Abstract
An in silico screen of the NIH Molecular Library Small Molecule Repository (MLSMR) of ∼350000 compounds and confirmatory bioassays led to identification of chaetochromin A (1) as an inhibitor of botulinum neurotoxin serotype A (BoNT A). Subsequent acquisition and testing of analogues of 1 uncovered two compounds, talaroderxines A (2) and B (3), with improved activity. These are the first fungal metabolites reported to exhibit BoNT/A inhibitory activity.
Keywords: in silico screen, botulinum neurotoxin serotype A, natural products, chaetochromin, talaroderxine, binding free energy, thermodynamic integration
Botulinum neurotoxins (BoNTs) are the etiologic agents of the neuroparalytic disorder botulism. BoNTs are among the most potent toxins known and are classified by the Centers for Disease Control and Prevention as one of the six highest risk threat agents for bioterrorism. The limitations of current therapies against botulism have prompted research for alternative therapeutics that could be used to treat pre- or postexposure BoNT intoxication, including studies to discover and develop small molecular weight inhibitors of BoNT.
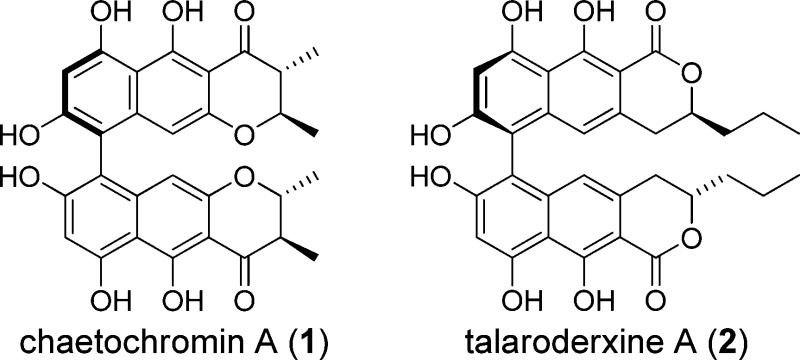
After demonstrating that a strategy of in silico screening, followed by tiered bioassay evaluation of screening hits, could successfully lead to confirmed inhibitors of botulinum neurotoxin serotype A (BoNT/A),1 we conducted a new round of in silico screening, this time using the NIH Molecular Library Small Molecule Repository (MLSMR) collection2 of ∼350000 compounds, for structures with likely binding affinity for the active site of BoNT/A. Among the first hits identified and confirmed active from that screen was chaetochromin A (1), a bis-naphtho-γ-pyrone produced by many species of Chaetomium(3) and more recently found in Fusarium spp.4 Herein, we report details of our in silico screen and the evaluation of chaetochromin A and several related compounds (2–6).
The in silico screen (Figure S1 in the Supporting Information) of the MLSMR was performed using Docking-based Virtual Screening Pipeline (DOVIS).5 DOVIS was used to systematically dock the MLSMR compounds to the catalytic binding site of the BoNTA protein target (PDB code 3C8A).6 The compounds were sorted by AutoDock 4.0 score,7 and the top 20000 compounds were chosen and minimized within a fixed BoNT/A catalytic site. After minimization, the compounds were rescored with three different scoring functions, AutoDock 4,7 LigScore 2,8 and X-Score.9 Three rescored lists were generated, with compounds sorted by corresponding docking score within each list. The top 500 compounds from each list were pooled for further analysis.
Two lists of compounds were generated at this point. The first, the consensus list, was populated with 300 compounds that appeared in all three rescored hit lists. The second, the MET-TOX list, consisted of 570 compounds that passed an imposed metabolism-toxicity (MET-TOX) filter, which contained mutagenic,10−10e hepatotoxic,11,11b and CYP-2D612,12b components. The consensus and MET-TOX lists were compared, and all unique compounds were retained in a combined list. The surviving compounds were visually inspected in the BoNT/A binding site. Using prior experience with this target, we removed compounds with unrealistic interactions or geometries, in particular, compounds incapable of coordination with the active site Zn2+.
Initially, a total of 100 chemically diverse compounds were selected from the combined list for testing (Figure S2 in the Supporting Information) for inhibition of the protease activity of BoNT/A LC using an HPLC-based assay.1 Chaetochromin A3,4,13,13b (MLS000563176, 1) was found to be the most active inhibitor in the group tested. A sample of chaetochromin A obtained from the NCI Repository (NSC 345647) for confirmatory testing exhibited only ∼50% inhibition at 100 μM, but subsequent HPLC-ELSD-MS analysis of this sample showed that it was <85% pure.
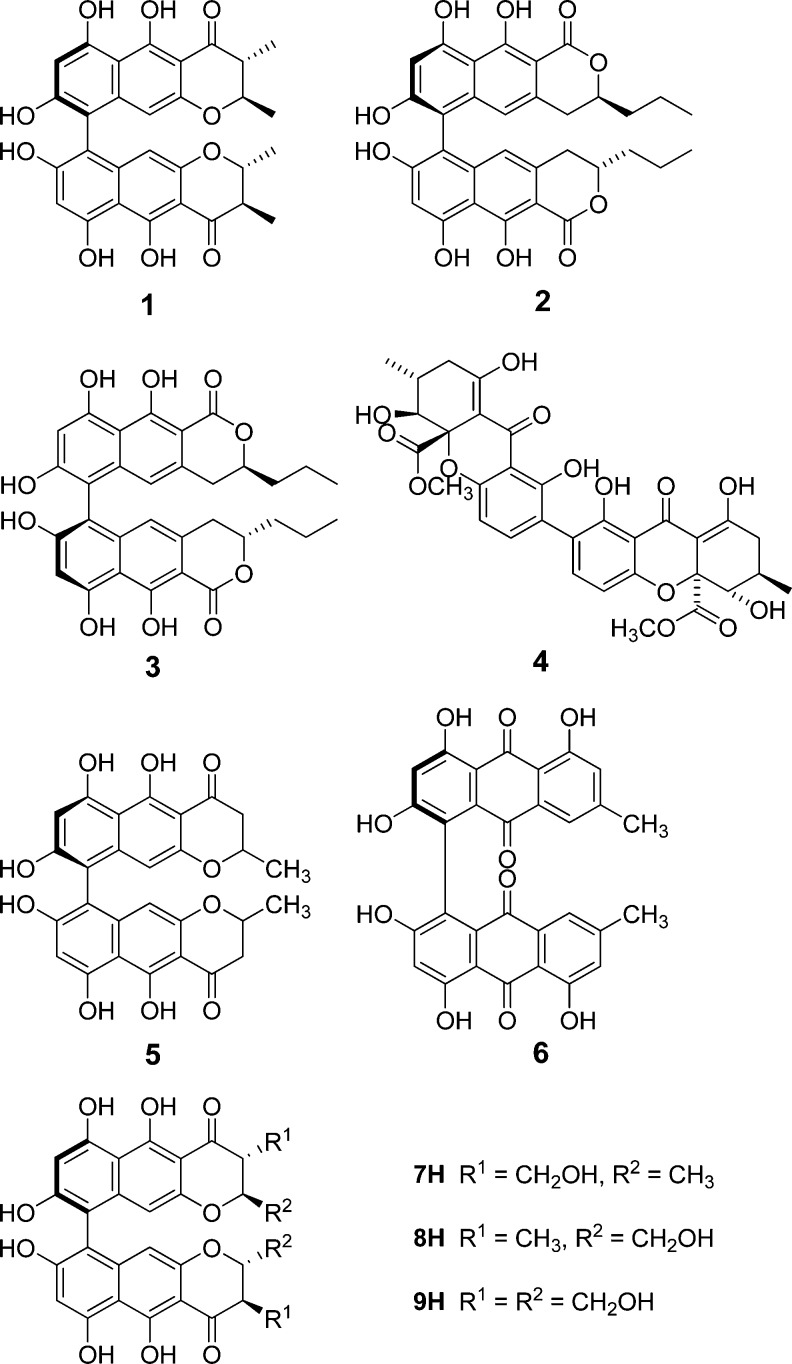
Samples of pure (≥95% by NMR) chaetochromin A (1) and several related compounds—talaroderxines A and B (2 and 3), secalonic acid A (4), cephalochromin (5), and skyrin (6)—were obtained from the fungal compound collection at the University of Iowa for comparative testing. Chaetochromin A (1) was isolated from solid-substrate fermentation cultures of Chaetomium arcuatum (syn. Chaetomium virescens) (NRRL 25243 = IMI 86456) isolated from a soil sample collected in Lucknow, India.14 Talaroderxines A and B (2 and 3) were obtained from liquid cultures of a coprophilous Delitschia sp. (JS 300) isolated from a sample of kangaroo dung collected in Australia. The talaroderxines were originally described as metabolites of Talaromyces derxii cultured from cultivated soil in Kurashiki, Japan,15 and, to our knowledge, have not been previously reported from any other source. Secalonic acid A (4)16,16b was obtained from a culture of Setophoma terrestris (NRRL 25008) obtained as a colonist of an Aspergillus flavus sclerotium that was buried in sandy field soil near Kilbourne, IL. Cephalochromin (5)13,13b,17,17b was obtained from a culture of Cosmospora sp. (syn. Acremonium butyri) (NRRL 28291) that was isolated as a colonist of the stromata of Hypoxylon sp. found on a hardwood log in Peoria, IL. Skyrin (6)18−18c was isolated from a culture of Geomyces pannorum (NRRL 22978) obtained as a colonist of an Aspergillus flavus sclerotium that was buried in sandy field soil near Kilbourne, IL. Compounds of this general type usually display axial chirality (i.e., atropisomerism) because of the high energy barrier to rotation at the bond linking the individual aromatic systems in each structure. The axial configurations of 1–3, 5, and 6 have been assigned and are reflected in the structures shown (Chart 1).
Chart 1.
Table 1 summarizes the results for both our primary HPLC-based protease assay and the tissue-based mouse phrenic nerve hemidiaphragm assay (MPNHDA). Not only was the activity of chaetochromin A (1), suggested by the in silico screen, confirmed, but talaroderxines A (2) and B (3) proved to be more active in the protease assay and showed considerable protection at 20 and 2 μM in the MPNHDA. While secalonic acid (4), cephalochromin (5), and skyrin (6) were roughly comparable to chaetochromin A in the protease assay, they were devoid of activity in the MPNHDA.
Table 1. Comparative Testing of Fungal Metabolitesa.
| BoNT/A
protease assay |
MPNHDAb |
||||
|---|---|---|---|---|---|
| % inhibition
(μM) |
paralytic
timed (μM) |
||||
| sample | 100 | 20 | IC50c (μM) | 20 | 2 |
| chaetochromin A (1) | 74.0 | 55.0 | 24.6 | 187.8e | 104.0 |
| talaroderxine A (2) | 96.0 | 85.0 | 3.7 | 197.0e | 188.7e |
| talaroderxine B (3) | 80.3 | 67.0 | 10.1 | 94.7f | 174.3e |
| secalonic acid (4) | 64.5 | 39.1 | 28.6 | 67.0 | NDg |
| cephalochromin (5) | 72.0 | 45.0 | 29.2 | 59.0 | NDg |
| skyrin (6) | 74.3 | 40.3 | 31.5 | 81.0 | 64.0 |
| CB 7969312h (control) | 98.0 | 96.2 | 2.1 | 300.0 | 300.0 |
Assays conducted as described in ref (1) and the Supporting Information.
Mouse phrenic nerve hemidiaphragm assay.
IC50 (50% inhibitory concentration) values were determined from nine concentrations of each compound using GraphPad Prism 4 (GraphPad Software, La Jolla, CA).
Average time to 50% loss of twitch tension (min); average value for BoNT/A toxin control, 62 min.
P value <0.01 (highly significant) in comparison with values recorded for the BoNT/A control; statistical analyses were performed using SigmaPlot 10 (Systat Software, San Jose, CA).
Solubility issues in the test medium at this concentration rendered this value unreliable.
Not determined.
A small molecule inhibitor described in ref (1).
In parallel with the biological testing, we performed virtual testing of some of these compounds using our lead optimization tools,19,20 including chaetochromin A (1), talaroderxine A (2), cephalochromin (5), and skyrin (6), along with three hypothetical side chain oxidized analogues of chaetochromin A (7H–9H). Secalonic acid was not included, due to substantial differences in its molecular scaffold relative to the bis-naphthopyrones.
Because no experimental structural data are available for BoNT/A-bound states of these compounds, we modeled them using AutoDock Vina.21 This docking algorithm preserved asymmetric carbon chirality but was insensitive to the higher order axial chirality of the compounds. Consequently, our modeled compounds docked with the BoNT/A active site in both P and M axial configurations. The docking score was not sufficient to rank-order closely related compounds and even less so to choose the P atropisomer over the M. Visual inspection revealed two representative consensus poses, one for each atropisomer, that had carbonyl and/or hydroxyl oxygens poised to coordinate active site Zn2+. Figure 1 illustrates the consensus poses for 1–3. However, only the P atropisomer was capable of coordinating Zn2+ by carbonyl and hydroxyl oxygens at the same time (Figure 1A,B). On the basis of this finding and on the fact that the original hit molecule, chaetochromin A (1), had the P configuration, we selected the pose with the P configuration for subsequent binding free energy calculations, excluding talaroderxine B (3).
Figure 1.
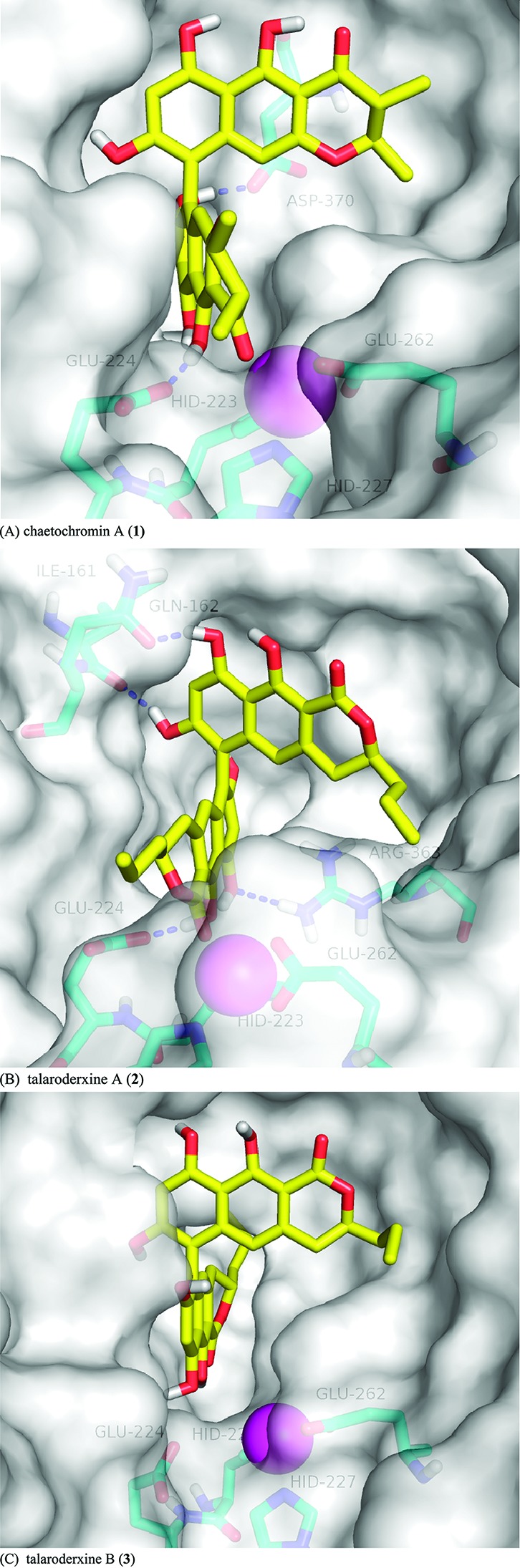
Proposed binding mode of (A) chaetochromin A (1), (B) talaroderxine A (2), and (C) talaroderxine B (3) in the active site of BoNT/A. The Zn2+ ion is shown as a magenta sphere, carbon atoms of the ligands are depicted in yellow, and those of the active site residues are in cyan. The molecular surface of the pocket is shown in a transparent light gray color. For clarity, only polar hydrogens are shown. Panels A and B depict the most populated cluster centers from SRTI-HREX simulations, whereas panel C is the result of docking.
Using the consensus pose with the P configuration, we rank-ordered the ligands by their relative binding free energies, which were computed using computationally demanding Single Reference Thermodynamic Integration (SRTI) approach augmented with Hamiltonian Replica Exchange (HREX).19,20 Because binding free energy predictions often depend on the initial structure of the complex, it was imperative that we employ a similar pose for all of the compounds as the consensus pose. For the same reason and because axial atropisomers do not readily interconvert in SRTI simulations, it was not possible to compare P and M atropisomers using only the relative binding free energy calculations.
Adequate sampling of the conformations of the numerous hydroxyl groups might be important for obtaining converged results. Indeed, each of the studied compounds has at least six hydroxyl and two carbonyl groups that can interact by forming internal hydrogen bonds. The formation of internal hydrogen bonds would reduce the number of favorable interactions with the BoNT/A active site and could affect the binding free energies. To ensure adequate sampling of the hydroxyl group rotations, we employed a specially designed nonphysical reference state (see the Supporting Information) that allowed for free rotation of the hydroxyl groups. Using this reference state in combination with the HREX option within SRTI enhanced conformational transitions of hydroxyl groups. Therefore, we expected HREX-SRTI to provide more accurate relative binding free energy predictions than conventional TI methods.
The results of the calculations are summarized in Table 2, with the representative complexes of the top scoring compounds shown in Figure 1A,B. Despite all of the precautions, our predictions had relatively large standard deviations that, in some cases, precluded our making a definitive prediction. Nevertheless, our calculations had sufficient precision to predict talaroderxine A (2) as a better binder than chaetochromin A (1). Interestingly, one of our designs (7H) was also predicted to be a better binder than chaetochromin A. This prediction remains to be tested experimentally, as the compound is as yet unknown. Although the precision of our calculations for the remaining compounds did not allow us to make definitive conclusions, the predicted changes in binding free energies appear to be in good agreement with experimentally determined IC50 values (Table 1).
Table 2. Predicted Binding Free Energies/Standard Deviations Relative to Chaetochromin Aa.
| compound | ΔΔG | SD |
|---|---|---|
| chaetochromin A (1) | 0.0 | 1.5 |
| talaroderxine A (2) | –4.7 | 2.1 |
| cephalochromin (5) | 1.4 | 1.5 |
| skyrin (6) | 0.2 | 1.1 |
| 7H | –2.6 | 1.3 |
| 8H | 0.7 | 1.6 |
| 9H | –0.3 | 2.8 |
The correlation between the predicted binding free energies and the observed IC50 values lends additional credibility to the consensus pose selected for calculations. In the absence of a crystallographic structure, we can use this validated pose for further lead optimization efforts.
We should note, however, that the higher activity of talaroderxine B relative to chaetochromin A might suggest that the consensus pose with the M configuration is also important (Figure 1C). Nevertheless, the observation that talaroderxine A (2) is more active than talaroderxine B (3) supports our emphasis on the P atropisomer for the present study.
Starting with the initial in silico screening hit chaetochromin A (1), we compared a small number of analogues and found that talaroderxines A (2) and B (3) were more potent in our primary HPLC-based and secondary mouse phrenic nerve hemidiaphragm assays. It is noteworthy that 1 and 2 share the same axial configuration and absolute configuration at the chiral centers they have in common; 3 has the same configuration at the chiral centers held in common, but the opposite axial chirality. The primary differences between 1 and 2 and 3 reside in the γ- versus α-pyrone and the methyl versus n-propyl substituents at the common chiral centers. The task before us now is to develop an adequate supply of 2 and 3, so that we can define their mechanism of action/mode of binding, assess their in vivo activity, and determine whether they are less toxic than chaetochromin A.22−26 Because talaroderxines A and B (2 and 3) are produced as a mixture in relatively low yields, this will be challenging but not insurmountable. These are the first fungal natural products reported to inhibit BoNT/A.
Acknowledgments
We are grateful to N. R. Torabazari for technical support with the MPNHDA.
Supporting Information Available
Experimental protocols for the bioassays, isolation of the fungal compounds, and molecular modeling and docking of the fungal metabolites into BoNT/A light chain. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by a grant awarded to L.A.S. from the Defense Threat Reduction Agency, JSTO-CBD Project Number 3.10037_07_RD_B. This work was also sponsored by the U.S. Department of Defense (DOD) High Performance Computing Modernization Program under the High Performance Computing Software Applications Institutes initiative. Research at the University of Iowa was supported by a grant from the National Science Foundation (CHE-11011847).
The opinions or assertions contained herein are the private views of the authors and not to be construed as reflecting the official views of the U.S. DOD.
The authors declare no competing financial interest.
Author Present Address
# NIH Chemical Genomics Center, National Human Genome Research Institute, NIH, Bethesda, Maryland 20892-3370.
Supplementary Material
References
- Roxas-Duncan V.; Enyedy I.; Montgomery V. A.; Eccard V. S.; Carrington M. A.; Lai H.; Gul N.; Yang D. C.; Smith L. A. Identification and biochemical characterization of small-molecule inhibitors of Clostridium botulinum neurotoxin serotype A. Antimicrob. Agents Chemother. 2009, 53, 3478–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH Molecular Libraries Small Molecule Repository (https://mlsmr.glpg.com/MLSMR_HomePage/centers.html): A collection of samples for high throughput biological screening distributed to the NIH Molecular Libraries Probe Production Centers Network. NCGC (NIH Chemical Genomics Center) is a member laboratory of this network.
- Sekita S.; Yoshihira K.; Natori S. Chaetochromin, a bis(naphthodihydropyran-4-one) mycotoxin from Chaetomium thielavioideum: Application of 13C-1H long-range coupling to the structure elucidation. Chem. Pharm. Bull. 1980, 28, 2428–2435. [Google Scholar]
- Singh S. B.; Zink D. L.; Bills G. F.; Teran A.; Silverman K. C.; Lingham R. B.; Felock P.; Hazuda D. J. Four novel bis-(naphtho-γ-pyrones) isolated from Fusarium species as inhibitors of HIV-1 integrase. Bioorg. Med. Chem. Lett. 2003, 13, 713–717. [DOI] [PubMed] [Google Scholar]
- Jiang X; Kumar K.; Hu X.; Wallqvist A.; Reifman J. DOVIS 2.0: An efficient and easy to use parallel virtual screening tool based on AutoDock 4.0. Chem. Cent. J. 2008, 2, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran C.; Rawat F.; Ludivico M. L.; Swaminathan S. Structure- and substrate-based inhibitor design for Clostridium botulinum neurotoxin serotype A. J. Biol. Chem. 2008, 283, 18883–18891. [DOI] [PubMed] [Google Scholar]
- Huey R.; Morris G. M.; Olson A. J.; Goodsell D. S. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007, 28, 1145–1152. [DOI] [PubMed] [Google Scholar]
- Krammer A.; Kirchoff P. D.; Jiang X.; Venkatachalam C. M.; Waldman M. LigScore: A novel scoring function for predicting binding affinities. J. Mol. Graphics Modell. 2005, 23, 395–407. [DOI] [PubMed] [Google Scholar]
- Wang R.; Lai L.; Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J. Comput.-Aided Mol. Des. 2002, 16, 11–26. [DOI] [PubMed] [Google Scholar]
- Accelrys (www.accelrys.com), Pipeline Pilot ADMET_EXT_Mutagen Component, was used to model the data: Kazius J.; McGuire R.; Bursi R.. Derivation and validation of toxicophores for mutagenicity prediction. J. Med. Chem. 2005, 48, 312–320. [DOI] [PubMed] [Google Scholar]
- Contrera J. F.; Matthews E. J.; Kruhlak N. L.; Benz R. D. In silico screening of chemicals for bacterial mutagenicity using electrotopological E-state indices and MDL QSAR software. Regul. Toxicol. Pharmacol. 2005, 43, 313–323. [DOI] [PubMed] [Google Scholar]
- Feng J.; Lurati L.; Ouyang H.; Robinson T.; Wang Y.; Yuan S.; Young S. S. Predictive toxicology: Benchmarking molecular descriptors and statistical methods. J. Chem. Inf. Comput. Sci. 2003, 1463–1470. [DOI] [PubMed] [Google Scholar]
- Helma C.; Cramer T.; Kramer S.; De Raedt L. Data mining and machine learning techniques for the identification of mutagenicity inducing substructures and structure activity relationships of noncongeneric compounds. J. Chem. Inf. Comput. Sci. 2004, 44, 1402–1411. [DOI] [PubMed] [Google Scholar]
- Xia X.; Maliski E., G.; Gallant P.; Rogers D. Classification of kinase inhibitors using a Bayesian model. J. Med. Chem. 2004, 47, 4463–4470. [DOI] [PubMed] [Google Scholar]
- Accelrys (www.accelrys.com), Pipeline Pilot ADMET_EXT_Hepatotoxic Component: Cheng A.; Dixon S. L.. In silico models for the prediction of dose-dependent human hepatotoxicity. J. Comput.-Aided Mol. Des. 2003, 17, 811–823. [DOI] [PubMed] [Google Scholar]
- Three hundred eighty-two of the compounds are from the original training data, with the remaining 54 from the validation data. However, the modeling methodology described in that paper was not used; instead, modified Bayesian learning, described inXia X.; Maliski E.; G.;Gallant P.; Rogers D. Classification of kinase inhibitors using a Bayesian model. J. Med. Chem. 2004, 47, 4463–4470was performed. [DOI] [PubMed] [Google Scholar]
- Accelrys (www.accelrys.com), Pipeline pilot ADMET_EXT_CYP2D6_Inhibitor Component: Susnow R., G.; Dixon S. L.. Use of robust classification techniques for the pediction of human cytochrome P450 2D6 inhibition. J. Chem. Inf. Comput. Sci. 2003, 43, 1308–1315. [DOI] [PubMed] [Google Scholar]
- Xia X.; Maliski E., G.; Gallant P.; Rogers D. Classification of kinase inhibitors using a Bayesian model. J. Med. Chem. 2004, 47, 4463–4470. [DOI] [PubMed] [Google Scholar]
- Koyama K.; Natori S. Chaetochromins B, C and D, bis(naphtho-gamma-pyrone) derivatives from Chaetomium gracile. Chem. Pharm. Bull. 1987, 35, 578–584. [DOI] [PubMed] [Google Scholar]
- Koyama K.; Natori S.; Iitaka Y. Absolute configurations of chaetochromin A and related bis(naphtha-γ-pyrone) mold metabolites. Chem. Pharm. Bull. 1987, 35, 4049–4055. [Google Scholar]
- Wicklow D. T.; Dowd P. F.; Gloer J. B.. Chaetomium mycotoxins with antiinsectan or antifungal activity. In Mycotoxin Contamination: Health Risk and Prevention Project; Proceedings of International Symposium of Mycotoxicology '99, Chiba, Japan. Mycotoxins: Supplement 99; Kumagai S., Ed.; Matsumoto Printing Co.: Tokyo, Japan, 2000; pp 267–271. [Google Scholar]
- Suzuki K.; Nozawa K.; Nakajima S.; Udagawa S.; Kawai K. Isolation and structures of antibacterial binaphtho-α-pyrones, talaroderxines A and B, from Talaromyces derxii. Chem. Pharm. Bull. 1992, 40, 1116–1119. [DOI] [PubMed] [Google Scholar]
- Steffens J. C.; Robeson D. J. Secalonic acid A, a vivotoxin in pink root-infected onion. Phytochemistry 1987, 26, 1599–1602. [Google Scholar]
- Howard C. C.; Johnstone R. A. W. Fungal metabolites. Part I. Stereochemical features and mass spectrometry of secalonic acids. J. Chem. Soc. Perkin Trans. I 1973, 2440–2444. [PubMed] [Google Scholar]
- Hegde V. R.; Miller J. R.; Patel M. G.; King A. H.; Puar M. S.; Horan A.; Hart R.; Yarborough R.; Gullo V. SCH 45752—An inhibitor of calmodulin-sensitive cyclic nucleotide phosphodiesterase activity. J. Antibiot. 1993, 46, 207–213. [DOI] [PubMed] [Google Scholar]
- Kock. I.; Draeger S.; Schulz B.; Elsässer B.; Tibor Kurtán T.; Kenéz A.; Antus S.; Pescitelli G.; Salvadori P.; Speakman J.-B.; Rheinheimer J.; Krohn K. Pseudoanguillosporin A and B: Two new isochromans isolated from the endophytic fungus Pseudoanguillospora sp. Eur. J. Org. Chem. 2009, 1427–1434. [Google Scholar]
- Gill M.; Gimenez A.; McKenzie R. W. Pigments of fungi, part 8. Bisanthraquinones from Dermocybe austroveneta. J. Nat. Prod. 1988, 51, 1251–1256. [Google Scholar]
- Antonowitz A.; Gill M.; Morgan P. M.; Yu J. Coupled anthraquinones from the toadstool Dermocybe icterinoides. Phytochemistry 1994, 37, 1679–1683. [Google Scholar]
- Koyama K.; Aida S.; Natori S. Supplemental observations on atropisomerism of fungal bis(naphtho-γ-pyrones). Chem. Pharm. Bull. 1990, 38, 2259–2261. [Google Scholar]
- Khavrutskii I. V.; Wallqvist A. Computing relative free energies of solvation using Single Reference Thermodynamic Integration augmented with Hamiltonian Replica Exchange. computing relative free energies of solvation using Single Reference Thermodynamic Integration augmented with Hamiltonian Replica Exchange. J. Chem. Theory Comput. 2010, 6, 3427–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavrutskii I. V.; Wallqvist A. Improved binding free energy predictions from Single-Reference Thermodynamic Integration augmented with Hamiltonian Replica Exchange. J. Chem. Theory Comput. 2011, 7, 3001–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K. Oral toxicity of chaetochromin, a new mycotoxin produced by Chaetomium virescens, to mice. Proc. Jpn. Assoc. Mycotoxicol. 1980, 28–29. [Google Scholar]
- Ito Y.; Ohtsubo K. Exencephaly in mice induced by feeding chaetochromin-containing diet. Proc. Jpn. Assoc. Mycotoxicol. 1982, 22–23. [Google Scholar]
- Ito Y.; Ohtsubo K. Teratogenicity of oral chaetochromin, a polyphenolic mycotoxin produced by Chaetomium spp., to mice embryo. Bull. Environ. Contam. Toxicol. 1987, 39, 299–303. [DOI] [PubMed] [Google Scholar]
- Koyama K.; Ominato K.; Natoria S.; Tashiro T.; Tsuruo T. Cytotoxicity and antitumor activities of fungal bis(naphtho-γ-pyrone) derivatives. J. Pharmacobio-Dyn. 1988, 11, 630–635. [DOI] [PubMed] [Google Scholar]
- Ohtsubo K. Myocardial and hepatic necrosis and bone marrow suppression in mice induced by single administration of chaetochromin, a polyphenolic mycotoxin produced by Chaetomium spp. J. Toxicol. Pathol. 1989, 2, 163–174. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.