Summary
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), and associated proteins (Cas) comprise the CRISPR-Cas system, which confers adaptive immunity against exogenic elements in many bacteria and most archaea. CRISPR-mediated immunization occurs through the uptake of DNA from invasive genetic elements such as plasmids and viruses, followed by its integration into CRISPR loci. These loci are subsequently transcribed and processed into small interfering RNAs that guide nucleases for specific cleavage of complementary sequences. Conceptually, CRISPR-Cas shares functional features with the mammalian adaptive immune system, while also exhibiting characteristics of Lamarckian evolution. Because immune markers spliced from exogenous agents are integrated iteratively in CRISPR loci, they constitute a genetic record of vaccination events and reflect environmental conditions and changes over time. Cas endonucleases, which can be reprogrammed by small guide RNAs have shown unprecedented potential and flexibility for genome editing, and can be repurposed for numerous DNA targeting applications including transcriptional control.
Introduction
Bacteria dominate many natural habitats, including inhospitable environments and challenging conditions that include predatory viruses. To survive, bacteria have developed many ways to fend off invaders (Labrie et al., 2010), including the recently described CRISPR system. CRISPR is an acronym for clustered regularly interspaced short palindromic repeats. CRISPR loci contain short, partially palindromic DNA repeats that occur at regular intervals and form loci that alternate repeated elements (CRISPR repeats) and variable sequences (CRISPR spacers) (Figure 1). These peculiar loci were first observed in 1987 (Ishino et al., 1987), but they received little attention until similar, idiosyncratic loci, were described in microbial genome drafts (Jansen et al., 2002). These loci are typically flanked by accompanying CRISPR-associated (cas) genes. Their biological role remained elusive until 2005, when three groups reported that the spacers were homologous to foreign genetic elements, including viruses and plasmids (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005). These reports lead to the hypothesis that CRISPRs might function as an immune system (Makarova et al., 2006). Evidence for CRISPR-mediated immune function quickly followed (Barrangou et al., 2007), and subsequent studies established that CRISPR-mediated defense involves sequence-specific, RNA-mediated (Brouns et al., 2008) targeting of mostly DNA (Marraffini and Sontheimer, 2008), and occasionally RNA (Hale et al., 2009). Since then, many studies have delved in the molecular underpinnings of the CRISPR-Cas system genetics, mechanisms, and applications.
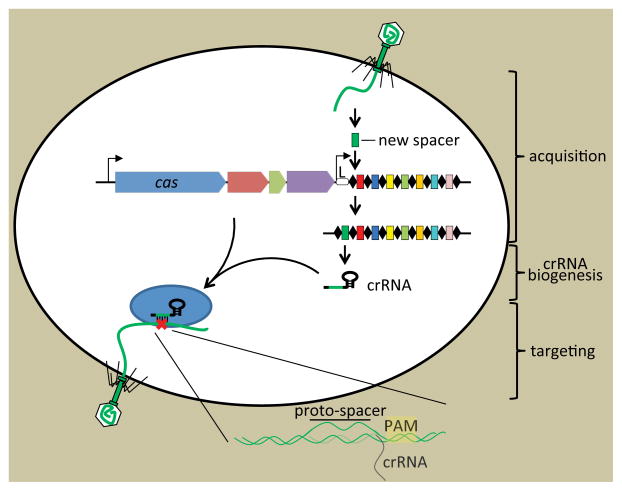
Figure 1. The three stages of CRISPR immunity.
CRISPR loci contain clusters of repeats (black diamonds) and spacers (colored boxes) that are flanked by a “leader” sequence (L) and CRISPR-associated (cas) genes. During adaptation new spacers derived from the genome of the invading virus are incorporated into the CRISPR array (Barrangou et al., 2007) by an unknown mechanism. The synthesis of a new Repeat is also required. During crRNA biogenesis a CRISPR precursor transcript is processed by Cas endoribonucleases within repeat sequences to generate small crRNAs (Brouns et al., 2008). During targeting the match between the crRNA spacer and target sequences (complementary protospacer) specifies the nucleolytic cleavage (red cross) of the invading nucleic acid (Garneau et al., 2010; Gasiunas et al., 2012; Jinek et al., 2012).
Generally, CRISPR-Cas systems and their elements are hypervariable and differ broadly in terms of occurrence, genes, sequences, number, and size across genomes. Indeed, CRISPR repeats can vary widely (23–55 nt), though they are typically 28–37 nt, and their partially palindromic nature allows them to form hairpin structures. Likewise, CRISPR spacers can also vary in size (21–72 nt), though they are typically 32–38 nt. Quantitatively, arrays that contain up to 588 repeats have been reported (in Haliangium ochraceum), but they are less than 50 units in most cases. Likewise, although up to 19 distinct loci have been reported in Methanocalcococcus, and 25 putative CRISPR loci have been suggested in Methanotorris igneus, organisms typically contain 1–2 CRISPR loci (Grissa et al., 2007). According to the database, CRISPRdb, CRISPRs occur in nearly half (1,126/2,480 ~45%) of bacterial genomes and the large majority (125/150 ~83%) of archaea (Grissa et al., 2007).
CRISPR-Cas systems have been classified into three major Types, namely Type I, Type II and Type III, and 12 subtypes, given their genetic content and structural and functional differences (Makarova et al., 2011a; Makarova et al., 2013). The core defining feature of CRISPR-Cas types and subtypes are the cas genes and the protein they encode, which are highly genetically and functionally diverse, illustrating the many biochemical functions that they carry throughout the different steps of CRISPR-mediated immunity. Noteworthy, the RNA recognition motif (RRM) is widespread in many Cas proteins, and most of the Cas families of proteins carry functional domains that interact with nucleic acids, such as DNA binding, RNA binding, helicase, and nuclease motifs (Makarova et al., 2002; Makarova et al., 2011a; Makarova et al., 2006; Makarova et al., 2011b; Makarova et al., 2013). Genetically, cas1 and cas2 universally occur across types and subtypes, whereas cas3, cas9 and cas10 have been defined as the signature genes for Type I, Type II and Type III, respectively. Phylogenetically, Type II systems have solely been identified in Bacteria, thus far, and there is a bias for Type I systems in bacteria, and Type III systems in archaea and hyperthermophiles.
Overall, CRISPR-Cas immune systems function in three steps. The first step is adaptation, in which new spacers are acquired from exogenous nucleic acid into the CRISPR locus. The adaptation step is followed by crRNA biogenesis, in which CRISPR arrays are transcribed and processed into small interfering CRISPR RNAs (crRNAs). The final step is targeting, in which crRNAs guide Cas nucleases for specific cleavage of homologous sequences (Figure 1). These steps have been described in a number of reviews published since 2008, including several recent extensive or focused detailed reviews (Barrangou, 2013; Barrangou and Horvath, 2012; Fineran and Charpentier, 2012; Marraffini, 2013; Reeks et al., 2013; Sorek et al., 2013; Westra et al., 2012; Wiedenheft et al., 2012). Comparative analyses have unraveled potential common ancestry between CRISPR-Cas system components and core elements that define mobile genetic elements (notably transposases), as well as other defense systems such as toxin-anti-toxin and restriction-modification systems (Makarova et al., 2013).
Here, we provide an overview of CRISPR-based adaptive immunity in bacteria and archaea and discuss the applications of CRISPR-Cas systems and their components.
CRISPR-Cas systems provide adaptive immunity
Although there are several innate immunity-like systems in bacteria, such as abortive infection, receptor mutation, and restriction-modification, the recently characterized CRISPR-Cas system has been described as an adaptive immune system, which provides specific and acquired immunization against exogenic mobile genetic elements.
The first biological evidence that CRISPR-Cas systems have a role in adaptive immunity was reported in 2007 when S. thermophilus CRISPR loci were shown to acquire novel spacers derived from the invasive phage DNA (Barrangou et al., 2007). The acquisition of phage DNA into the leader end of a CRISPR array led to sequence-specific, inheritable immunity against phages bearing homologous sequences. Only a small proportion of the population gained CRISPR encoded immunity, but this immunization, albeit infrequent, provided a high level of resistance (Barrangou et al., 2007; Deveau et al., 2008; Levin et al., 2013). Several studies of the S. thermophilus system have shown that most areas of the viral genome are targeted, including both DNA strands, coding and non-coding sequences, and all transcription modules. Nevertheless, a recent study showed that sampling of the viral genome is biased (Paez-Espino et al., 2013), which might be due to DNA structural or composition features. There is a danger that CRISPR systems can target host sequences. Indeed, self-targeting is a very rare and lethal event(Paez-Espino et al., 2013). Given this danger, CRISPRs must have a way to distinguish self from non-self, which is yet to be characterized. Several subsequent studies in S. thermophilus confirmed this adaptive immunity phenomenon, and characterized genetic elements involved (Deveau et al., 2008; Horvath et al., 2008). A study in 2010 showed this CRISPR-Cas system can also vaccinate cells against plasmid uptake by adaptive spacer acquisition (Garneau et al., 2010). Noteworthy, spacers were acquired against antibiotic resistance genes, which could prevent the uptake of any plasmid bearing complementary sequences.
Several studies of model CRISPR-Cas systems have also shown adaptive spacer uptake in E. coli Type I-E systems (Datsenko et al., 2012; Swarts et al., 2012; Yosef et al., 2012) from plasmid exposure, as well as in a Type III system from Sulfolobus solfataricus (Erdmann and Garrett, 2012). Interestingly, in these studies, a phenomenon described as “priming” showed that the first immunization events influence subsequent acquisition events, again supporting the adaptive nature of these processes.
The sequence in the exogenous nucleic acid element corresponding to a CRISPR spacer has been defined as a protospacer (Deveau et al., 2008), which is flanked by a system-specific highly conserved CRISPR motif, subsequently renamed protospacer adjacent motif (PAM) (Mojica et al., 2009) (Figure 2). PAMs are typically 2–5 nt highly conserved sequence motifs immediately flanking one side of the protospacer (within 1–4 nt of one extremity). These motifs have been identified in several Type I systems (where they primarily consist of 2–3nt motifs occurring on the 5′ end of the protospacer, Figure 2A) and many Type II systems (where they range between 2–5nt and primarily occur on the 3′ end of the protospacer, Figure 2B), while they are yet to be characterized in Type III systems (Mojica et al., 2009; Sorek et al., 2013). PAMs have been implicated in both immunization (sampling of the exogenous DNA for spacer uptake) (Paez-Espino et al., 2013) and targeting (because PAM mutations preclude target cleavage) (Gasiunas et al., 2012; Jiang et al., 2013a; Jinek et al., 2012; Sapranauskas et al., 2011; Sashital et al., 2012). This is consistent with the preferential mutation of the PAM by viruses to escape CRISPR immunity (Semenova et al., 2011; Sun et al., 2013).
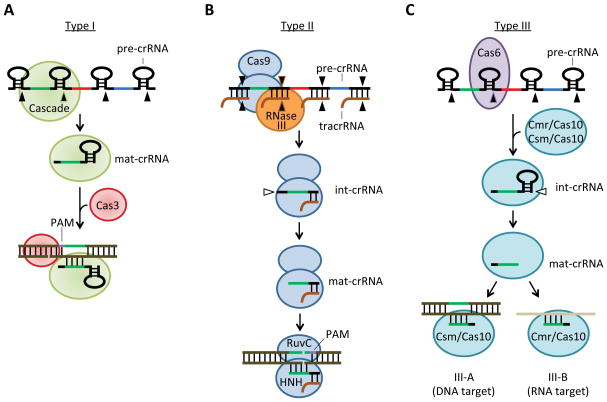
Figure 2. Mechanism of crRNA biogenesis and targeting in the three Types of CRISPR-Cas systems.
Black arrowheads, primary processing sites of the crRNA precursor (pre-crRNA) to liberate intermediate crRNAs (int-crRNA). White arrowhead, further processing of the int-crRNA to yield mature crRNAs (mat-crRNA). Green line, target sequence (same sequence as crRNA spacer). Purple line, PAM (Mojica et al., 2009). (A) In Type I systems, primary processing of the pre-crRNA is achieved by the Cas6 endoribonuclease within the Cascade complex (Brouns et al., 2008). Cleavage occurs at the base of the stem-loop formed by the repeat RNA to release mat-crRNAs. The Cascade recruits the Cas3 nuclease to nick the DNA strand complementary to the proto-spacer, immediately downstream of the region of interaction with the crRNA spacer (Sinkunas et al., 2013). (B) In Type II systems primary processing requires the annealing of the tracrRNA to the repeat sequences of the pre-crRNA and the subsequent cleavage of the dsRNA by the host RNase III (Deltcheva et al., 2011). Primary processing occurs in the contex of Cas9 and it is followed by the trimming of the 5′-end repeat and spacer sequences of the int-crRNA to yield mat-crRNAs. Target cleavage requires the crRNA, the tracrRNA and the RuvC and HNH domains of Cas9, each of which cleaves one DNA strand of the proto-spacer region, 3-nt upstream of the PAM (Gasiunas et al., 2012; Jinek et al., 2012). (C) In Type III systems Cas6 cleaves the pre-crRNA to generate int-crRNAs that are incorporated into a Cmr/Cas10 or Csm/Cas10 complex, where further maturation occurs through the trimming of 3′-end sequences (Hale et al., 2012; Hale et al., 2009). While genetic evidence indicates that III-A subTypes cleave target DNA sequences (Hatoum-Aslan et al., 2014), biochemical data suggests that subType III-B cleave RNA molecules (Hale et al., 2009).
Although we still do not fully understand how spacers are acquired and how new CRISPR repeats are synthesized, Cas1 and Cas2 are involved. Spacer acquisition in Type I systems require overexpression of cas1 and cas2 (Yosef et al., 2012), and acquisition is lost when cas1 and cas2 are mutated (Datsenko et al., 2012). This is consistent with the sequence-based co-evolutionary patterns that couple cas1 to CRISPR repeat sequences (Horvath et al., 2009). Furthermore, cas1 and cas2 (and the proteins they encode) are dispensable for other CRISPR-related processes such as crRNA biogenesis and targeting (Brouns et al., 2008; Deltcheva et al., 2011; Hatoum-Aslan et al., 2014; Sapranauskas et al., 2011).
Biochemical studies of Cas1 and Cas2 have provided insights into their functional roles. Cas1 binds tightly to dsDNA in a sequence-independent manner and both Cas1 and Cas2 are metal-dependent endonucleases (Beloglazova et al., 2008; Nam et al., 2012; Samai et al., 2010). Cas1 can cleave dsDNA into short fragments that might serve as precursors for novel spacers (Han et al., 2009; Wiedenheft et al., 2009). Because Cas1 also has the ability to resolve Holliday junctions and thus promote DNA integration and recombination events, it is a noteworthy candidate for both novel spacer sampling and integration into the repeat-spacer array.
Clearly, much of the mechanisms involved in spacer acquisition are unknown. The molecular details involved in and responsible for recognizing non-self DNA, scanning for candidate protospacers, generating a novel repeat, and polarized integration of the new repeat-spacer unit into the CRISPR array are yet to be determined.
Lamarckian vs Darwinian evolution
CRISPR loci that are modified to provide immunity in one generation can be passed on to the next generation. This heritability of acquired characteristics, together with the patterns of CRISPR-Cas evolution, provide strong examples of Lamarckian evolution. (Koonin and Makarova, 2013). Indeed, as aforementioned, there is a non-random bias in novel spacer sampling and acquisition, which provides a phenotypic trait (phage resistance) that is beneficial to the immunized host, and passed on to the surviving cell line. This contrasts with the random and subsequently selected for nature of events assigned to Darwinian phenomena. CRISPR loci are examples of bona fide Lamarckian evolution, especially given the direct link between the acquired genotype (novel spacer integrated into the host genome) and the corresponding phenotypic change (the ability to resist complementary sequences), which is the essence of the Lamarckian paradigm (Koonin and Wolf, 2009). Furthermore, the Lamarckian argument is supported by the fact that environmental cues (phage attack), trigger heritable genetic changes (spacer integration) that provide adaptation (resistance to the predatory phage). Lastly, the natural co-existence of a diversity of CRISPR genotypes (Paez-Espino et al., 2013; Sun et al., 2013) supports the mathematically-predicted Lamarckian nature of CRISPR genotype evolution in mixed populations (Haerter and Sneppen, 2012). We should point out that some aspects of CRISPR-Cas systems are more consistent with Darwinian evolution, notably the increased fitness of the variants that have acquired novel spacers (Childs et al., 2012), and more precise insights into the type of evolution that CRISPR systems provide to bacteria will be available as the molecular mechanisms of spacer acquisition are elucidated.
RNA-guided Cas nucleases protect the host by destroying the invader’s genome
The classification of CRISPR-Cas systems into three types primarily reflects the different cas gene content of these systems. Since the Cas proteins are responsible for crRNA biogenesis and the recognition and destruction of the invading nucleic acid, it is not surprising that each CRISPR type displays a unique molecular mechanism of action (Figure 2).
Types I and III systems exhibit the most similarities (Figure 2A and C). Both systems rely on the Cas6 family of endoribonucleases to cleave the repeat sequences of the crRNA precursor to generate small crRNAs. Cas6 cleavage occurs typically 8 nucleotides upstream of the 5′ end of the spacer sequence, generating crRNAs with 8 nucleotides of repeat sequence at their 5′ end, known as the crRNA tag (Carte et al., 2008; Hatoum-Aslan et al., 2011). In Type III CRISPR-Cas systems, however, crRNAs are further trimmed at the 3′ end by an unknown nuclease (Hale et al., 2008; Hatoum-Aslan et al., 2011) (Figure 2C). Targeting in both Types is facilitated by a large, multi-subunit ribonucleoprotein complex. The targeting complex in Type I systems is called Cascade (CRISPR-associated complex for antiviral defense) (Brouns et al., 2008), which is formed by Cse1, Cse2, Cas7, Cas5, Cas6e subunits in E. coli (Type IE) and Csy1, Csy2, Csy3 and Cas6f in Pseudomonas aeruginosa (Type IF) (Wiedenheft et al., 2011). Nucleolytic cleavage of the target DNA is carried out by a Cas3 family exonuclease, which is not a member of the complex (Sinkunas et al., 2013) (Figure 2A). Two sequences in the target are essential for cleavage, the PAM and “seed” sequences. In the E. coli Type IE system the PAM sequence consists of the three-nucleotide motif AAG (Savitskaya et al., 2013; Swarts et al., 2012; Yosef et al., 2012), is located immediately upstream of the protospacer and is recognized by the Cascade complex, through direct contacts between Cse1 and the target DNA (Sashital et al., 2012). Because the PAM sequences are absent from the repeats, this requirement provides a mechanism for tolerance to ‘self’ sequences that are homologous, i.e. the spacers in the CRISPR array. The “seed” sequence is a 6- to 8-nucleotide sequence within the guide crRNA which is strictly complementary to the target sequence, (Semenova et al., 2011; Wiedenheft et al., 2011), and enthalpically drives complementary base-pairing between the crRNA and the target, and likely R-loop formation. It is hypothesized that the first event in target recognition are transient, short-lived protein:DNA interactions between the Cascade complex and the PAM sequence. This is followed by an attempt to hybridize the crRNA:DNA seed sequences, most likely the rate-limiting step of the targeting reaction. Evidence for this model comes from studies of Cas9 targeting (Sternberg et al., 2014) (see below). The PAM and seed requirements for cleavage are readily exploited by mutant phages containing modifications of either of these sequences, which prevent cleavage and CRISPR immunity (Barrangou et al., 2007; Deveau et al., 2008).
The different Type III CRISPR-Cas systems seem to differ in the target nucleic acid: while genetic data indicates that Type III-A CRISPR-Cas systems cleave DNA molecules in vivo (Marraffini and Sontheimer, 2008), Type III-B CRISPR-Cas systems cleave RNA targets in vitro (Hale et al., 2009) (Figure 2C). In both cases, targeting requires a large ribonucleoprotein complex, the Cas10-Csm complex in Type III-A (Hatoum-Aslan et al., 2013) and the CMR complex in Type III-B (Hale et al., 2009; Staals et al., 2013; Zhang et al., 2012). Both targeting complexes possess the signature subunit Cas10, which most likely harbors the nucleolytic activity of the complex (Hatoum-Aslan et al., 2014; Ramia et al., 2013).
In contrast to Types I and III systems, Type II systems require minimal Cas machinery for immunity (Figure 2B). For crRNA biogenesis, these systems utilize a trans-encoded CRISPR RNA (tracrRNA), a small RNA that shares partial complementarity with CRISPR repeats (Deltcheva et al., 2011). Pairing between the tracrRNA and the precursor crRNA generates a double-stranded substrate that is cleaved by the host-encoded RNase III to liberate the small crRNAs. These are further trimmed at their 3′ end by an unknown nuclease. Cleavage of the DNA target in Type II systems is carried out by a single large multidomain protein, Cas9. This is an RNA-guided double-stranded DNase with two independent nuclease domains, HNH and RuvC, each of which cleaves one strand of the target DNA (Gasiunas et al., 2012; Jinek et al., 2012). Recent studies have established that formation of a complex with crRNA:tracrRNA drives Cas9 conformation changes that direct target DNA binding and cleavage, in a PAM-dependent manner (Jinek et al., 2014; Nishimasu et al., 2014; Sternberg et al., 2014).
Comparison between CRISPR-Cas and the mammalian adaptive immune systems
To defend the host successfully, adaptive immune systems need to develop three essential features: specificity, diversity, and memory. Specificity is required to recognize the exogenous structures and organisms that need to be attacked, and distinguish them from those that are to be tolerated. Diversity in the immune response is necessary to defend the host from a large variety of pathogens and/or malignant elements, as to ensure survival to exposure from a broad range of invasive agents. Finally, the generation of a memory of past infections allows a more efficient reaction upon recurrent infections, and allows the host to rapidly mount a response upon re-exposure. Both the adaptive branch of the mammalian immune system (Murphy, 2012) and CRISPR-Cas immunity have these key features. However, the molecular and cellular mechanisms that enable these features are different. Below, we contrast these differences; for a more detailed comparison between both immune systems we refer the readers to a recent review on this topic (Goren et al., 2012).
A fundamental distinction between both systems is the molecular nature of their target. While mammalian immunity has to deal with a wide range of exogenous agents and types of invasive cellular organisms that often display variable structural elements, the principal infectious agents of prokaryotes are viruses and plasmids. In addition, whereas mammalian immunity need to survey and protect distinct tissues within the host, unicellular prokaryotes need to attack the cytoplasmic nucleic acids of the invader; i.e. within the context of a single cell with a short lifespan. As a consequence of these differences, the molecules that provide specificity to detect these targets are different. In the mammalian adaptive immune system, antibodies (proteins) specifically recognize and bind three-dimensional antigens, namely portions of proteins, sugar or nucleic acids present in the organisms and/or malignant cells that need to be recognized. The proteinaceous nature of the antibodies allows them to circulate throughout the body as free molecules or at the surface of lymphocytes. The equivalent of the antibody/antigen pair in CRISPR immunity are the crRNA and the protospacer sequence within the invading genome, which interact within the boundaries of the prokaryotic cell. The mechanisms to achieve specificity are also fundamentally different. While B cells undergo several rounds of maturation to select, in a Darwinian fashion, the clones expressing the fittest antibodies for a particular antigen, CRISPR-Cas immune systems directly acquire the protospacer sequences to produce a corresponding crRNA. This relatively simple mechanism is possible due to the one-dimensional nucleic acid interaction required to execute CRISPR immunity and sets the basis for the Lamarckian adaptation of bacteria to their viruses provided by this immune system.
The specificity of the immune response is required to prevent the attack of ‘self’ elements and ‘non-self’ commensal organisms. In mammals, the central tolerance mechanisms ensure that newly developing B cells producing anti-self antibodies and T cells recognizing MHC-presented self-peptides with high affinity are eliminated in the bone marrow and in the thymus, respectively. Peripheral tolerance mechanisms, on the other hand, inactivate mature lymphocytes to promote tolerance to ‘non-self’ elements, such as the gut commensal microbiota and innocuous dietary proteins. In the CRISPR immune response, tolerance to ‘self’ is required to prevent the targeting of the spacer sequences within the CRISPR array, which are, by nature, complementary to the crRNAs they encode. This is achieved through recognition by the Cas machinery of features in the spacer flanking sequences that are not present in the flanking sequences of bona-fide protospacers (see above). Another mechanism for tolerance to ‘self’ is required to prevent the acquisition of spacer sequences from the bacterial chromosome. Whether and how CRISPR-Cas systems distinguish self DNA from, for example, phage DNA during the acquisition phase is yet to be determined. Tolerance to ‘non-self’ has not been observed for CRISPR immunity so far. Beneficial plasmids and prophages (bacteriophages that integrate into the host chromosome) could be considered commensal elements of bacteria and an immune response towards these elements would be detrimental for the host. Experimental evidence, however, indicates that an active CRISPR-Cas system and its target cannot co-exist in the same cell (Jiang et al. 2013b) and therefore tolerance of plasmids (Jiang et al., 2013b) and prophages (Edgar and Qimron, 2010; Nozawa et al., 2011) is not possible. Perhaps the regulation of CRISPR immunity at the transcriptional level (Pul et al., 2010; Westra et al., 2010; Yosef et al., 2011) could provide an ‘on-off’ switch for the tolerance of prokaryotic foreign genetic elements.
In mammalian immune systems, diversity is achieved through the generation of many different antibodies through V(D)J recombination and somatic hypermutation of light chain genes. This ensures that virtually any target can be recognized. After the infection is clear, a subpopulation of the lymphocytes that produce the most effective antibodies become memory cells, ready to proliferate and attack the pathogen during subsequent exposures. As mentioned previously, CRISPR-Cas systems acquire spacers directly from the invading genome, at the same time capturing and storing the diversity and memory of the CRISPR immune response into the CRISPR array. It is suspected that any viral and plasmid DNA sequence can be acquired, as long as they are adjacent to a proper PAM sequence, and in principle the CRISPR immune response can deal with the great diversity of prokaryotic viruses and plasmids. It is yet to be established, however, whether CRISPR immunity can also defend the host from RNA phages (in this case the acquisition of spacers will require reverse transcription), which represent a substantial proportion of the prokaryotic viruses (Zinder, 1980). Importantly, in contrast to the antibody diversity and memory generated by the mammalian immune system during the life span of the individual, most of the spacers acquired during CRISPR immunity are inherited by the progeny after bacterial duplication (though some can be lost, presumably during DNA replication of the repeat sequences through polymerase slippage). The inheritable nature of CRISPR-Cas systems is a major advantage for the host’s progeny, and a major difference between these two immune systems. A final contrast regards how memory is generated in both systems. In mammals, memory lymphocytes can rise after either infection or vaccination. For CRISPR, it remains to be determined whether spacers can be acquired and expressed during infection, before the infecting phage kills the cell, or from occasional defective viruses that inject their DNA into the prokaryotic host (a process analogous to vaccination).
Applications of CRISPR-Cas systems
The genetic polymorphism of cas genes, together with the functional diversity of the proteins they encode, and in combination with the features and natural functions of CRISPR-Cas systems, have set the stage for a broad array of applications, both exploiting their native roles, and by re-purposing their functional features.
Using CRISPR diversity for genotyping
Originally, the first application of CRISPR loci was exploiting CRISPR repeat occurrence and number diversity to type Mycobacterium and Yersinia (Pourcel et al., 2005) isolates, as defined by spacer oligo typing (spoligotyping) (Groenen et al., 1993). Spacer content was subsequently used for genotyping bacteria of industrial importance, and providing insights into the common origin of strains as defined by the conservation of ancestral spacers (Barrangou, 2012). Indeed, polarized spacer addition at the leader end of the CRISPR locus provides a genetic tape record of immunization events, and a basis for genetic tracking of the genetic trajectory of a strain. CRISPR loci have been used for genotyping of several bacteria including Mycobacterium, Yersinia, Corynebacterium, Pseudomonas, Legionella, Streptococci, Escherichia, Salmonella, and Lactobacillus, among others (Barrangou and Horvath, 2012; Shariat et al., 2013). This approach can also be used to determine the relatedness of strains that are of pathogenic interest, notably in food outbreak investigations such as E. coli and Salmonella (Shariat et al., 2013).
Beyond single strain phylogeny, CRISPR loci provide a basis for probing complex ecological populations and determine the diversity of both host and virus populations in complex systems, as documented in natural habitats (Andersson and Banfield, 2008; Heidelberg et al., 2009; Held and Whitaker, 2009; Tyson and Banfield, 2008) and human samples (Pride et al., 2011). Extensive analyses of CRISPR sequences using metagenomic deep sequencing technologies can also provide critical insights into co-evolutionary dynamics, and the evolutionary trajectories of both host and virus populations, as the arms race unfolds (Levin et al., 2013; Paez-Espino et al., 2013; Sun et al., 2013). Indeed, CRISPR has been shown to be a major force shaping viral genome evolution in some systems, in which SNPs, INDELs and recombinations specifically allow viruses to circumvent CRISPR-encoded adaptive immunity in the host. Nevertheless, the potential of CRISPR for broad genotyping or epidemiological surveys, clinical diagnostics, and industrial surveillance will necessitate validation across a diverse set of genera and species of interest.
Building resistance to viruses in industrial cultures
From an industrial standpoint, there is tremendous need of and potential for building phage resistance in industrially relevant hosts that historically have been, or could be at risk of phage attacks. Keeping in mind the absurd scale of some industrial bioprocessing plants that rely on bacteria to generate bioproducts of interest (therapeutics, enzymes) and foods (dairy products), the risk of phage attack and impact of phage outbreaks can equate to financially catastrophic and significant losses. The original discovery of CRISPR adaptive immunity in the dairy starter culture Streptococcus thermophilus has already been harnessed to obtain phage-resistant mutants to formulate dairy starter cultures with improved sustainability and lifespan for perennial exploitation in the food industry (Barrangou and Horvath, 2012). Noteworthy, it has been shown that iterative build up of viral resistance is efficient, using a set of genetically diverse phages to optimize the breadth and depth of phage resistance (Barrangou et al., 2013; Barrangou and Horvath, 2012). Practically, the unique set of acquired spacers can also serve as a genetic tag for valuable proprietary strains. Notwithstanding early adoption of CRISPR-based lytic phage resistance in dairy cultures, native systems may be broadly used in other industrial bacteria. Also, the ability to readily package and heterologously transfer CRISPR-Cas systems in prokaryotes (Sapranauskas et al., 2011) and even eukaryotes (Cong et al., 2013; Mali et al., 2013b) illustrate the widespread potential of CRISPR-mediated viral resistance, either naturally, or using genetic engineering given the ability to design and synthetize spacers to re-program Cas nucleases (Sapranauskas et al., 2011). The viral resistance upside is illustrated by a recent report showing excision of HIV-1 provirus in human (Ebina et al., 2013), further highlighting the broad promise of this technology for viral immunity beyond the natural hosts of CRISPR-Cas systems.
Beyond viral resistance, CRISPR-mediated adaptive immunity has potential to vaccinate bacterial strains against the uptake and dissemination of undesirable genetic elements such as plasmids that carry antibiotic resistance genes (Garneau et al., 2010) and virulence traits (Bikard et al., 2012). Likewise, because CRISPR can be used to target mobile genetic elements, there is potential to use CRISPR to target transposons and other mobile elements that contribute to genome plasticity as to ensure the maintenance, integrity and stability of the genomes of industrial workhorses widely used in the biotechnology, pharmaceutical and bioproducts industries.
Genome Engineering
The early models for CRISPR-Cas systems postulated that interference was RNA-mediated and protein-dependent, akin to the RNA interference mechanism of eukaryotes (Makarova et al., 2006). It was later established that DNA is the primary target of CRISPR-Cas systems (Marraffini and Sontheimer, 2008), and that interference occurs through sequence-specific target DNA cleavage (Garneau et al., 2010; Marraffini and Sontheimer, 2008). These findings arguably laid the foundation for the subsequent use of these systems as programmable nucleases, with many inherent biotechnological applications. The detailed characterization of CRISPR immunity that followed, particularly of the Type II, Cas9-mediated immunity (Deltcheva et al., 2011; Gasiunas et al., 2012; Jinek et al., 2012; Sapranauskas et al., 2011), led to the realization of the potential for this enzyme in genome engineering. 2013 has seen an explosion of reports that use the RNA-guided programmable feature of Cas9 for the development of powerful technologies for genome editing, genetic screens and modulation of gene expression (Pennisi, 2013), which we describe briefly below.
Engineering genomes with Cas9
While Type I and III CRISPR-Cas systems provide RNA-guided nuclease activity, this is achieved in the context of a large, multimeric, crRNA-Cas ribonucleoprotein complex (Figure 2) that complicates the development of a molecular tool. In contrast, Type II systems only rely on a single endonuclease which generates predictable dsDNA breaks into the target sequence, namely Cas9 (Sapranauskas et al., 2011). In truth, Cas9 requires the participation of an “enabling RNA”, tracrRNA, as well as RNaseIII, to load the crRNA guide (Deltcheva et al., 2011) (see above and Figure 2B) and the crRNA and tracrRNA to cleave its target (Gasiunas et al., 2012; Jinek et al., 2012). Both of these requirements can be bypassed by the use of a chimeric RNA guide, also known as single-guide RNA (sgRNA): a fusion between the tracrRNA and crRNA that maintains the guide properties of the crRNA as well as the tracrRNA:crRNA secondary structures that are critical for Cas9 cleavage (Jinek et al., 2012). This technological improvement was fundamental towards establishing Cas9 as an ideal RNA-guided dsDNA nuclease, and has led many researchers to exploit it as a genetic engineering tool, in the same way ZFNs and TALENs have been used in the past (Wood et al., 2011).
In a bacterial host, Cas9 cleavage of genomic sequences leads to cell death, presumably due to the introduction of chromosomal lesions that can only be repaired with high fidelity, thus re-introducing the target (Bikard et al., 2012; Jiang et al., 2013a). The absence of target mutations that could escape Cas9 cleavage are thought to be the consequence of the lack of non-homologous end joining (NHEJ) repair mechanisms in most bacteria (Shuman and Glickman, 2007). This phenomenon has been exploited to use Cas9 as a powerful and efficient counter-selection tool to introduce specific mutations in bacteria, essentially using CRISPR-based self targeting for directed mutagenesis (Bikard et al., 2012). In contrast, dsDNA breaks in mammalian DNA are partially repaired by the INDEL-generating NHEJ pathway (Figure 3) (Deriano and Roth, 2013). Therefore expression of S. pyogenes Cas9 and an appropriate sgRNA can be used to introduce INDELs and knock-out virtually any gene, provided that it contains a the corresponding PAM sequence (GG in this case, or CC on the complementary strand) (Cong et al., 2013; Jinek et al., 2013). The high efficiency of Cas9-mediated INDEL generation has been used to perform forward genetic screens in which cells expressing the nuclease are transduced with a lentivirus expressing a library of sgRNAs that target each gene of the cell. Analysis of the sgRNA sequences present in cells screened or selected for a particular phenotype is used to identify the candidate genes responsible for this phenotype (they are presumably mutated) (Shalem et al., 2013; Wang et al., 2013b). In addition, dsDNA breaks stimulate homologous recombination in mammalian cells (Smih et al., 1995), thus the introduction of a suitable template that can recombine with the target sequence can be employed to generate site-directed, single-nucleotide substitutions (Figure 3) (Cong et al., 2013; Mali et al., 2013b). Moreover, the co-expression of Cas9 and several RNA guides can be exploited for the generation of multiple mutations in one single multiplexing step (Cong et al., 2013; Wang et al., 2013a).
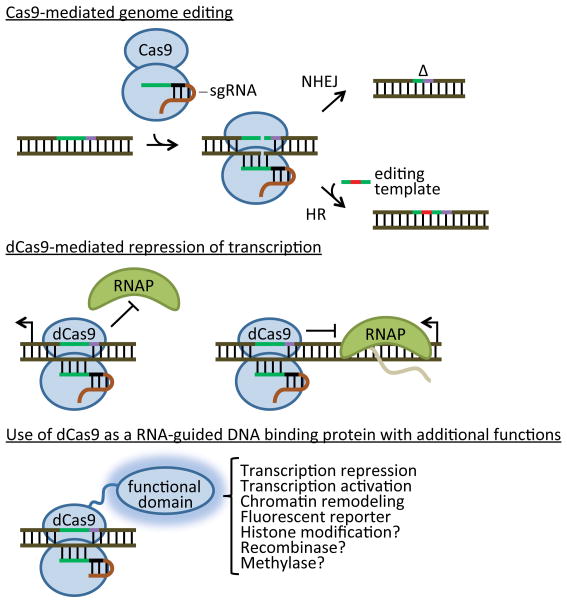
Figure 3. Cas9-based genetic applications.
Wild-Type Cas9 loaded with a single guide RNA (sgRNA) generates dsDNA breaks that can be used to introduce target mutations. Chromosomal breaks can be repaired by non-homologous end joining (NHEJ), creating indels (Δ) that introduce knock-out frameshift mutations. If a sequence homologous to the Cas9 target is provided (the editing template; either linear dsDNA or a short oligonucleotide), the break can be repaired by homologous recombination (Cong et al., 2013; Mali et al., 2013b). In this case, site-specific mutations in the editing template are can also be incorporated in the genome. A catalytically dead Cas9 (dCas9, containing mutations in both the RuvC and HNH actives sites) can be used as an RNA-guided DNA binding protein that can repress both transcription initiation when bound to promoter sequences or transcription elongation when bound to the template strand within an open reading frame (Bikard et al., 2013; Qi et al., 2013). dCas9 can also be fused to different functional domains to bring enzymatic activities and/or reporters to specific sites of the genome (Gilbert et al., 2013b).
An unresolved issue for this technology is the frequency of off-target effects, i.e. the probability of Cas9 cleavage of a sequence partially homologous to the sgRNA. While this undesirable effects are known to occur (Fu et al., 2013), researchers have developed strategies to overcome them with the use of a RuvC active site mutation that converts Cas9 into a nickase, optimally exploited using two paired guides that direct nicking at two adjacent sites only in the intended target sequence (Mali et al., 2013a; Ran et al., 2013). Despite this caveat, the use of Cas9-directed genome editing has been applied very successfully in a broad range of hosts and cell lines, including in human cells, mice, rats, zebrafish, plants, flies, nematodes, yeast, bacteria and many other organisms (Pennisi, 2013), creating a revolution in molecular biology arguably comparable to the introduction of restriction endonucleases.
Control of gene expression using dCas9
With the mutation of both the RuvC and HNH nucleolytic active sites (Gasiunas et al., 2012; Jinek et al., 2012), Cas9 can be converted into a RNA-guided dsDNA binding protein, known as dCas9. This feature has been exploited to repress or activate gene expression in bacteria and mammalian cells. In both organisms, repression can be achieved by directing dCas9 to bind promoter sequences, thus interfering with RNA polymerase (RNAP) transcription initiation, or bind sequences within the open reading frame (particularly the template strand) and block transcription elongation (Figure 3) (Bikard et al., 2013; Qi et al., 2013). Activation of gene expression, on the other hand, requires the fusion of Cas9 to an activation domain. In bacteria, Cas9 can be fused to the omega (ω) subunit of RNAP (rpoZ) in an ΔrpoZ background in Escherichia coli. The dCas9-ω fusion can then be directed to promoter sequences using an appropriate guideRNA to recruit RNAP and activate transcription (Bikard et al., 2013). In mammalian cells, dCas9 can be fused to transcriptional activators such as VP64 and the p65 activator domain (Figure 3) (Gilbert et al., 2013a; Maeder et al., 2013; Mali et al., 2013a). The addition of other functional domains to dCas9 can expand the uses of the programmable DNA binding capability of this enzyme. For example, the KRAB and Kox1 chromatin-modifying domains can be used to attain stronger repression (Gilbert et al., 2013a), and GFP can be used to visualize specific loci within the chromosome (Chen et al., 2013; Malina et al., 2013). The fusion of other domains, such as recombinases, methylases and histone-modifying enzymes will undoubtedly create new opportunities for this technology (Figure 3).
A perspective on the future of CRISPR immunity
There are many useful applications for CRISPR-Cas immunity and its biological significance is no less exciting. For example, phages can acquire CRISPR-Cas systems (in their genome) to specifically target host defense systems, an evolutionary puzzling turn of events (Seed et al., 2013). There are also phages carrying genes that inhibit CRISPR immunity (Bondy-Denomy et al., 2013). In the pathogen Francisella novicida, Cas9 can repress an endogenous lipoprotein gene to promote pathogenesis by preventing the host proinflammatory response against this lipoprotein (Sampson et al., 2013). CRISPR-Cas systems have also been shown to acts as barriers against horizontal gene transfer (Bikard et al., 2012; Jiang et al., 2013b; Zhang et al., 2013), a fact that highlights the importance of CRISPR immunity in the evolution of prokaryotes. One may accordingly wonder: what is next for the CRISPR field?
Undoubtedly, the translation of Cas9-based genome editing technologies into medical applications, most notably gene therapy, has the potential to make a significant impact on human health (provided that the off-target effects are reduced to acceptable levels). Recent advances in understanding the biochemical nature of Cas9 targeting (Sternberg et al., 2014), together with structural insights into Cas9:sgRNA complex formation (Jinek et al., 2014; Nishimasu et al., 2014) will lead to the development of improved molecular biology tools. In addition, a cursory look at the CRISPR intellectual property landscape, publication pace, and citation rate reflect a widespread awareness of the potential of CRISPR-Cas systems and suggests that they will rapidly be routinely used in many, if not most, molecular biology laboratories. Therefore the accelerating pace of industrial exploitation, commercialization and financial investment(s) is setting the stage for a sustainable CRISPR revolution.
In addition to the potential of CRISPR-Cas9 for genome surgery in eukaryotic cell lines, the native role of CRISPR-Cas systems in providing adaptive immunity has a dramatic impact on microbial communities and their predators in most habitats (Andersson and Banfield, 2008; Heidelberg et al., 2009; Held and Whitaker, 2009; Tyson and Banfield, 2008), and their true impact is yet to be defined on a global basis. If adaptive immunity in characterized model systems is representative of the overall impact of CRISPR-Cas systems on the host-virus population co-evolutionary dynamics and genome trajectories, their role in the composition, evolution and ecology of microbial communities in nature is and will continue to be exciting. We also anticipate that additional applications and novel insights will stem from further characterization of CRISPR model systems, their genetic elements, functional modules and mechanism of action.
Highlights.
CRISPR-Cas systems provide DNA-encoded and RNA-mediated adaptive immunity in bacteria
Immunization occurs through uptake of DNA from invasive genetic elements for sequence-specific vaccination
Sequence-specific immunity is mediated by small interfering RNAs that guide endonucleases for interference via DNA cleavage
RNA-guided CRISPR nucleases have important applications, most notably for the engineering of eukaryotic genomes.
Acknowledgments
RB is supported by start up funds from North Carolina State University. LAM is supported by the Searle Scholars Program, the Rita Allen Scholars Program, an Irma T. Hirschl Award, a Sinsheimer Foundation Award and a NIH Director’s New Innovator Award (1DP2AI104556-01). We are grateful to Daniel Mucida (The Rockefeller University) for critical reading of the manuscript. The authors would like to thank their many colleagues and collaborators in the CRISPR field for their insights into these fantastic molecular systems.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- Barrangou R. RNA-mediated programmable DNA cleavage. Nat Biotechnol. 2012;30:836–838. doi: 10.1038/nbt.2357. [DOI] [PubMed] [Google Scholar]
- Barrangou R. CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip Rev RNA. 2013;4:267–278. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Coute-Monvoisin AC, Stahl B, Chavichvily I, Damange F, Romero DA, Boyaval P, Fremaux C, Horvath P. Genomic impact of CRISPR immunization against bacteriophages. Biochem Soc Trans. 2013;41:1383–1391. doi: 10.1042/BST20130160. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Horvath P. CRISPR: new horizons in phage resistance and strain identification. Annu Rev Food Sci Technol. 2012;3:143–162. doi: 10.1146/annurev-food-022811-101134. [DOI] [PubMed] [Google Scholar]
- Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W, et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs LM, Held NL, Young MJ, Whitaker RJ, Weitz JS. Multiscale model of CRISPR-induced coevolutionary dynamics: diversification at the interface of Lamarck and Darwin. Evolution. 2012;66:2015–2029. doi: 10.1111/j.1558-5646.2012.01595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Roth DB. Modernizing the Nonhomologous End-Joining Repertoire: Alternative and Classical NHEJ Share the Stage. Annu Rev Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Qimron U. The Escherichia coli CRISPR system protects from lambda lysogenization, lysogens, and prophage induction. J Bacteriol. 2010;192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann S, Garrett RA. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol. 2012;85:1044–1056. doi: 10.1111/j.1365-2958.2012.08171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202–209. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013a;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013b doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren M, Yosef I, Edgar R, Qimron U. The bacterial CRISPR/Cas system as analog of the mammalian adaptive immune system. RNA Biol. 2012;9:549–554. doi: 10.4161/rna.20177. [DOI] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen PM, Bunschoten AE, van Soolingen D, van Embden JD. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- Haerter JO, Sneppen K. Spatial structure and Lamarckian adaptation explain extreme genetic diversity at CRISPR locus. MBio. 2012;3:e00126–00112. doi: 10.1128/mBio.00126-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV, 3rd, Graveley BR, Terns RM, et al. Essential Features and Rational Design of CRISPR RNAs that Function with the Cas RAMP Module Complex to Cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Lehmann K, Krauss G. SSO1450 - a CAS1 protein from Sulfolobus solfataricus P2 with high affinity for RNA and DNA. FEBS Lett. 2009;583:1928–1932. doi: 10.1016/j.febslet.2009.04.047. [DOI] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc Natl Acad Sci USA. 2011;108:21218–21222. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Samai P, Marraffini LA. Genetic Characterization of Antiplasmid Immunity through a Type III-A CRISPR-Cas System. J Bacteriol. 2014;196:310–317. doi: 10.1128/JB.01130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Samai P, Maniv I, Jiang W, Marraffini LA. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J Biol Chem. 2013;288:27888–27897. doi: 10.1074/jbc.M113.499244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Nelson WC, Schoenfeld T, Bhaya D. Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS One. 2009;4:e4169. doi: 10.1371/journal.pone.0004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol. 2009;11:457–466. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]
- Horvath P, Coute-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, Barrangou R. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int J Food Microbiol. 2009;131:62–70. doi: 10.1016/j.ijfoodmicro.2008.05.030. [DOI] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013a;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Maniv I, Arain F, Wang Y, Levin BR, Marraffini LA. Dealing with the Evolutionary Downside of CRISPR Immunity: Bacteria and Beneficial Plasmids. PLoS Genet. 2013b;9:e1003844. doi: 10.1371/journal.pgen.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science. 2014 doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS. CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013;10:679–686. doi: 10.4161/rna.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- Levin BR, Moineau S, Bushman M, Barrangou R. The population and evolutionary dynamics of phage and bacteria with CRISPR-mediated immunity. PLoS Genet. 2013 doi: 10.1371/journal.pgen.1003312. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and Bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011a;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011b;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. The basic building blocks and evolution of CRISPR-cas systems. Biochemical Soc Trans. 2013;41:1392–1400. doi: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malina A, Mills JR, Cencic R, Yan Y, Fraser J, Schippers LM, Paquet M, Dostie J, Pelletier J. Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev. 2013;27:2602–2614. doi: 10.1101/gad.227132.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA. CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial Pathogens. PLoS Pathog. 2013;9:e1003765. doi: 10.1371/journal.ppat.1003765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Murphy KTPWMJC. Janeway’s immunobiology. New York: Garland Science; 2012. [Google Scholar]
- Nam KH, Ding F, Haitjema C, Huang Q, DeLisa MP, Ke A. Double-stranded endonuclease activity in Bacillus halodurans clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas2 protein. J Biol Chem. 2012;287:35943–35952. doi: 10.1074/jbc.M112.382598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa T, Furukawa N, Aikawa C, Watanabe T, Haobam B, Kurokawa K, Maruyama F, Nakagawa I. CRISPR inhibition of prophage acquisition in Streptococcus pyogenes. PLoS One. 2011;6:e19543. doi: 10.1371/journal.pone.0019543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K, Stahl B, Barrangou R, Banfield JF. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat Commun. 2013;4:1430. doi: 10.1038/ncomms2440. [DOI] [PubMed] [Google Scholar]
- Pennisi E. The CRISPR craze. Science. 2013;341:833–836. doi: 10.1126/science.341.6148.833. [DOI] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Pride DT, Sun CL, Salzman J, Rao N, Loomer P, Armitage GC, Banfield JF, Relman DA. Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time. Genome Res. 2011;21:126–136. doi: 10.1101/gr.111732.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramia NF, Tang L, Cocozaki AI, Li H. Staphylococcus epidermidis Csm1 is a 3′-5′ exonuclease. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeks J, Naismith JH, White MF. CRISPR interference: a structural perspective. Biochem J. 2013;453:155–166. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samai P, Smith P, Shuman S. Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1552–1556. doi: 10.1107/S1744309110039801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitskaya E, Semenova E, Dedkov V, Metlitskaya A, Severinov K. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol. 2013;10:716–725. doi: 10.4161/rna.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed KD, Lazinski DW, Calderwood SB, Camilli A. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature. 2013;494:489–491. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2013 doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariat N, Sandt CH, Dimarzio MJ, Barrangou R, Dudley EG. CRISPR-MVLST subtyping of Salmonella enterica subsp enterica serovars Typhimurium and Heidelberg and application in identifying outbreak isolates. BMC Microbiol. 2013;13:254. doi: 10.1186/1471-2180-13-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smih F, Rouet P, Romanienko PJ, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Staals RH, Agari Y, Maki-Yonekura S, Zhu Y, Taylor DW, van Duijn E, Barendregt A, Vlot M, Koehorst JJ, Sakamoto K, et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014 doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CL, Barrangou R, Thomas BC, Horvath P, Fremaux C, Banfield JF. Phage mutations in response to CRISPR diversification in a bacterial population. Environ Microbiol. 2013;15:463–470. doi: 10.1111/j.1462-2920.2012.02879.x. [DOI] [PubMed] [Google Scholar]
- Swarts DC, Mosterd C, van Passel MW, Brouns SJ. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–2007. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013a;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic Screens in Human Cells Using the CRISPR/Cas9 System. Science. 2013b doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Pul U, Heidrich N, Jore MM, Lundgren M, Stratmann T, Wurm R, Raine A, Mescher M, Van Heereveld L, et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol. 2010;77:1380–1393. doi: 10.1111/j.1365-2958.2010.07315.x. [DOI] [PubMed] [Google Scholar]
- Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema J, Waghmare S, Zhou K, Barendregt A, Westphal W, Heck A, Boekema E, Dickman M, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Kiro R, Edgar R, Qimron U. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proc Natl Acad Sci USA. 2011;108:20136–20141. doi: 10.1073/pnas.1113519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. Structure and Mechanism of the CMR Complex for CRISPR-Mediated Antiviral Immunity. Mol Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ. Processing-Independent CRISPR RNAs Limit Natural Transformation in Neisseria meningitidis. Mol Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder ND. Portraits of viruses: RNA phage. Intervirology. 1980;13:257–270. doi: 10.1159/000149133. [DOI] [PubMed] [Google Scholar]