Abstract
The relationship between enzyme inhibition and antimicrobial potency of adenine-based NAD+-dependent DNA ligase (LigA) inhibitors was investigated using a strain of the Gram-negative pathogen Haemophilus influenzae lacking its major AcrAB-TolC efflux pump and the Gram-positive pathogen Streptococcus pneumoniae. To this end, biochemical inhibitors not mediating their antibacterial mode of action (MOA) via LigA were removed from the analysis. In doing so, a significant number of compounds were identified that acted via inhibition of LigA in S. pneumoniae but not in H. influenzae, despite being inhibitors of both isozymes. Deviations from the line correlating antimicrobial and biochemical potencies of LigA inhibitors with the correct MOA were observed for both species. These deviations, usually corresponding to higher MIC/IC50 ratios, were attributed to varying compound permeance into the cell.
Keywords: permeance, LigA, mode of action, antimicrobial, biochemical potency, efflux
NAD+-dependent DNA ligases (LigA) are essential for the formation of phosphodiester bonds at single-strand breaks in double-stranded DNA, such as those that occur as part of DNA repair and ligation of Okazaki fragments during DNA replication.1,2 There is genetic evidence in a wide range of bacterial species that LigA is essential for growth.3 Because the human counterpart of this class of enzymes is ATP-dependent, this has led to the expectation that bacterial-specific inhibitors could be identified, which could serve as starting points for the development of antibacterials for clinical use.4,5 Several laboratories have screened their compound libraries for LigA inhibitors with enzyme assays, identified improved analogues with antimicrobial activity, and shown that this activity was mediated by inhibition of LigA. In this way, pyridochromanones6 and pyridopyrimidines7 both provided in vitro validation of LigA; that is, these compounds inhibited bacterial growth by means of chemical inhibition of this intracellular target.
Recently, a class of adenine-based LigA inhibitors was identified that resulted from a high-throughput screen of AstraZeneca's proprietary compound library using a biochemical assay. Iterative chemistry efforts led to sufficient improvement of antimicrobial potency and biophysical properties such that adequate systemic exposure could be achieved, resulting in efficacy in animal models of infection.3 This major step forward validated LigA as an antibacterial target in vivo. The analysis of the relationship between enzyme inhibition and antimicrobial potency described here focused on the initial chemistry effort that improved the antibacterial activity of the original screening hit.8
All compounds were tested for biochemical potency against Haemophilus influenzae LigA3 in an enzyme assay, and 320 compounds showed an inhibitory concentration at which activity was reduced by 50% (IC50) of less than 50 μM. The antimicrobial activity of these actives was quantified as minimum inhibitory concentration (MIC)9 using H. influenzae HIN101 that lacks activity of the multidrug efflux transporter AcrABTolC via which adenosines to various extents are extruded (see below).3,10 There were 219 compounds with an MIC of 64 μg/mL and lower. To assess the mode of action (MOA), the MICs were compared between parental strain H. influenzae HIN101 and an isogenic strain overexpressing LigA, H. influenzae HIN102.3 If the antibacterial activity were mediated via LigA, the MIC would be higher in the overexpressing strain. Because a 4-fold difference reliably could be measured, this was the minimal elevation to conclude that the MOA in the parental strain was via LigA. For 54 less potent compounds, the higher concentration could not be tested due to the fixed maximum test concentration, 64 μg/mL, in which case the result was inconclusive. For 56 compounds, the antimicrobial activity remained unchanged, indicating that the MOA in the test strain was mediated by a target other than LigA. It should be noted that in typical target-based drug discovery projects, these orphaned compounds are abandoned since they have antimicrobial activity not related to biochemical inhibition of the target of interest. These could, however, serve as new drug discovery starting points, in particular upon discovery of their cellular target.
In summary, 109 compounds with a MOA via LigA in H. influenzae were identified. This is not a trivial result. Adenine-based LigA inhibitors are rapidly reversible inhibitors that are competitive with NAD+.3 Their potency is determined in biochemical assays containing NAD+ at a low multiple of its Km of around 1–10 μM. Experimentally determined intracellular NAD+ concentrations (1–10 mM11) are orders of magnitude higher, suggesting that outcompeting NAD+ at these levels with a reversible inhibitor would be virtually impossible, in particular since a large fraction of metabolic flux via LigA needs to be inhibited before bacterial growth is halted.12 On the other hand, evolutionary considerations have led to the assumption that substrate concentrations in the vicinity of a molecular target would be in the range of an enzyme's Km.13 The ability to identify NAD+-competitive inhibitors of LigA, which apparently reach sufficiently high intracellular concentrations to mediate their antimicrobial action via this target, seems to support this last prediction.
Because there is precedence for antimicrobial compounds having different MOA in Gram-positive and Gram-negative species,14 a similar analysis was performed with Streptococcus pneumoniae using a LigA-overexpressing strain, S. pneumoniae SPN102, isogenic with the parental strain, S. pneumoniae SPN100.3 This resulted in 203 antimicrobial compounds of which 169 displayed a MOA via LigA, 14 did not, and 20 were inconclusive. Of these 169, 99 compounds were antimicrobial and acted via LigA in both species. Therefore, the relative number of compounds that were shown not to act via the desired target was four times higher (56 vs 14) in H. influenzae than in S. pneumoniae. This indicated that a significant number of compounds were antimicrobial against both species but only acted via LigA in S. pneumoniae, despite being bona fide inhibitors of both isozymes. In the examples shown in Table 1, replacement of the ribose moiety with tetrahydrofuran and large substitutions at either the adenine 2′-hydroxyl, the ribose 5′-carbon, or the 3′-hydroxyl altered the MOA in H. influenzae, yet left that in S. pneumoniae unchanged. For these compounds, the loss of the correct MOA in H. influenzae was correlated with a loss of potency in the enzyme inhibition assay and elevated MIC, whereas for S. pneumoniae, both potency in the enzyme inhibition assay and MIC were retained. This pattern was also apparent from antimicrobial activities against the overexpression strains of both species (Table 2). Increased cellular target levels in the overexpression strain HIN102 caused diminished antimicrobial potency showing that in the parental strain HIN101 the MOA was via LigA. The MOA of that same compound in the overexpression strain, however, was not necessarily clear. The maximum increase in MIC upon overexpression was at least 16- and 64-fold for H. influenzae and S. pneumonia, respectively3 (Table 2), and this activity in the overexpression strain was potentially mediated via inhibition of LigA (hence “inconclusive” in Table 2). However, this again implied that adenosines that showed a lower fold increase caused an antimicrobial effect via a different mechanism when present at higher concentrations in the overexpression strain, even though the MOA in the parental strain was via LigA. Determination of the alternative target(s) in each species will be required to explain the increased number of compounds that do not act via LigA in H. influenzae vs that in S. pneumoniae. However, this finding emphasizes that MOA determinations cannot be extrapolated between Gram-positive and Gram-negative species.14
Table 1. Examples of Substitutions in Adenine-Based LigA Inhibitors Resulting in an Alternative MOA in H. influenzae but Not in S. pneumoniae.
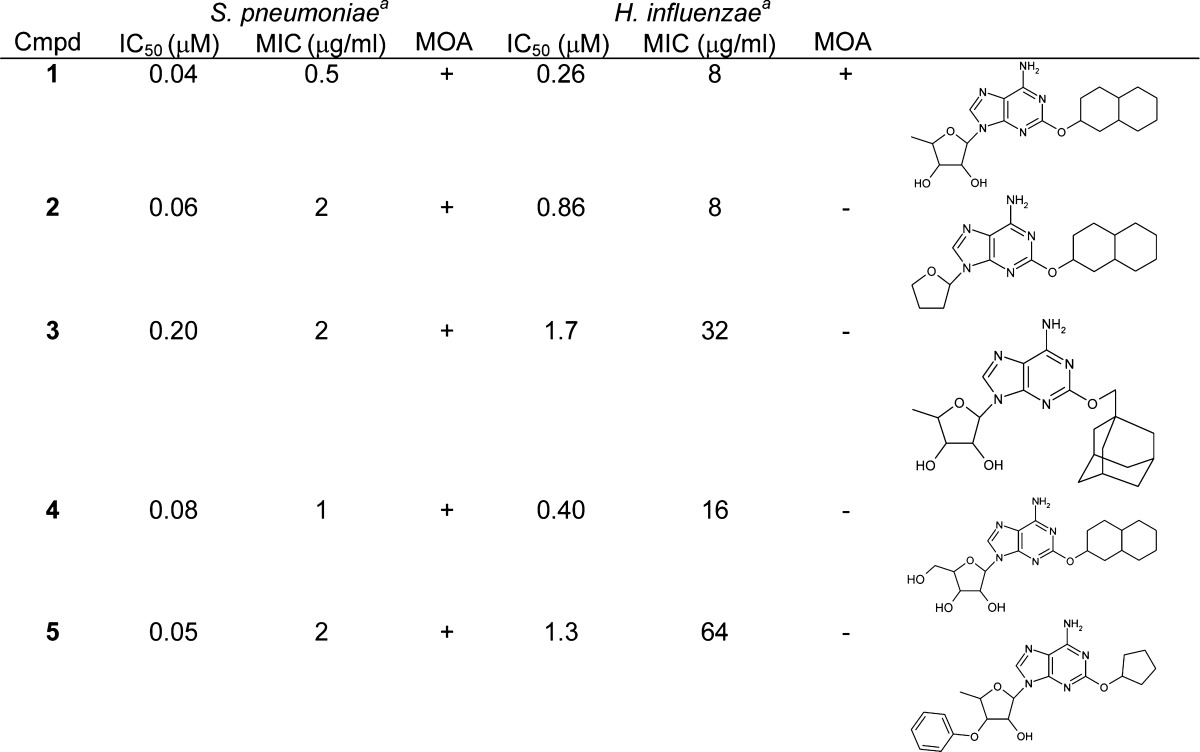
MIC and MOA obtained in the test strain S. pneumoniae SPN100 and H. influenzae HIN101; MOA via LigA (+) or via another yet undetermined target (−).
Table 2. Examples of MOA Determination in Both Parental (Hin101 and Spn100) and Overexpression (Hin102 and Spn102) Strains of H. influenzae and S. pneumoniae.
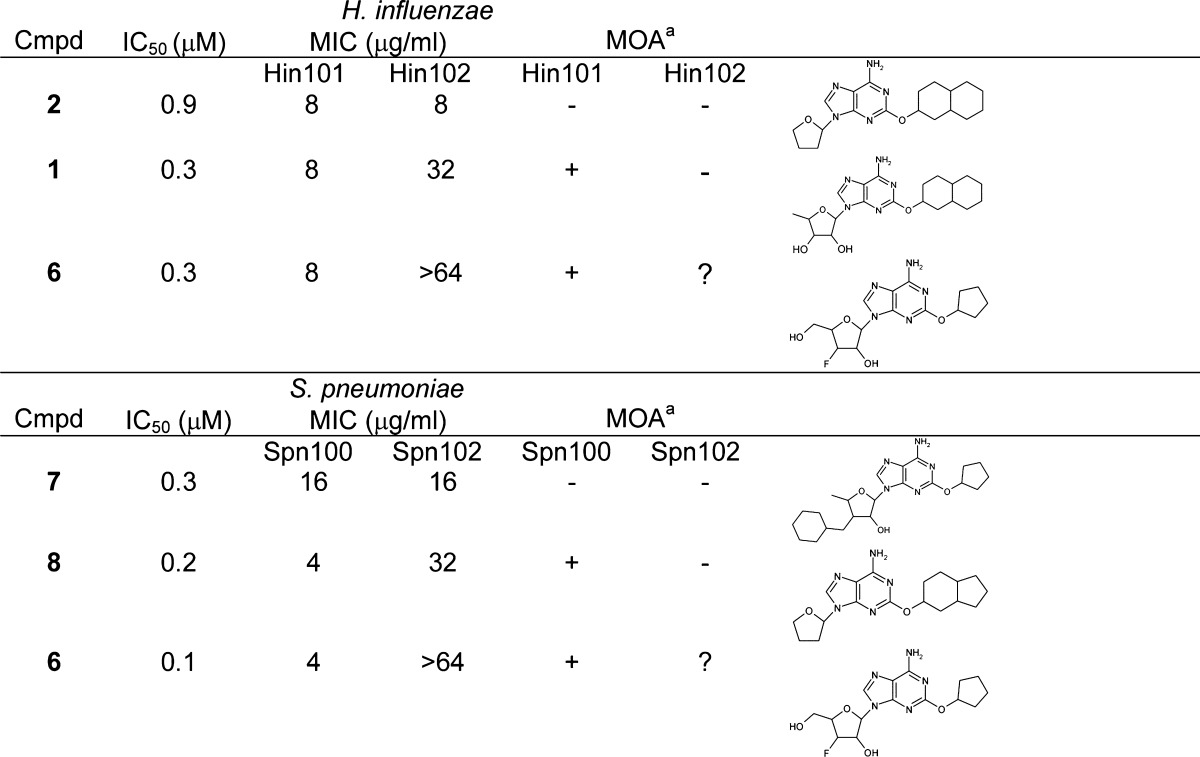
MOA via LigA (+), via another yet undetermined target (−), or an inconclusive result (?).
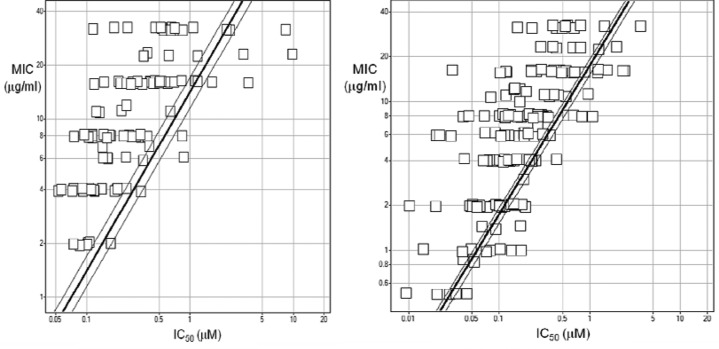
In order for a rapidly reversible, biochemical inhibitor of an intracellular target to result in inhibition of cell growth, it needs to reach a sufficiently high intracellular concentration, as required by its biochemical potency, in a critical amount of time. In doing so, its rate of permeation also requires to compensate for continuous compound dilution, a consequence of, at least initially, uninterrupted cell enlargement and division. If permeant properties vary among biochemically equipotent inhibitors, then a linear relationship between IC50 and MIC is not to be expected. Rather, a triangular pattern relating IC50 to MIC should result, in which a range of MICs is attained by biochemically equivalent inhibitors. This is exactly what was observed for LigA inhibitors (Figure 1). For example, a biochemical potency of IC50 equal to 100 nM yielded MICs as low as 1 μg/mL and as high as 32 μg/mL, and conversely, among compounds with an MIC of 8 μg/mL, IC50 values varied from 50 nM to 1 μM (Figure 1). The minimum MICs observed for given IC50 values lie along the hypotenuse of the triangular relationship and indicate a lower limit where MIC is dominated by biochemical potency. These limits were fitted empirically for LigA inhibitors in H. influenzae and S. pneumoniae, using simple least-squares regression weighted by the MIC/IC50 ratio for each compound, and constrained to proportionality between MIC and IC50. The line is thus formed by compounds with the best permeance characteristics while accounting for experimental variation. In both strains, it was found that the line corresponded to a “best-case” MIC/IC50 ratio of about 15 (mol/mol, assuming a MW of 500). Thus, all LigA inhibitors with this biochemical mode of inhibition and optimum permeance characteristics are expected to exhibit MICs no better than about 15 times the IC50.
Figure 1.
Correlation between biochemical and antimicrobial potency of (left) 109 adenine-based inhibitors of H. influenzae LigA against H. influenzae HIN101, a strain lacking the AcrB efflux pump subunit, and (right) 169 adenine-based inhibitors of S. pneumoniae LigA against S. pneumoniae SPN100, which mediate their antibacterial MOA via this target. The triangular pattern is characterized by its hypotenuse, shown as a line displaying a linear relationship between the two parameters when compound permeation is thought not to limit antimicrobial potency. Parallel lines indicate a 95% confidence interval.
Reversible biochemical inhibitors resulting from target-based screening efforts may have low permeance that is gradually improved through iterative chemistry efforts and may lead to an MIC even when the permeant properties are suboptimal. Isolation of resistant mutants using these compounds is expected to yield strains with mutations that reduce either biochemical potency or permeance. In this way, membrane proteins with even the slightest ability to move inhibitors to the extracellular fluids may be identified as efflux pumps. The relevance of such a finding is ultimately determined by the balance of permeation and biochemical potency of the clinical drug for which biochemical and antimicrobial potency, as well as MOA, need to be followed throughout the drug discovery process. Ideally, its datum would reside on the hypotenuse, having an MIC that is primarily predicted by its biochemical potency. Consequentially, mutations resulting in increased efflux pump activity are less likely to occur, and resistance frequencies may be significantly lower. Alternatively, a drug may have lower MICs but not as low as the biochemical potency would predict, suggesting permeation as a bottleneck. Although a more potent antimicrobial, use of such compound could lead to higher clinical resistance frequencies.
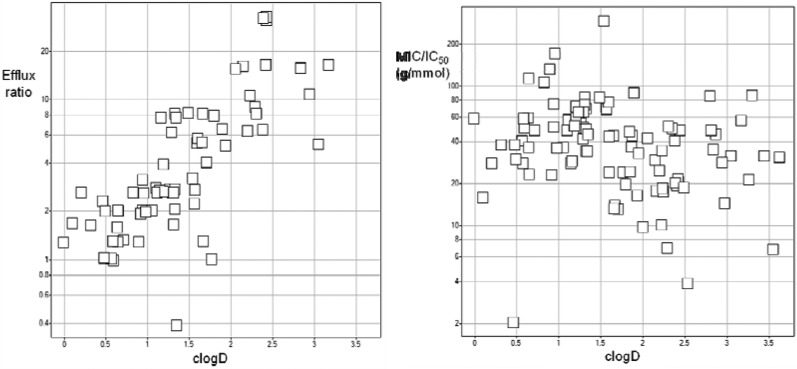
The 99 compounds that acted via LigA in both species are physicochemically a relatively homogeneous group of neutral molecules with a molecular weight ranging from 320 to 490 and clog D(15) ranging from 0 to 3.5 (see the Supporting Information). Compound entry into wild-type bacteria is the result of compound permeation overwhelming efflux. The efflux ratio in H. influenzae (MICwild-type/MICHin101)16 was strongly dependent on the hydrophobicity of the compound, increasing approximately 10-fold when clog D varied from 0.5 to 2.5 (Figure 2). None of the 27 compounds with clog D < 1.2 showed efflux, defined as an MICwild-type/MICHin101 ≥ 4,15 whereas, conversely, all 18 compounds with clog D > 1.8 were effluxed. This is in-line with the hypothesis that compound polarity is required to permeate through the hydrophilic porins and reach the periplasm of Gram-negative pathogens.17 Most of these compounds have a fractional polar surface area (FPSA) <0.4,18 following the hypothesis of Manchester et al.16 that substitution with privileged groups not detrimental to enzyme inhibition may be required to avoid efflux low at higher FPSA.
Figure 2.
Correlation between clog D(15) and the efflux ratio (i.e., MICwild-type/MICHin101) (left) and the MICHin101/IC50 ratio (right) in H. influenzae using the data set of 99 compounds with a MOA via LigA in both S. pneumoniae and H. influenzae (29 indeterminate values were excluded from the left graph).
Although the MIC/IC50 ratios of both species showed a much weaker correlation, the trend seemed as steep as the above-mentioned efflux ratio and showed a 10-fold lowering of the MIC/IC50 upon an increase of clog D from 0.5 to 2.5 (Figure 2). This is similar to that described for azaindoles in S. pneumoniae(19) and presumably reflects that, in the absence of efflux, permeation into the periplasm is relatively fast as compared to permeation through the inner membrane, which can be promoted by increasing hydrophobicity.
The described analysis of adenine-based LigA inhibitors shows that antimicrobial activity results from the combination of biochemical potency and compound permeance. Determination of the MOA is essential to confirm the relevance of the biochemical potency and the cellular compartment in which the compound mediates its effect; extrapolation of the MOA to either other compounds within the set or other pathogens is likely to confound the analysis. Hydrophobicity aids permeance through the cytosolic membrane yet reduces permeation through the porins in the outer membrane of Gram-negative pathogens. The increasing need for antibacterial agents to combat Gram-negative infections20 makes solving this conundrum a high priority.
Experimental Procedures
The bacterial strains used and the determination of LigA IC50 values and MIC have been described previously.3,9 IC50 values were determined by interpolating percentages inhibition measured at 2-fold serially dilutions of compound concentration; the geometric mean of the results (N ≥ 2) is the reported IC50. The MOA of compounds using overexpression assays was determined by measuring MICs in duplicate on different days. An at least 4-fold difference between overexpression and test strain on both test occasions led to the conclusion that the MOA in the test strain was LigA; otherwise, it was considered incompatible with that target. The reported MIC is the geometric mean of the two. Reproducibility of the MIC determination was high, leading to an estimated false positive rate of the duplicate test for each species of less than 0.1%. Compounds that were inactive against the overexpression strain, that is, MIC > 64 ug/mL, were scored as 128 μg/mL. If these compounds displayed a geometric mean MIC against the test strain of ≤32 μg/mL, a 4-fold difference was reached, and it was concluded that these acted via NAD+-dependent LigA; otherwise, the result was considered inconclusive.
Glossary
Abbreviations
- FPSA
fractional polar surface area
- H. influenzae
Haemophilus influenzae
- IC50
compound concentration at which enzyme activity is inhibited by 50%
- LigA
DNA ligase
- MIC
minimum inhibitory concentration
- S. pneumoniae
Streptococcus pneumoniae
Supporting Information Available
Table listing the above-mentioned 99 compounds with MOA via LigA in both H. influenzae and S. pneumoniae and their associated physiochemical and biological data. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
§ Department of Biochemistry and Howard Hughes Medical Institute, Brandeis University, Waltham, Massachusetts 02452, United States.
The authors declare the following competing financial interest(s): All authors are, or were, employees of AstraZeneca..
Supplementary Material
References
- Lehman I. R. DNA ligase: Structure, mechanism and function. Science 1974, 186, 790–797. [DOI] [PubMed] [Google Scholar]
- Shuman S. DNA Ligases: Progress and Prospects. J. Biol. Chem. 2009, 284, 17365–17369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S. D.; Eakin A. E.; Buurman E. T.; Newman J. V.; Gao N.; Huynh H.; Johnson K. D.; Lahiri S.; Shapiro A.; Walkup G. K.; Yang W.; Stokes S. Novel bacterial NAD+-dependent DNA ligase Inhibitors with broad spectrum potency and antibacterial efficacy in vivo. Antimicrob. Agents Chemother. 2011, 55, 1088–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal J. M. DNA and RNA ligases: structural variations and shared mechanisms. Curr. Opin. Struct. Biol. 2008, 18, 96–105. [DOI] [PubMed] [Google Scholar]
- Wilkinson A.; Day J.; Bowater R. Bacterial DNA ligases. Mol. Microbiol. 2001, 40, 1241–1248. [DOI] [PubMed] [Google Scholar]
- Brötz-Oesterhelt H.; Knezevic I.; Bartel S.; Lampe T.; Warnecke-Eberz U.; Ziegelbauer K.; Habich D.; Labischinski H. Specific and potent inhibition of NAD+ -dependent DNA Ligase by pyridochromanones. J. Biol. Chem. 2003, 278, 39435–39442. [DOI] [PubMed] [Google Scholar]
- Meier T. I.; Yan D.; Peery R. B.; McAllister K. A.; Zook C.; Peng S.; Zhao G. Identification and characterization of an inhibitor specific to bacterial NAD+-dependent DNA ligases. FEBS J. 2008, 275, 5258–5271. [DOI] [PubMed] [Google Scholar]
- Cavero-Tomas M.; Gowravarum M.; Huyhn H.; Ni H.; Stokes S.. Substituted adenines and the use thereof. WO-2006040558.
- Clinical Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard, 8th ed.; Clinical Laboratory Standards Institute: Wayne, PA, 2009; M07-A8, Vol. 29, No. 2. [Google Scholar]
- Sanchez L.; Pan W.; Vinas M.; Nikaido H. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J. Bacteriol. 1997, 179, 6855–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B. D.; Kimball E. H.; Gao M.; Osterhout R.; Osterhout S. J.; Rabinowitz J. D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nature Chem. Biol. 2009, 5, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korycka-Machala M.; Rychta E.; Brzostek A.; Sayer H. R.; Rumijowska-Galewicz A.; Bowater R. P.; Dziadek J. Evaluation of NAD+-dependent DNA ligase of mycobacteria as a potential target for antibiotics. Antimicrob. Agents Chemother. 2007, 51, 2888–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A.Structure and Mechanism in Protein Science; W. H. Freeman and Company: New York, NY, 1999. [Google Scholar]
- Buurman E. T.; Johnson K. D.; Kelly R. K.; MacCormack K. Different modes of action of naphthyridones in Gram-positive and Gram-negative bacteria. Antimicrob. Agents Chemother. 2006, 50, 385–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. J.; Buttar D.; Cumming J. G.; Davis A. M.; Norinder U.; Rodgers S. L. Automated QSAR with a hierarchy of global and local models. Mol. Inf. 2011, 30, 960–972. [DOI] [PubMed] [Google Scholar]
- Manchester J. I.; Buurman E. T.; Bisacchi G. S.; McLaughlin R. E. Molecular determinants of AcrB-mediated bacterial efflux: Implications for drug discovery. J. Med. Chem. 2012, 55, 2532–2537. [DOI] [PubMed] [Google Scholar]
- O'Shea R.; Moser H. E. Physicochemical properties of antibacterial compounds: Implications for drug discovery. J. Med. Chem. 2008, 51, 2871–2878. [DOI] [PubMed] [Google Scholar]
- Ertl P; Rohde B.; Selzer P. Calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000, 43, 3714–3717. [DOI] [PubMed] [Google Scholar]
- Manchester J. I.; Dussault D. D.; Rose J. A.; Boriack-Sjodin P. A.; Uria-Nickelsen M.; Ioannidis G.; Bist S.; Fleming P.; Hull K. G. Discovery of a novel azaindole class of antibacterial agents targeting the ATPase domains of DNA gyrase and topoisomerase IV. Bioorg. Med. Chem. Lett. 2012, 10.1016/j.bmcl.2012.05.128. [DOI] [PubMed] [Google Scholar]
- Siegel R. E. Emerging Gram-negative antibiotic resistance: Daunting challenges, declining sensitivities and dire consequences. Respir. Care 2008, 53, 471–479. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.