-, 7-, 13-, and 19-month-old SAMP1 mice fed a control or ubiquinol-10-supplemented diet (n=3–5). *p<0.05; **p<0.01. AMPK, AMP-activated protein kinase; CREB, cAMP response element-binding protein; LKB1, liver serine/threonine kinase B1; cAMP, cyclic adenosine monophosphate.
Ubiquinol-10 increased the deacetylation status of PGC-1α by SIRT1 activation and stimulated AMPK phosphorylation in HepG2 cells
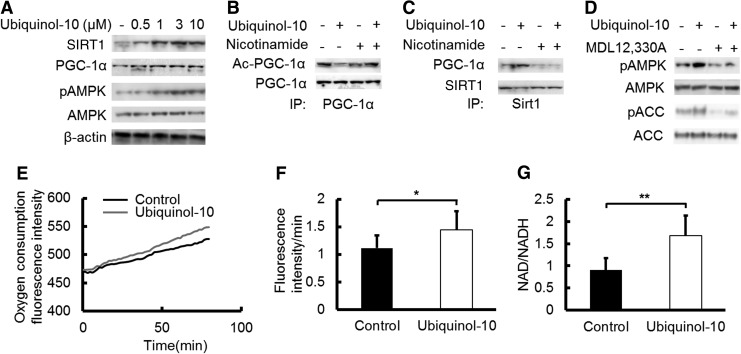
To confirm that ubiquinol-10 can activate SIRT1, PGC-1α, and AMPK signaling pathways, we examined its effect on these protein levels in human hepatoma HepG2 cells. SIRT1, PGC-1α, and phosphorylated AMPK were increased in the ubiquinol-10-treated HepG2 cells in a dose-dependent manner (Fig. 6A), with the expression of SIRT1, PGC-1α, and phosphorylated AMPK increasing with a low ubiquinol-10 dose (≤3 μM). The PGC-1α acetylation levels were decreased by ubiquinol-10, and the SIRT1 inhibitor nicotinamide reversed the effect of SIRT1 (Fig. 6B). Based on these observations, we next sought to determine whether SIRT1 was capable of directly interacting with PGC-1α. As shown in Figure 6C, a SIRT1 antibody co-immunoprecipitated with PGC-1α. Ubiquinol-10 increased the levels of co-immunoprecipitated PGC-1α, while nicotinamide reversed this effect (Fig. 6C). Since ubiquinol-10 may activate AMPK by increasing cAMP production, we next treated HepG2 cells with ubiquinol-10 in the presence of the adenylyl cyclase (AC) inhibitor MDL-12,330A (Fig. 6D). MDL-12,330A inhibited the ability of ubiquinol-10 to increase the phosphorylation of both AMPK and the AMPK substrate acetyl-CoA carboxylase (ACC), which is a marker of AMPK activity. These findings indicate that cAMP-AMPK signaling activation is essential for ubiquinol-10 to exert its effects.
FIG. 6.
Ubiquinol-10 increased mitochondrial metabolic activity by promoting deacetylation of PGC-1α by SIRT1. (A) HepG2 cells were treated with the indicated ubiquinol-10 doses for 48 h. The cell lysates (10 μg) were analyzed by western blot. HepG2 cells were treated with 0.5, 1, 3, and 10 μM ubiquinol-10. (B) Immunoblot analysis of PGC-1α deacetylation in HepG2 cells after treatment with or without 3 μM ubiquinol-10 in the presence of the SIRT1 inhibitor nicotinamide (20 mM). (C) Molecular interactions of SIRT1 and PGC-1α were determined after treatment with or without ubiquinol-10 in the presence of nicotinamide. (D) Phosphorylation of AMPK and AMPK substrate ACC was determined after treatment with or without ubiquinol-10 (3 μM) in the presence of the AC inhibitor MDL-12,330A (50 μM). (E) Oxygen consumption was measured in HepG2 cells with or without ubiquinol-10 (n=10). (F) Oxygen consumption rate (rate of fluorescence intensity/min) is shown in the panel (n=10). (G) ELISA analysis to determine NAD and NADH levels in HepG2 cells with or without ubiquinol-10 (n=6). *p<0.05; **p<0.01. AC, adenylate cyclase; ACC, acetyl-CoA carboxylase.
Ubiquinol-10 increased mitochondrial metabolic activity
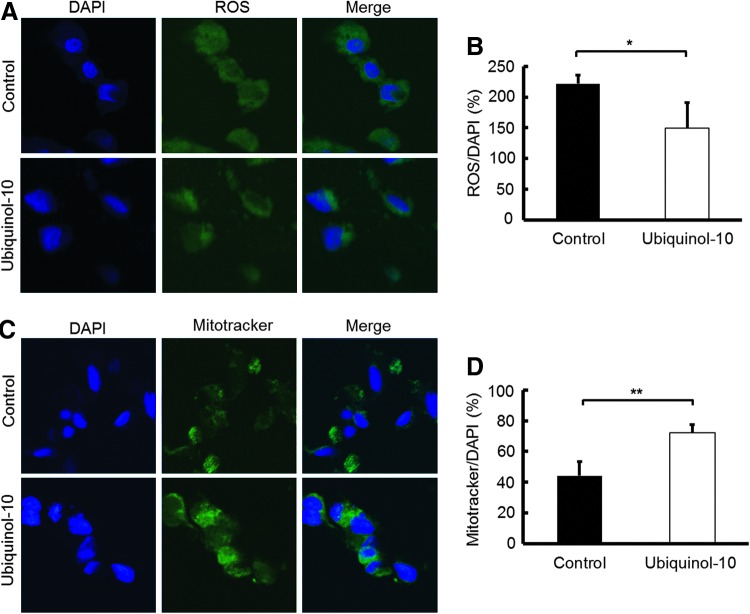
To determine the effect of ubiquinol-10 on mitochondrial metabolic activity, HepG2 cells were treated with ubiquinol-10 and the oxygen consumption rate was determined. Ubiquinol-10 treatment resulted in a significantly increased rate of oxygen consumption (Fig. 6E, F). The NADH and NAD concentrations were also measured in ubiquinol-10-treated HepG2 cells. The NAD/NADH ratio was significantly increased after ubiquinol-10-treatment (Fig. 6G). We next measured the total ROS production using specific fluorescent probes and found that the total ROS levels were significantly decreased by ubiquinol-10-treatment (Fig. 7A, B). The possibility that the effect of ubiquinol-10 might be associated with mitochondria biogenesis was investigated further in HepG2 cells by labeling mitochondria with Mitotracker. HepG2 cells treated with ubiquinol-10 had higher numbers of mitochondria compared with control cells (Fig. 7C, D). Together, these data provide biochemical evidence that ubiquinol-10 increases the amount of mitochondria and regulates mitochondrial metabolic activity.
FIG. 7.
Ubiquinol-10 increased mitochondrial metabolic activity. (A) ROS levels were measured in HepG2 cells with or without ubiquinol-10 using laser fluorescence confocal microscopy (X200). DAPI was used to indicate nuclei (blue) and ROS (green). (B) Quantification analysis of ROS fluorescence intensity of various formulations in (A) using ZEN 2011 software (LSM700; Carl Zeiss, Jena, Germany). Three visual fields from each well were randomly chosen to analyze blue (DAPI) and green (ROS) fluorescence and to calculate the ROS/DAPI ratio (n=5). (C) HepG2 cells with or without ubiquinol-10 were incubated with Mitotracker green to stain mitochondria, and then examined by laser fluorescence confocal microscopy (X200). DAPI was used to indicate nuclei (blue). (D) Quantification of Mitotracker/DAPI is shown in the panel (n=5). *p<0.05; **p<0.01. ROS, reactive oxygen species.
Discussion
A widely accepted hypothesis of how aging leads to age-related diseases is that oxidative damage accumulates in tissues (17). In mammals, increased resistance to oxidative stress that can be provided by antioxidants is consistently associated with delayed aging, resistance to age-related diseases, and enhanced longevity (60). Ubiquinol-10 is regarded as one of the most important antioxidants. Ubiquinol-10 biosynthesis decreases with age and its deficit in tissues is believed to be associated with degenerative changes appearing over the course of aging (5, 22). Our experimental evidence indicates that aging, indeed, results in an increase in oxidative damage to DNA, proteins, and lipids and a decrease in the ratio of GSH:GSSG in the liver (Fig. 1). Therefore, we speculate that ubiquinol-10-mediated modulation of the antioxidant defense system may play a central role in reducing oxidative stress in tissues, and, in turn, retard the effects of aging.
Sirtuins are proposed to be regulators of aging and age-related diseases (7, 56). SIRT1, the most studied member of the family, plays an important role in several processes ranging from cell cycle regulation to energy homeostasis. In this study, we found that age-related decreases in SIRT1 levels were suppressed by dietary ubiquinol-10 supplementation (Fig. 2), which also decelerated the progression of age-related diseases and senescence (Fig. 3 and Supplementary Fig. S1), but no life span extension was observed (Supplementary Fig. S1F). This latter finding may be because Sirt1 has so many important functions in mammalian physiology and metabolism, and global up-regulation of SIRT1 can exert opposing effects, or, alternatively, global up-regulation may, indeed, slow overall aging, but has no effect on life spans in the mouse strains tested.
SIRT3, the most studied mitochondrial sirtuin, deacetylates a number of mitochondrial proteins and might also play a role in regulating ATP production (42, 55). A large body of evidence suggests that CR reduces the age-associated accumulation of oxidative damage of proteins, lipids, and DNA by enhancing the activity of mitochondrial antioxidant defense systems (29, 71). Some analyses showed that CR increases SIRT3 levels, which resulted in lower ROS production and decreased amounts of oxidative damage to mitochondria (62). SIRT1 expression has also been reported to be positively regulated by CR (14) and exercise (72). Interestingly, SIRT3 expression was shown to be regulated by the activity of the ERRα-dependent coactivator PGC-1α that binds to the SIRT3 promoter (16), while SIRT1 was convincingly shown to deacetylate PGC-1α to increase its potential to activate transcription (38). With ubiquinol-10 supplementation, SIRT1 protein levels increased in liver tissue, resulting in increased mitochondrial biogenesis in response to activated PGC-1α-dependent effects on mitochondrial biogenesis and respiration after its activation by SIRT3. Thus, SIRT1 activates SIRT3 by promoting ERRα-dependent PGC-1α activation (Fig. 8). Here, we found that age-related decreases in SIRT3 levels were suppressed by ubiquinol-10 supplementation (Fig. 2).
FIG. 8.
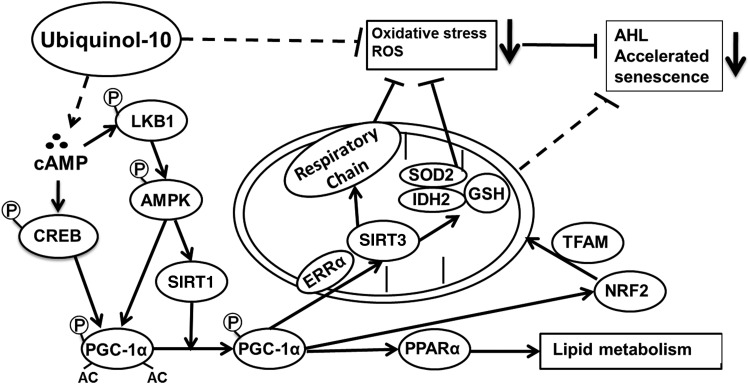
Possible mechanism of the anti-aging effects of ubiquinol-10 supplementation. In response to ubiquinol-10, increased cAMP levels activate SIRT1 and PGC-1α by increasing the levels of phosphorylated CREB, LKB1, and AMPK. SIRT1 deacetylates and activates PGC-1α that can then induce Sirt3, which activates SOD2, IDH2, and mitochondria genes though NRF2 and TFAM. Activated SOD2, IDH2, and mitochondria genes produced an increased ratio of GSH:GSSG that decreases ROS levels and increases the activity of mitochondrial electron transport chain complexes I and IV. Together, these effects protect tissues from oxidative stress and prevent the accelerated senescence seen in SAMP1 mice.
We demonstrated that ubiquinol-10 supplementation suppressed age-related decreases and acetylation of SOD2 (Figs. 2B and 4B), which is a major mitochondrial antioxidant enzyme. Decreased SOD2 activity causes elevated ROS production, mitochondrial membrane potential loss, and early cell apoptosis (27). SIRT3-dependent SOD2 deacetylation increased the specific activity of SOD2 and its ROS-scavenging capacity. We also confirmed that ubiquinol-10 supplementation suppressed age-related acetylation of IDH2 (Fig. 4C). Data from other groups showed that IDH2 activation is induced in response to increased ROS levels and enhances the activity of the glutathione antioxidant defense system in mitochondria, and that SIRT3 could enhance deacetylation and activation of IDH2 (62). Hence, SIRT3 may modulate oxidative damage in the liver by enhancing the activity of mitochondrial antioxidant defense systems. The capacity of PGC-1α to promote mitochondrial biogenesis is related to its actions on the transcription of NRF1 and NRF2, both of which are up-regulated by PGC-1α. NRF1 and NRF2 are key trans-acting elements in mitochondrial biogenesis that regulate the transcription of several nuclear-encoded genes, including TFAM, which is itself a vital coordinator of the transcription and replication of the mitochondrial genome. Here, we demonstrated that ubiquinol-10 affects the anti-oxidative system by activating expression of genes related to mitochondrial function, including SIRT1, SIRT3, PGC-1α, SOD2, IDH2, NRF2, and TFAM in the liver and that these effects likely arise from an enhancement of mitochondrial activity (Figs. 2, 4, and 8).
The primary function of mitochondria is ATP production, which is conducted by mitochondrial RC complexes. Mitochondrial complex I is a transmembrane protein complex that oxidizes matrix NADH to NAD+, and reduces membrane-bound ubiquinone-10 to ubiquinol-10. We found that complex I and IV activity decreased during aging, and that ubiquinol-10 supplementation suppressed this decrease (Fig. 4D, E). The activity of mitochondrial electron transfer is suggested to depend on the concentration of ubiquinone-10 in the inner mitochondrial membrane (25, 47). Therefore, the suppression of age-related decreases in complex I activity after ubiquinol-10 supplementation may be due to an increase either in the inner mitochondrial membrane CoQ10 concentration or in the complex I protein expression levels. Since the complex I activity did not depend on the presence of ubiquinone (MS141; MitoSciences, Eugene, OR), ubiquinol-10 supplementation likely affects complex I expression levels. In addition, age-related decreases in complex I (NADH dehydrogenase [ubiquinone] 1 β subcomplex subunit 8, NDUFB8) and complex IV (mitochondrially encoded cytochrome c oxidase I, MTCO1) protein levels were suppressed in mitochondria fractions from SAMP1 mice fed a ubiquinol-10-supplemented diet (Supplementary Fig. S3). Ubiquinol-10 supplementation also inhibited the reduction in mtDNA levels that was seen during aging for mice fed a control diet (Fig. 4F). Taken together, these results show that ubiquinol-10 may slow the progression of age-related diseases, including hearing loss by enhancing mitochondrial activity and mitochondrial antioxidant defense system activity and increasing ATP production (Figs. 3 and 4). Previous studies showed that the mean ABR hearing threshold at low frequencies was significantly lower in ubiquinone-10-supplemented control mice (61, 62). Similar to SAMP1 mice, ubiquinone-10 suppressed AHL in C57BL/6J mice by increasing antioxidant activities via SIRT3 activation.
While ubiquinol-10 suppressed age-related decreases in the expression of SIRT1, SIRT3, and PGC-1α, its direct effect remains unclear. cAMP plays a key role as an intracellular second messenger for transduction events that occur in response to a number of extracellular signals (30). We observed that age-related decreases in cAMP levels were suppressed by ubiquinol-10 supplementation (Fig. 5A). The CREB is probably one of the best understood phosphorylation-dependent transcription factors. Increases in either calcium or cAMP levels can induce the activation of CREB, which leads to the transcription of target genes that carry cAMP-responsive elements in their promoters (30). A recent study indicated that increases in cAMP production were responsible for AMPK activation (41). Activated AMPK triggers both phosphorylation and deacetylation of PGC-1α, with the latter being catalyzed by SIRT1. SIRT1 activation follows AMPK-dependent increases in NAD+. Phosphorylation of Thr 172 in the activation loop of AMPK is required for AMPK activation, and several groups have demonstrated that the LKB1 directly mediates this event (58). LKB1 is well-documented as the critical upstream kinase required for AMPK activation. The effect of cAMP is proposed to involve protein kinase A (PKA) phosphorylation and activation. In addition, genetic and biochemical findings indicate that LKB1 exerts its effects by phosphorylation related to PKA. Aging might result in lower levels of AMPK phosphorylation due to age-related decreases in cAMP. In our experiment, ubiquinol-10 supplementation lessened the age-related decrease in the ratio of phosphorylated AMPK/AMPK and phosphorylated LKB1/LKB1 (Fig. 5). However, we cannot exclude the possibility that ubiquinol-10 may also induce AMPK phosphorylation through calmodulin-dependent protein kinase kinase and intracellular calcium. Taken together, ubiquinol-10 may induce some pathways that activate mitochondrial function by increasing cAMP levels and activating CREB, LKB1, and AMPK (Fig. 8). Nevertheless, as discussed earlier, the mechanism of ubiquinol-10 action other than mitochondria activation should be considered to explain in greater detail its beneficial effect on aging. For example, the ubiquinol-dependent antioxidant system of the plasma membrane was suggested to be important for preventing the accumulation of oxidative damage and regulating the externally initiated ceramide signaling pathway (37). In fact, antioxidant activity of CoQ10 in liver plasma membrane was confirmed in aged rats fed a polyunsaturated fatty acids plus CoQ10 diet (4). Furthermore, a recent study showed that ubiquinol in endothelial cell Golgi compartments may be an important cofactor to maintain endothelial nitric oxide synthase in a coupled conformation that is required to produce physiological nitric oxide (NO), an important mediator of cardiovascular homeostasis, and, eventually, quench leaking-uncoupled electrons (34).
In summary, we propose that ubiquinol-10 may induce pathways that activate SIRT1 and PGC-1α by increasing cAMP levels and activating CREB and AMPK. SIRT1 can activate SIRT3 via ERRα-dependent activity of PGC-1α. SIRT3 modulates oxidative damage in the liver by enhancing the activity of mitochondrial antioxidant defense systems, resulting in significantly delayed senescence and age-related diseases in SAMP1 mice fed a diet supplemented with ubiquinol-10. We postulate that this may be a mechanism of aging retardation promoted by ubiquinol-10, which suggests that ubiquinol-10 may be useful for treating age-related diseases. In future studies, we will use Sirt1−/− and Sirt3−/− mice to decipher the direct effect of ubiquinol-10 supplementation other than mitochondria activation. In SAMP1 mice and various cell lines, we can determine the effect of ubiquinol-10 on other tissues, including adipose and muscle tissues.
Materials and Methods
Animals and cell culture
Methods involving animals and cell culture are described in detail in the Supplementary Data.
Measurement of oxidative damage
Oxidative damage to lipids (malondialdehyde) and proteins (protein carbonyls) were measured by western blot analysis. DNA oxidation levels (AP sites) were determined using a DNA damage quantification kit (Dojindo, Rockville, MD) according to the manufacturer's instructions. GSH and GSSG concentrations were determined with the GSSG/GSH Quantification Kit (Dojindo) according to the manufacturer's instructions.
Real-time reverse transcription-polymerase chain reaction analysis
Total RNA was extracted from the livers of 2-, 7-, 13-, and 19-month-old SAMP1 mice using TRIzol Reagent (Invitrogen, New York, NY), followed by treatment with DNA-Free (Applied Biosystems, Foster City, CA) to remove contaminating DNA and then subjected to reverse transcription using an Omniscript RT kit (Applied Biosystems, New York, NY) with random primers. Quantitative real-time RT-PCR analysis was carried out using an ABI PRISM 7500 Sequence Detection System with SYBR Green (Takara Bio, Tokyo, Japan). We assessed the stability of some genes and found that glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin are the most stable genes in our experiment and, as such, were suitable for use as reference genes. The forward and reverse primer sequences are listed in Table 1.
Table 1.
Primer Sequences
| Gene | Primer sequences (5′–3′) | Length (bp) |
|---|---|---|
| Pgc-1α | Forward primer TCACCACCGAAATCCTTA | 178 |
| Reverse primer GGTGTCTGTAGTGGCTTGAT | ||
| Sirt1 | Forward primer CCTTGGAGACTGCGATGTTA | 158 |
| Reverse primer GTGTTGGTGGCAACTCTGAT | ||
| Sirt3 | Forward primer TACAGGCCCAATGTCACTCA | 168 |
| Reverse primer ACAGACCGTGCATGTAGCTG | ||
| Pparα | Forward primer GCGTACGGCAATGGCTTTAT | 56 |
| Reverse primer GAACGGCTTCTTCAGGTTCTT | ||
| mtDNA (mt-Nd2, NADH dehydrogenase) | Forward primer CCTATCACCCTTGCCATCAT | 193 |
| Reverse primer GAGGCTGTTGCTTGTGTGAC | ||
| nDNA (Pecam 1) | Forward primer ATGGAAAGCCTGCCATCATG | 235 |
| Reverse primer TCCTTGTTGTTCAGCATCAC | ||
| Gapdh | Forward primer TGCACCACCAACTGCTTAG | 177 |
| Reverse primer GGATGCAGGGATGATGTTC | ||
| β-Actin | Forward primer GACAGGATGCAGAAGGAGATTACT | 142 |
| Reverse primer TGATCCACATCTGCTGGAAGGT |
mtDNA quantitation
To quantify the amount of mtDNA per nuclear genome, we used the following primers. To quantify nDNA, we used a primer set that detects the platelet/endothelial cell adhesion molecule 1 (Pecam 1) gene on chromosome 6. Quantification of the relative copy number of mtDNA to nDNA was carried out using analysis of the difference in threshold amplification between mtDNA and nDNA (ΔΔC(t) method) using real-time PCR. The forward and reverse primer sequences are listed in Table 1.
Mitochondrial complex activity assay
To determine the activity of mitochondrial complex I, a microplate assay kit for complex I activity (MitoSciences) was used with 50 μg mitochondrial proteins. Complex I was immunocaptured on microplates, and the activity was determined from the oxidation of NADH to NAD+. Complex I activity was measured by the increase in absorbance at 450 nm and expressed as the change in absorbance per minute per microgram of protein. The cytochrome c oxidase (COX) enzyme (Complex IV) was immunocaptured within the microplate wells and determined colorimetrically by absorbance changes at 550 nm.
Cyclic AMP measurement
Cyclic AMP levels were determined using a cyclic AMP chemiluminescent kit (Cell Signaling Technology, Danvers, MA) according to the manufacturer's instructions.
Measurement of ATP levels
ATP was extracted from livers using the AMERIC-ATP kit (AMERIC, Tokushima, Japan). Extracts (10 μl) were added to 90 μl luciferase reagent and measured immediately. Based on the maximum light intensity of the resulting bioluminescence measured in a luminometer (Luminescencer PSN AB2200; ATTO, Tokyo, Japan), a calibration curve with varying ATP dilutions was also prepared. Data were normalized as ATP μmol/g protein.
Measurement of ROS
The oxidative stress levels in HepG2 cells were measured by a commercial kit according to the manufacturer's instructions (Total ROS/Superoxide Detection kit; Enzo Life Sciences, New York, NY). HepG2 cells were sedimented by centrifugation at 400 g for 5 min and then washed. The washed cells were incubated with 1 μM ROS/Superoxide Detection Mix for 60 min at 37°C. Green and blue fluorescence intensities were measured using a microplate fluorescence reader (BioTek, Winooski, VT) at excitation/emission wavelengths of 488/520 and 350/470 nm, respectively.
Mitochondria staining
MitoTracker® Green FM (Cell Signaling Technology) was added into the culture media at a final concentration of 200 nM. HepG2 cells were incubated for 30 min, and then visualized by fluorescence microscopy (Axiovert 200 M; Carl Zeiss, Jena, Germany).
Respirometric assay
For respirometric analysis, oxygen consumption was measured using a commercial kit according to the manufacturer's instructions (MitoXpress®-Xtra; Luxcel, Cork, Ireland). The MitoXpress oxygen probe was added to the culture media at a final concentration of 3 μM. Samples were then covered with 100 μl of pre-warmed (37°C) heavy mineral oil and read kinetically for 1 h at 30°C on a fluorescence plate reader (SpectraMax Gemini; Molecular Devices, Sunnyvale, CA) at excitation/emission wavelengths of 380/650 nm, with a measurement cycle of 1 min.
NAD/NADH analysis
NAD and NADH were analyzed independently in extracts of whole cells (1×106) prepared as previously described (57). NAD and NADH concentrations were determined using a Fluorimetric NAD/NADH Ratio Assay Kit (AAT Bioquest, Inc., Sunnyvale, CA).
Statistical analysis
We used the StatView software package (Abacus Concepts, Berkeley, CA) for data analysis. Real-time PCR, western blot, and ELISA results are presented as the mean±SD, body weight, degree of senescence, and food intake results are presented as the mean±SE. Nonparametric Mann–Whitney tests were used for in vivo results, as a normal distribution could not be assumed and Kaplan–Meier curves were used for animal survival. Body weights were compared by repeated measures ANOVAs. A p-value less than 0.05 was considered significant.
Supplementary Material
Abbreviations Used
- ABR
auditory brainstem response
- AC
adenylate cyclase/adenylyl cyclase
- ACC
acetyl-CoA carboxylase
- AHL
age-related hearing loss
- AMPK
AMP-activated protein kinase
- AP
apurinic/apyrimidinic
- cAMP
cyclic adenosine monophosphate
- CoQ9
coenzyme Q9
- CoQ10
coenzyme Q10
- COX
cytochrome c oxidase
- CR
calorie restriction
- CREB
cAMP response element binding protein
- ERRα
estrogen-related receptor alpha
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- IDH2
isocitrate dehydrogenase 2
- LKB1
liver serine/threonine kinase B1
- mtDNA
mitochondrial DNA
- NDH
NADH dehydrogenase
- nDNA
nuclear DNA
- NO
nitric oxide
- NRF2
nuclear factor erythroid 2-related factor 2
- PCR
polymerase chain reaction
- PGC-1α
peroxisome proliferator-activated receptor γ coactivator 1α
- PKA
protein kinase A
- PPARα
peroxisome proliferator-activated receptor-alpha
- RC
respiratory chain
- ROS
reactive oxygen species
- RT-PCR
reverse transcription-polymerase chain reaction
- SAMP1
senescence-accelerated mouse prone 1
- SIRT1
sirtuin1
- SIRT3
sirtuin3
- SOD2
superoxide dismutase 2
- TFAM
mitochondrial transcription factor A
Acknowledgments
The authors thank Masayasu Kitagawa, Taizo Kawabe, Masanori Kato, and Shuka Matsumoto (Frontier Biochemical & Medical Research Laboratories, Kaneka Corporation) for determining the mitochondria concentrations of CoQ9 or CoQ10 and preparing the mouse diet. They also thank Kiyoshi Matsumoto (Research Center for Human and Environmental Science, Shinshu University) for technical assistance and care of mice. This research was supported in part by Grant-In-Aid 2012 from the Japanese Coenzyme Q Association.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Aberg F, Appelkvist EL, Dallner G, and Ernster L. Distribution and redox state of ubiquinones in rat and human tissues. Arch Biochem Biophys 295: 230–234, 1992 [DOI] [PubMed] [Google Scholar]
- 2.Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, and Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab 8: 333–341, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barja G. Updating the mitochondrial free radical theory of aging: an integrated view, key aspects, and confounding concepts. Antioxid Redox Signal 19: 1420–1445, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bello RI, Gomez-Diaz C, Buron MI, Alcain FJ, Navas P, and Villalba JM. Enhanced anti-oxidant protection of liver membranes in long-lived rats fed on a coenzyme Q10-supplemented diet. Exp Gerontol 40: 694–706, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Beyer RE, Burnett BA, Cartwright KJ, Edington DW, Falzon MJ, Kreitman KR, Kuhn TW, Ramp BJ, Rhee SY, Rosenwasser MJ, et al. Tissue coenzyme Q (ubiquinone) and protein concentrations over the life span of the laboratory rat. Mech Ageing Dev 32: 267–281, 1985 [DOI] [PubMed] [Google Scholar]
- 6.Buffenstein R, Edrey YH, Yang T, and Mele J. The oxidative stress theory of aging: embattled or invincible? Insights from non-traditional model organisms. Age (Dordr) 30: 99–109, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai W, Ramdas M, Zhu L, Chen X, Striker GE, and Vlassara H. Oral advanced glycation endproducts (AGEs) promote insulin resistance and diabetes by depleting the antioxidant defenses AGE receptor-1 and sirtuin 1. Proc Natl Acad Sci USA 109: 15888–15893, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, and Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458: 1056–1060, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carling D, Zammit VA, and Hardie DG. A common bicyclic protein kinase cascade inactivates the regulatory enzymes of fatty acid and cholesterol biosynthesis. FEBS Lett 223: 217–222, 1987 [DOI] [PubMed] [Google Scholar]
- 10.Chabi B, Ljubicic V, Menzies KJ, Huang JH, Saleem A, and Hood DA. Mitochondrial function and apoptotic susceptibility in aging skeletal muscle. Aging Cell 7: 2–12, 2008 [DOI] [PubMed] [Google Scholar]
- 11.Chiba Y, Shimada A, Kumagai N, Yoshikawa K, Ishii S, Furukawa A, Takei S, Sakura M, Kawamura N, and Hosokawa M. The senescence-accelerated mouse (SAM): a higher oxidative stress and age-dependent degenerative diseases model. Neurochem Res 34: 679–687, 2009 [DOI] [PubMed] [Google Scholar]
- 12.Crowley VE, Yeo GS, and O'Rahilly S. Obesity therapy: altering the energy intake-and-expenditure balance sheet. Nat Rev Drug Discov 1: 276–286, 2002 [DOI] [PubMed] [Google Scholar]
- 13.Donmez G, Wang D, Cohen DE, and Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell 142: 320–332, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Finkel T, Deng CX, and Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature 460: 587–591, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fridovich I. Mitochondria: are they the seat of senescence? Aging Cell 3: 13–16, 2004 [DOI] [PubMed] [Google Scholar]
- 16.Giralt A, Hondares E, Villena JA, Ribas F, Diaz-Delfin J, Giralt M, Iglesias R, and Villarroya F. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. J Biol Chem 286: 16958–16966, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haigis MC. and Yankner BA. The aging stress response. Mol Cell 40: 333–344, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hekimi S, Lapointe J, and Wen Y. Taking a “good” look at free radicals in the aging process. Trends Cell Biol 21: 569–576, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, and Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun 1: 3, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jo SH, Son MK, Koh HJ, Lee SM, Song IH, Kim YO, Lee YS, Jeong KS, Kim WB, Park JW, Song BJ, and Huh TL. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J Biol Chem 276: 16168–16176, 2001 [DOI] [PubMed] [Google Scholar]
- 21.Kaeberlein M, Kirkland KT, Fields S, and Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol 2: E296, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalen A, Norling B, Appelkvist EL, and Dallner G. Ubiquinone biosynthesis by the microsomal fraction from rat liver. Biochim Biophys Acta 926: 70–78, 1987 [DOI] [PubMed] [Google Scholar]
- 23.Kubo H, Fujii K, Kawabe T, Matsumoto S, Kishida H, and Hosoe K. Food content of ubiquinol-10 and ubiquinone-10 in the Japanese diet. J Food Comp Anal 21: 199–210, 2008 [Google Scholar]
- 24.Lenaz G. Quinone specificity of complex I. Biochim Biophys Acta 1364: 207–221, 1998 [DOI] [PubMed] [Google Scholar]
- 25.Lenaz G, Battino M, Castelluccio C, Fato R, Cavazzoni M, Rauchova H, Bovina C, Formiggini G, and Parenti Castelli G. Studies on the role of ubiquinone in the control of the mitochondrial respiratory chain. Free Radic Res Commun 8: 317–327, 1990 [DOI] [PubMed] [Google Scholar]
- 26.Lenaz G, Fato R, Castelluccio C, Genova ML, Bovina C, Estornell E, Valls V, Pallotti F, and Parenti Castelli G. The function of coenzyme Q in mitochondria. Clin Investig 71: S66–S70, 1993 [DOI] [PubMed] [Google Scholar]
- 27.Liu C, Ogando D, and Bonanno JA. SOD2 contributes to anti-oxidative capacity in rabbit corneal endothelial cells. Mol Vis 17: 2473–2481, 2011 [PMC free article] [PubMed] [Google Scholar]
- 28.Marchi S, Giorgi C, Suski JM, Agnoletto C, Bononi A, Bonora M, De Marchi E, Missiroli S, Patergnani S, Poletti F, Rimessi A, Duszynski J, Wieckowski MR, and Pinton P. Mitochondria-ros crosstalk in the control of cell death and aging. J Signal Transduct 2012: 329635, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masoro EJ. Caloric restriction and aging: an update. Exp Gerontol 35: 299–305, 2000 [DOI] [PubMed] [Google Scholar]
- 30.Mayr B. and Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol 2: 599–609, 2001 [DOI] [PubMed] [Google Scholar]
- 31.Menardo J, Tang Y, Ladrech S, Lenoir M, Casas F, Michel C, Bourien J, Ruel J, Rebillard G, Maurice T, Puel JL, and Wang J. Oxidative stress, inflammation, and autophagic stress as the key mechanisms of premature age-related hearing loss in SAMP8 mouse Cochlea. Antioxid Redox Signal 16: 263–274, 2012 [DOI] [PubMed] [Google Scholar]
- 32.Miyamae T, Seki M, Naga T, Uchino S, Asazuma H, Yoshida T, Iizuka Y, Kikuchi M, Imagawa T, Natsumeda Y, Yokota S, and Yamamoto Y. Increased oxidative stress and coenzyme Q10 deficiency in juvenile fibromyalgia: amelioration of hypercholesterolemia and fatigue by ubiquinol-10 supplementation. Redox Rep 18: 12–19, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohr D, Bowry VW, and Stocker R. Dietary supplementation with coenzyme Q10 results in increased levels of ubiquinol-10 within circulating lipoproteins and increased resistance of human low-density lipoprotein to the initiation of lipid peroxidation. Biochim Biophys Acta 1126: 247–254, 1992 [DOI] [PubMed] [Google Scholar]
- 34.Mugoni V, Postel R, Catanzaro V, De Luca E, Turco E, Digilio G, Silengo L, Murphy MP, Medana C, Stainier DY, Bakkers J, and Santoro MM. Ubiad1 is an antioxidant enzyme that regulates eNOS activity by CoQ10 synthesis. Cell 152: 504–518, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J 417: 1–13, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navarro A. and Boveris A. The mitochondrial energy transduction system and the aging process. Am J Physiol Cell Physiol 292: C670–C686, 2007 [DOI] [PubMed] [Google Scholar]
- 37.Navas P, Villalba JM, and de Cabo R. The importance of plasma membrane coenzyme Q in aging and stress responses. Mitochondrion 7Suppl: S34–S40, 2007 [DOI] [PubMed] [Google Scholar]
- 38.Nemoto S, Fergusson MM, and Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha}. J Biol Chem 280: 16456–16460, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Ochoa JJ, Pamplona R, Ramirez-Tortosa MC, Granados-Principal S, Perez-Lopez P, Naudi A, Portero-Otin M, Lopez-Frias M, Battino M, and Quiles JL. Age-related changes in brain mitochondrial DNA deletion and oxidative stress are differentially modulated by dietary fat type and coenzyme Q(1)(0). Free Radic Biol Med 50: 1053–1064, 2011 [DOI] [PubMed] [Google Scholar]
- 40.Okamoto T, Matsuya T, Fukunaga Y, Kishi T, and Yamagami T. Human serum ubiquinol-10 levels and relationship to serum lipids. Int J Vitam Nutr Res 59: 288–292, 1989 [PubMed] [Google Scholar]
- 41.Omar B, Zmuda-Trzebiatowska E, Manganiello V, Goransson O, and Degerman E. Regulation of AMP-activated protein kinase by cAMP in adipocytes: roles for phosphodiesterases, protein kinase B, protein kinase A, Epac and lipolysis. Cell Signal 21: 760–766, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onyango P, Celic I, McCaffery JM, Boeke JD, and Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA 99: 13653–13658, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, Ke H, Rehmann H, Taussig R, Brown AL, Kim MK, Beaven MA, Burgin AB, Manganiello V, and Chung JH. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 148: 421–433, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu X, Brown K, Hirschey MD, Verdin E, and Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab 12: 662–667, 2010 [DOI] [PubMed] [Google Scholar]
- 45.Quiles JL, Ochoa JJ, Battino M, Gutierrez-Rios P, Nepomuceno EA, Frias ML, Huertas JR, and Mataix J. Life-long supplementation with a low dosage of coenzyme Q10 in the rat: effects on antioxidant status and DNA damage. Biofactors 25: 73–86, 2005 [DOI] [PubMed] [Google Scholar]
- 46.Quiles JL, Pamplona R, Ramirez-Tortosa MC, Naudi A, Portero-Otin M, Araujo-Nepomuceno E, Lopez-Frias M, Battino M, and Ochoa JJ. Coenzyme Q addition to an n-6 PUFA-rich diet resembles benefits on age-related mitochondrial DNA deletion and oxidative stress of a MUFA-rich diet in rat heart. Mech Ageing Dev 131: 38–47, 2010 [DOI] [PubMed] [Google Scholar]
- 47.Rauchova H, Battino M, Fato R, Lenaz G, and Drahota Z. Coenzyme Q-pool function in glycerol-3-phosphate oxidation in hamster brown adipose tissue mitochondria. J Bioenerg Biomembr 24: 235–241, 1992 [DOI] [PubMed] [Google Scholar]
- 48.Rodgers JT, Lerin C, Gerhart-Hines Z, and Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett 582: 46–53, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossi DJ, Jamieson CH, and Weissman IL. Stems cells and the pathways to aging and cancer. Cell 132: 681–696, 2008 [DOI] [PubMed] [Google Scholar]
- 50.Safarinejad MR, Safarinejad S, and Shafiei N. Effects of the reduced form of coenzyme Q10 (ubiquinol) on semen parameters in men with idiopathic infertility: a double-blind, placebo controlled, randomized study. J Urol 188: 526–531, 2012 [DOI] [PubMed] [Google Scholar]
- 51.Saitoh Y, Hosokawa M, Shimada A, Watanabe Y, Yasuda N, Murakami Y, and Takeda T. Age-related cochlear degeneration in senescence-accelerated mouse. Neurobiol Aging 16: 129–136, 1995 [DOI] [PubMed] [Google Scholar]
- 52.Saitoh Y, Hosokawa M, Shimada A, Watanabe Y, Yasuda N, Takeda T, and Murakami Y. Age-related hearing impairment in senescence-accelerated mouse (SAM). Hear Res 75: 27–37, 1994 [DOI] [PubMed] [Google Scholar]
- 53.Schmelzer C. and Doring F. Micronutrient special issue: Coenzyme Q(10) requirements for DNA damage prevention. Mutat Res 733: 61–68, 2012 [DOI] [PubMed] [Google Scholar]
- 54.Schmelzer C, Kubo H, Mori M, Sawashita J, Kitano M, Hosoe K, Boomgaarden I, Doring F, and Higuchi K. Supplementation with the reduced form of Coenzyme Q10 decelerates phenotypic characteristics of senescence and induces a peroxisome proliferator-activated receptor-alpha gene expression signature in SAMP1 mice. Mol Nutr Food Res 54: 805–815, 2010 [DOI] [PubMed] [Google Scholar]
- 55.Schwer B, North BJ, Frye RA, Ott M, and Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol 158: 647–657, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sebastian C, Satterstrom FK, Haigis MC, and Mostoslavsky R. From sirtuin biology to human diseases: an update. J Biol Chem 287: 42444–42452, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seo BB, Kitajima-Ihara T, Chan EK, Scheffler IE, Matsuno-Yagi A, and Yagi T. Molecular remedy of complex I defects: rotenone-insensitive internal NADH-quinone oxidoreductase of Saccharomyces cerevisiae mitochondria restores the NADH oxidase activity of complex I-deficient mammalian cells. Proc Natl Acad Sci USA 95: 9167–9171, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shackelford DB. and Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer 9: 563–575, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shin J, Zhang D, and Chen D. Reversible acetylation of metabolic enzymes celebration: SIRT2 and p300 join the party. Mol Cell 43: 3–5, 2011 [DOI] [PubMed] [Google Scholar]
- 60.Sohal RS. and Weindruch R. Oxidative stress, caloric restriction, and aging. Science 273: 59–63, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Someya S, Xu J, Kondo K, Ding D, Salvi RJ, Yamasoba T, Rabinovitch PS, Weindruch R, Leeuwenburgh C, Tanokura M, and Prolla TA. Age-related hearing loss in C57BL/6J mice is mediated by Bak-dependent mitochondrial apoptosis. Proc Natl Acad Sci USA 106: 19432–19437, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, and Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143: 802–812, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stocker R. and Suarna C. Extracellular reduction of ubiquinone-1 and −10 by human Hep G2 and blood cells. Biochim Biophys Acta 1158: 15–22, 1993 [DOI] [PubMed] [Google Scholar]
- 64.Takeda T. [Senescence-accelerated mouse (SAM): with special reference to age-associated pathologies and their modulation]. Nihon Eiseigaku Zasshi 51: 569–578, 1996 [DOI] [PubMed] [Google Scholar]
- 65.Takeda T, Hosokawa M, and Higuchi K. Senescence-accelerated mouse (SAM): a novel murine model of senescence. Exp Gerontol 32: 105–109, 1997 [DOI] [PubMed] [Google Scholar]
- 66.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, and Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell 40: 893–904, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomasetti M, Littarru GP, Stocker R, and Alleva R. Coenzyme Q10 enrichment decreases oxidative DNA damage in human lymphocytes. Free Radic Biol Med 27: 1027–1032, 1999 [DOI] [PubMed] [Google Scholar]
- 68.Toyama K, Sugiyama S, Oka H, Iwasaki Y, Sumida H, Tanaka T, Tayama S, Jinnouchi H, Matsui K, and Ogawa H. Rosuvastatin combined with regular exercise preserves coenzyme Q10 levels associated with a significant increase in high-density lipoprotein cholesterol in patients with coronary artery disease. Atherosclerosis 217: 158–164, 2011 [DOI] [PubMed] [Google Scholar]
- 69.Voss P. and Siems W. Clinical oxidation parameters of aging. Free Radic Res 40: 1339–1349, 2006 [DOI] [PubMed] [Google Scholar]
- 70.Wada H, Goto H, Hagiwara S, and Yamamoto Y. Redox status of coenzyme Q10 is associated with chronological age. J Am Geriatr Soc 55: 1141–1142, 2007 [DOI] [PubMed] [Google Scholar]
- 71.Weindruch R, Walford RL, Fligiel S, and Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr 116: 641–654, 1986 [DOI] [PubMed] [Google Scholar]
- 72.Williams CB. and Gurd BJ. Skeletal muscle SIRT1 and the genetics of metabolic health: therapeutic activation by pharmaceuticals and exercise. Appl Clin Genet 5: 81–91, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiao B, Sanders MJ, Underwood E, Heath R, Mayer FV, Carmena D, Jing C, Walker PA, Eccleston JF, Haire LF, Saiu P, Howell SA, Aasland R, Martin SR, Carling D, and Gamblin SJ. Structure of mammalian AMPK and its regulation by ADP. Nature 472: 230–233, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan J, Fujii K, Yao J, Kishida H, Hosoe K, Sawashita J, Takeda T, Mori M, and Higuchi K. Reduced coenzyme Q10 supplementation decelerates senescence in SAMP1 mice. Exp Gerontol 41: 130–140, 2006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.