Abstract
There is intense interest in the development of small molecule inhibitors of the acetyl-lysine-reading bromodomain protein module. These inhibitors represent a way of interfering therapeutically in epigenetic processes, and there are currently two bromodomain inhibitors in clinical trials. The success of these compounds rests on safety aspects of epigenetic target modulation being addressed.
Epigenetic processes are currently the focus of intense interest in medicinal chemistry and drug discovery.1 The term “epigenetics” can be defined as involving heritable changes in gene expression or phenotype that are stable between cell divisions but which do not involve changes in the underlying DNA sequence.1 Such changes include methylation of DNA and post-translational modification (PTM) of histones, the core proteins around which nuclear DNA is packaged (Figure 1A). The most prevalent PTMs are methylation and acetylation of histone lysines, often on the exposed histone protein tails. However, an array of other modifications has been identified, including amino acid phosphorylation and addition of proteins such as ubiquitin. Consequently, it has been proposed that the specific pattern of histone PTMs, or marks, might represent a histone code, in which a certain pattern of marks regulates gene expression, leading to specific downstream effects. While probably an oversimplification, this concept has proved inspirational and led to the idea of writer, reader, and eraser proteins that modulate and interpret the histone code.
Figure 1.
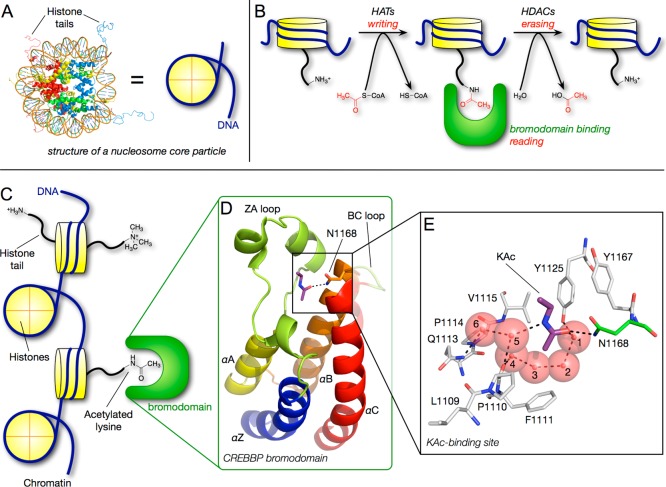
(A) Histones are the core proteins around which nuclear DNA is packaged. Histone H2A, yellow; histone H2B, red; histone H3, blue; and histone H4, green (PDB ID: 1KX5). (B) Histone acetyl-transferase (HAT) enzymes add an acetyl group to histone lysines and are viewed as “writers”, bromodomains bind to and “read” lysine acetylation, and histone deacetylases (HDACs) act as “erasers” and remove histone lysines. (C) Bromodomains are a KAc-recognition domain that act as “readers” of lysine acetylation state. (D) Bromodomains form a characteristic four-helix bundle comprising helices αZ, αA, αB, and αC. (E) Recognition of the KAc residue occurs via a direct hydrogen bond between the acetyl carbonyl oxygen atom and the N1168 (in CREBBP). A second interaction occurs between the acetyl carbonyl oxygen atom and the phenol of Y1125 (in CREBBP) via one of the structured water molecules.
Acetylation of histone lysines and the involvement of this PTM in increased gene transcription are well-established phenomena.2 The neutralization of the positively charged lysine ε-amino group, resulting from acetylation, reduces the electrostatic interaction with the negatively charged DNA phosphates, giving a more relaxed chromatin structure that is associated with transcriptionally active genes. Acetylation of histone lysine residues is achieved by histone acetyl-transferase (HAT) enzymes (writers), and acetyl groups are removed by histone deacetylases (HDACs, erasers) (Figure 1B). Two HDAC inhibitors, Vorinostat and Romidepsin, are in clinical use for the treatment of cutaneous T-cell lymphoma, demonstrating the possibility of treating cancer by modifying cellular lysine acetylation. In addition to manipulating chromatin structure, acetylated lysine (KAc) residues affect gene transcription through interaction with bromodomain-containing proteins (BCPs), a KAc-recognition domain that reads the lysine acetylation state (Figure 1C).3
Named after the Drosophila gene brahma, where they were first identified,4 bromodomains are ∼110 amino acid modules that exist as part of larger protein architectures. Many of these proteins regulate gene transcription and include HATs, methyltransferases, and transcriptional coactivators. X-ray crystallography reveals that bromodomains form a characteristic four-helix bundle comprising helices αZ, αA, αB, and αC (Figure 1D).3 The KAc residue binds in a defined pocket at one end of the helical bundle. This pocket is mainly hydrophobic in nature but typically contains four crystallographically well-ordered water molecules that form the base of the KAc-binding pocket (Figure 1E). Recognition of the KAc residue involves a direct hydrogen bond between the acetyl carbonyl oxygen atom and the NH2 group of a well-conserved asparagine amide. A second interaction occurs between the acetyl carbonyl oxygen atom and the phenol of a conserved tyrosine via one of the structured water molecules (Figure 1E). Crystallographic analysis coupled with bioinformatics studies suggest that these interactions are likely similar in most of the 61 bromodomains. Consequently, their ability to distinguish between different protein binding partners is thought to arise from sequence diversity in the ZA and BC loops, which bind the residues adjacent to KAc. Despite mediating a protein–protein interaction, this defined pocket represents a tractable target for small molecules.5 As BCPs have been associated with a number of diseases, including cancers, inflammation, neurological indications, and HIV,6 there has been significant interest in developing molecular tools to enable validation of BCPs as therapeutic targets.
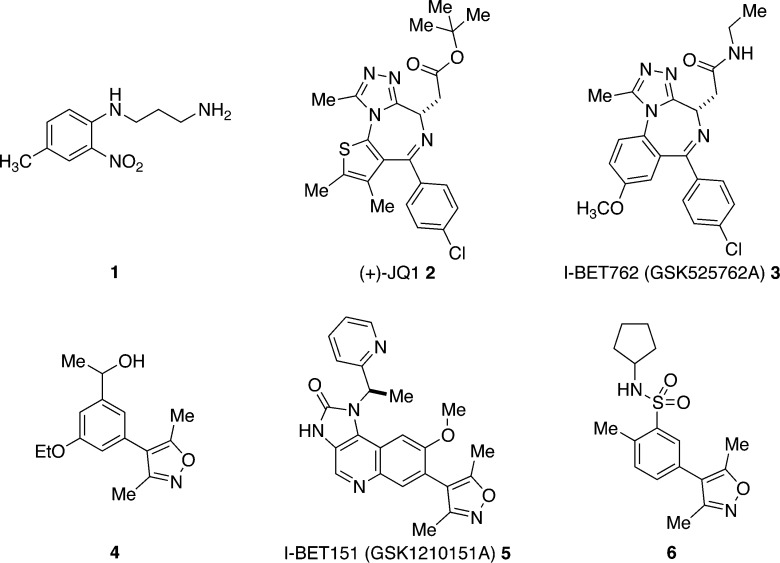
Pioneering work by Zhou et al. identified compound 1 (Figure 2) as the first small molecule ligand for a bromodomain (PCAF).7 Subsequently, the search for potent and selective bromodomain ligands was catalyzed by the publication of two methyltriazolodiazepine-based ligands (2 and 3, Figure 2) for the bromodomain and extra C-terminal domain (BET) family of BCPs.8,9 This class of BCPs comprises the tandem bromodomain-containing BRD2–4 and the testis-specific BCP, BRDT. (+)-JQ1 (2, Figure 2), reported by Filippakopoulos et al., is based on compounds disclosed in a patent from Mitsubishi Pharmaceuticals. This compound is a high-affinity BET bromodomain ligand with an IC50 value of 77 nM for the first bromodomain of BRD4 [BRD4(1)]. I-BET762 (3) was discovered by researchers at GlaxoSmithKline (GSK) using a phenotype screen for small molecule up-regulators of apolipoprotein A1 (ApoA1), which is involved in protection from atherosclerosis progression and anti-inflammatory effects.9 I-BET762 has IC50 values of 32.5–42.5 nM for the BET bromodomains. Both (+)-JQ1 and I-BET762 show good selectivity for the BET bromodomains over other bromodomain families. X-ray crystal structures show that both (+)-JQ1 and I-BET762 bind to the KAc-binding pocket of BRD4(1) and occupy part of the peptide-binding groove.
Figure 2.
Chemical structures of the bromodomain ligands 1–6.
A second chemotype, based on the 3,5-dimethylisoxazole KAc mimic, has also yielded potent and selective bromodomain ligands.10−12 Hewings et al. reported a small, ligand-efficient, 4-aryl-3,5-dimethylisoxazole derivative (4) with ∼5 μM affinity for BRD4(1).10 Dawson et al. reported another 4-aryl-3,5-dimethylisoxazole derivative (I-BET151, 5), which shows affinity of ∼200–790 nM for the BET bromodomains.11 Researchers at GSK have reported a second set of 4-aryl-3,5-dimethylisoxazole derivatives (e.g., 6) that are also potent and selective BET bromodomain inhibitors.12 The availability of X-ray structures of ligands bound to the BET bromodomains has enabled a structure-based design approach to the optimization of the 3,5-dimethylisoxazole-based ligands, and a good understanding of SAR is beginning to emerge.
(+)-JQ1, I-BET762, and I-BET151 have been used to probe the cellular function of the BET bromodomains in cancer and inflammation. (+)-JQ1 evokes differentiation, growth arrest, and apoptosis in patient-derived cell lines from the aggressive, BRD4-dependent, and lethal nuclear protein in testis (NUT) midline carcinoma (NMC). In patient-derived NMC mouse xenografts, (+)-JQ1 treatment led to tumor reduction and increased overall survival.8 (+)-JQ1 has also been used to demonstrate that BRD4 is potentially a therapeutic target for acute myeloid leukemia (AML).13 I-BET151 (5) is effective against xenograft leukemia models containing mixed lineage leukemia (MLL) gene fusions.11 Interestingly, the effects of compound 2 and 5 are due, at least in part, to down-regulation of the oncogene Myc.14 I-BET762 shows anti-inflammatory activity as a result of down-regulating pro-inflammatory genes. Additionally, Zhang et al. have shown that a (+)-JQ1 analogue suppressed NF-κB-dependent transcription of pro-inflammatory genes in kidney cells.15
GSK have recently announced a clinical trial that will investigate the use of I-BET762 to treat NMC. Given the BRD4-dependent nature of these cancers and the effects of (+)-JQ1 in patient-derived NMC mouse xenografts, this is a logical indication to be targeted by BET bromodomain inhibitors. Interestingly, the methyltriazolodiazepine-derived I-BET762 has been selected as the clinical candidate, although the 3,5-dimethylisoxazole-based I-BET151 showed better oral bioavailability and a longer half-life.16 Additionally, it has recently emerged that a compound developed by ResVerlogix, RVX-208, which is an ApoA1 up-regulator, functions by inhibiting the BET bromodomains. This compound is in phase 2 clinical trials for the treatment of both coronary atherosclerotic plaque and atherosclerotic cardiovascular disease. Approval of I-BET762 and RVX-208 as drugs would represent a very positive outcome for the bromodomain field. However, these clinical trails must address some important questions that face both bromodomain inhibitors and epigenetic medicine in general. There is concern over the safety of drugs, such as HDAC inhibitors, that alter the pattern of histone modification, given the potential heritability of these changes. Inhibition of reader domains might superficially seem to be a safer approach, if it is assumed that these compounds only block protein–histone interactions and do not alter the pattern of histone marks. However, many BCPs are part of catalytic proteins that do effect histone modification (e.g., the bromodomain-containing HAT, cAMP response element-binding protein binding protein [CREBBP]) or interact with other proteins that do. In the case of NMC, there is currently no effective treatment; hence, any improvement in patient survival or quality of life is to be welcomed. However, if bromodomains are to be targeted for the treatment of nonterminal diseases, then several questions concerning safety must be addressed. First, it must be established whether epigenetic modifications can be safely altered in the long term and whether inheritance of these alterations poses problems. Second, inhibition of BET bromodomain–histone interactions affects multiple genes, and it is unclear whether targeting this point in a pathway will result in toxic side effects. Furthermore, current BET bromodomain inhibitors show little selectivity between the members of the BET bromodomain family or between the first and the second bromodomains of each BET protein. It might be that selectivity between these bromodomains is required to give compounds with an acceptable side effect profiles.
Bromodomains represent exciting new therapeutic targets for which druglike small molecule ligands can be developed. Recent work in this field demonstrates that small molecules targeted at the KAc-binding pocket of bromodomains can effectively inhibit protein–protein interactions. Additionally, it seems that at least for oncology indications, targeting an epigenetic reader domain is a feasible therapeutic strategy. It is exciting that bromodomain ligands potentially present a new approach for manipulating targets that have previously seemed undruggable. For example, it has proved difficult to develop inhibitors of the Myc oncoprotein; however, it seems that at least some of the effects of BRD4 bromodomain inhibitors are as a result of modulating Myc expression. These tantalizing new opportunities warrant the development of further probe compounds that selectively and potently inhibit the bromodomain-KAc interaction. The success of bromodomain inhibitors as drugs will ultimately rely on the questions over long-term safety of these compounds being resolved. These questions can only be answered by the continued development of small molecule bromodomain inhibitors. Given the current interest in this field, it seems certain that potent and selective small molecule inhibitors of more bromodomains will be disclosed soon. These compounds will likely provide insight into the use of bromodomain inhibitors as drugs and the fundamental biology of these fascinating protein modules.
Acknowledgments
I am grateful to David Hewings, Laura Jennings, Tim Rooney, Prof. Chris Schofield, and Brian Wilson for critical reading of the manuscript. I apologize to authors whose work has not been cited due to space constraints.
I thank Cancer Research UK, EPSRC, GSK, Pfizer Neusentis, and UCB for funding my laboratory's work on bromodomain inhibitors.
The authors declare no competing financial interest.
References
- Arrowsmith C. H.; Bountra C.; Fish P. V.; Lee K.; Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discovery 2012, 11, 384–400. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Acetylation: A regulatory modification to rival phosphorylation?. EMBO J. 2000, 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P.; Knapp S. The bromodomain interaction module. FEBS Lett. 2012, 586, 2692–2704. [DOI] [PubMed] [Google Scholar]
- Haynes S. R.; Dollard C.; Winston F.; Beck S.; Trowsdale J.; Dawid I. B. The bromodomain: A conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992, 20, 2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidler L. R.; Brown N.; Knapp S.; Hoelder S. Druggability Analysis and Structural Classification of Bromodomain Acetyl-lysine Binding Sites. J. Med. Chem. 2012, 10.1021/jm300346w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C.-W.; Tough D. F. Bromodomains: A new target class for small molecule drug discovery. Drug Discovery Today: Ther. Strategies 2012, 10.1016/j.ddstr.2011.12.002. [DOI] [Google Scholar]
- Zeng L.; Li J.; Muller M.; Yan S.; Mujtaba S.; Pan C.; Wang Z.; Zhou M.-M. Selective small molecules blocking HIV-1 Tat and coactivator PCAF association. J. Am. Chem. Soc. 2005, 127, 2376–2377. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P.; Qi J.; Picaud S.; Shen Y.; Smith W. B.; Fedorov O.; Morse E. M.; Keates T.; Hickman T. T.; Felletar I.; Philpott M.; Munro S.; McKeown M. R.; Wang Y.; Christie A. L.; West N.; Cameron M. J.; Schwartz B.; Heightman T. D.; La Thangue N.; French C. A.; Wiest O.; Kung A. L.; Knapp S.; Bradner J. E. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E.; Jeffrey K. L.; Schaefer U.; Beinke S.; Dewell S.; Chung C.-W.; Chandwani R.; Marazzi I.; Wilson P.; Coste H.; White J.; Kirilovsky J.; Rice C. M.; Lora J. M.; Prinjha R. K.; Lee K.; Tarakhovsky A. Suppression of inflammation by a synthetic histone mimic. Nature 2010, 468, 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewings D. S.; Wang M.; Philpott M.; Fedorov O.; Uttarkar S.; Filippakopoulos P.; Picaud S.; Vuppusetty C.; Marsden B.; Knapp S.; Conway S. J.; Heightman T. D. 3,5-Dimethylisoxazoles Act As Acetyl-lysine-mimetic Bromodomain Ligands. J. Med. Chem. 2011, 54, 6761–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M. A.; Prinjha R. K.; Dittmann A.; Giotopoulos G.; Bantscheff M.; Chan W.-I.; Robson S. C.; Chung C.-W.; Hopf C.; Savitski M. M.; Huthmacher C.; Gudgin E.; Lugo D.; Beinke S.; Chapman T. D.; Roberts E. J.; Soden P. E.; Auger K. R.; Mirguet O.; Doehner K.; Delwel R.; Burnett A. K.; Jeffrey P.; Drewes G.; Lee K.; Huntly B. J. P.; Kouzarides T. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011, 478, 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamborough P.; Diallo H.; Goodacre J. D.; Gordon L.; Lewis A.; Seal J. T.; Wilson D. M.; Woodrow M. D.; Chung C.-W. Fragment-based discovery of bromodomain inhibitors part 2: Optimization of phenylisoxazole sulfonamides. J. Med. Chem. 2012, 55, 587–596. [DOI] [PubMed] [Google Scholar]
- Zuber J.; Shi J.; Wang E.; Rappaport A. R.; Herrmann H.; Sison E. A.; Magoon D.; Qi J.; Blatt K.; Wunderlich M.; Taylor M. J.; Johns C.; Chicas A.; Mulloy J. C.; Kogan S. C.; Brown P.; Valent P.; Bradner J. E.; Lowe S. W.; Vakoc C. R. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011, 478, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore J. E.; Issa G. C.; Lemieux M. E.; Rahl P. B.; Shi J.; Jacobs H. M.; Kastritis E.; Gilpatrick T.; Paranal R. M.; Qi J.; Chesi M.; Schinzel A. C.; McKeown M. R.; Heffernan T. P.; Vakoc C. R.; Bergsagel P. L.; Ghobrial I. M.; Richardson P. G.; Young R. A.; Hahn W. C.; Anderson K. C.; Kung A. L.; Bradner J. E.; Mitsiades C. S. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.; Liu R.; Zhong Y.; Plotnikov A. N.; Zhang W.; Zeng L.; Rusinova E.; Gerona-Nevarro G.; Moshkina N.; Joshua J.; Chuang P. Y.; Ohlmeyer M.; He J. C.; Zhou M.-M. Down-regulation of NF-κB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J. Biol. Chem. 2012, 10.1074/jbc.M112.359505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal J.; Lamotte Y.; Donche F.; Bouillot A.; Mirguet O.; Gellibert F.; Nicodeme E.; Krysa G.; Kirilovsky J.; Beinke S.; McCleary S.; Rioja I.; Bamborough P.; Chung C.-W.; Gordon L.; Lewis T.; Walker A. L.; Cutler L.; Lugo D.; Wilson D. M.; Witherington J.; Lee K.; Prinjha R. K. Identification of a novel series of BET family bromodomain inhibitors: binding mode and profile of I-BET151 (GSK1210151A). Bioorg. Med. Chem. Lett. 2012, 22, 2968–2972. [DOI] [PubMed] [Google Scholar]