Abstract
The evolutionary conservation of T lymphocyte subsets bearing αβ T cell antigen receptors (TCRs) using invariant α-chains is indicative of unique and important functions. Amongst these T lymphocytes, Natural Killer (NK) T cells that express an invariant TCRα chain and recognize lipid antigens presented by the non-classical MHC class I molecule CD1d are probably the most studied. However, a new population of evolutionarily conserved T cells with another invariant TCRα rearrangement was recently characterized. These cells, which are very abundant in humans, tend to reside in mucosal tissues and were therefore named mucosal-associated invariant T cells or MAIT cells. Until recently little was known about MAIT cells, however several recent advances in our understanding of MAIT cell characteristics and functions secures their upcoming rise to fame in the immunology field and in clinical practice.
Introduction
T lymphocytes that express an αβ T cell antigen receptor (TCR) are usually categorized into either CD4+ or CD8+ subsets that recognize peptide antigens presented by class II or class I molecules of the major histocompatibility complex (MHC), respectively. However, mature T cells that express αβ TCRs yet lack expression of CD4 and CD8 also exist in mice and humans. Analysis of TCR expression by the peripheral human CD4− CD8− double negative (DN) αβ T cells demonstrated, as early as 1993, that these cells preferentially expressed a very limited TCR repertoire (1). Two essentially invariant TCRα rearrangements were consistently identified in the αβ DN population of several individuals. These results suggested that αβ DN T cells might recognize a limited spectrum of antigens potentially presented by non-polymorphic MHC molecules. The first TCRα rearrangement identified corresponded to an invariant rearrangement between TRAV10 (Vα24) and TRAJ18 (Jα18) gene segments with no N region diversity. Today, we know that this unique TCRα chain, when combined with a limited number of Vβ chains in mice or with TRBV25 (Vβ11) in humans, is expressed by iNKT cells, which recognize various lipid and glycolipid antigens presented by the non-MHC-encoded and non-polymorphic molecule, CD1d (2).
The second TCRα rearrangement frequently identified in DN αβ T cells used the human TRAV1-2 (Vα7.2) and TRAJ33 (Jα33) gene segments (TRAV1 (Vα19) and TRAJ33 (Jα33) in mice), with two variable amino acids encoded in the V-J junction. In 1999, a seminal study by the Lantz laboratory ascribed this unique TCRα rearrangement to a new subset of T cells (3). These cells were found to be conserved between mammalian species and were enriched within the gut lamina propria, an observation which led to their denomination of muscosal-associated invariant T (MAIT) cells. MAIT cells were found to be restricted by MR1 (4), a monomorphic class I-related MHC molecule encoded outside of the MHC region with extremely high sequence conservation among mammals in its ligand-binding groove (5).
Altogether, this stringent evolutionary conservation suggested an important and perhaps non-redundant function(s) fulfilled by MAIT cells. However, further detailed characterization of the MAIT-MR1 axis remained hampered by the lack of tools for the identification of MAIT cells ex vivo, and especially by the unknown nature of the MAIT antigen(s). In the past 2 years, these major hurdles have been overcome, revealing even more surprising discoveries. First, although MAIT cells appear to be relatively rare in mice, they constitute a very significant population of T cells in humans, accounting for between 1% and 10% of T cells in peripheral blood, and representing a predominant population of T cells in the human liver and mucosal tissues (6–9). Second, MR1 molecules were shown to present a completely new and unexpected class of antigens to MAIT cells, vitamin B metabolites (10). These findings reveal a new facet of immunity that is, to date, largely unexplored and would predict that MAIT cells are key players in immunity.
MAIT cell identification and phenotype
Until recently, most work on MAIT cells had relied on the study of a few isolated MAIT clones and hybridomas as well as TCR transgenic mice expressing the invariant MAIT TCR. While useful, these tools had obvious limitations. A new monoclonal antibody directed against the human TRAV1-2 TCR chain, when used in combination with anti-CD161 or anti-IL-18Rα, was shown to be sufficient to identify MAIT cells within the CD3+ CD4− compartment (6). Furthermore, with the identification of at least one MAIT antigen (see below), MR1-Ag tetramers have been developed that enable the characterization and tracking of MAIT cells ex vivo, both in mice and humans (7).
In humans, MAIT subsets are mostly DN or CD8+ (expressing mostly the homodimeric form of CD8αα), with a minor subset being CD4+ cells. MAIT cells express an effector memory phenotype as well as high levels of the NK receptor CD161, of ABCD1 (MDR, a membrane transporter that efflux xenobiotics), and of the receptors for IL-12, IL-18 and IL-23. Human MAIT cells also express high amounts of CC-chemokine receptor 6 (CCR6) and CXC-chemokine receptor 6 (CXCR6), which are involved in cell trafficking to peripheral tissues, particularly the intestine and the liver. In addition, they express the transcription factors PLZF and RoRγt. Upon TCR stimulation by the MR1-Ag complex, human MAIT cells produce IFNγ, TNFα and IL-2 and also IL-17 after PMA + ionomycin stimulation (6–9). Furthermore, they were recently shown to be cytotoxic, killing bacteria-infected epithelial cells (11).
In conventional naïve mouse strains MAIT cells are very rare. As such, their phenotypical and functional characterization in unmanipulated animals has yet to be fully described. However, a recent study showed that MAIT cells could proliferate to make up ≈25% of all αβ T cells in the lungs after infection with the intracellular bacteria Franciscella Tularensis (12), arguing that, like in humans, MAIT cells can constitute a very significant proportion of T cells in mice.
To circumvent the problem of low MAIT cell number in unchallenged mice, three independent lines of TRAV1-TRAJ33 TCR transgenic mice have been generated (6, 13, 14). Although the same TRAV1-TRAJ33 TCR α chain was used in each case, the genetic elements used to control transgene expression were different. The MAIT cells generated in these transgenic animals differ greatly in terms of cell surface phenotype and function (6, 13, 14). Therefore, it remains unclear which, if any, of these models actually recapitulate MAIT cell development in wildtype mice. It is also unclear to what extent MAIT cell development in mice reflects MAIT cell development in humans and whether mouse MAIT cells are functionally and phenotypically equivalent to MAIT cells in humans. The strong phylogenic conservation of the MAIT-MR1 axis would argue that it should be the case, but it remains to be formally demonstrated.
MAIT reactivity and antigens
In 2010, two publications described the antimicrobial activity of human and mouse MAIT cells (15, 16). MAIT cells were shown to react to antigen presenting cells (APCs) infected with a wide variety of, although not all, bacteria and yeasts, but not viruses (15, 16). This reactivity required MR1 expression on APCs and did not seem to involve the main innate immune pathways (15). These results suggested that MAIT cells might be responsive to a conserved microbe-derived product presented by MR1 molecules. This product was resistant to protease digestion and did not co-purify with conventional lipids (17, 18). By manipulating the refolding of the MR1 protein produced under various culture conditions followed by mass spectrometry analysis of MR1-complexed ligands, the groups of McCluskey and Rossjohn revealed that MR1 could present vitamin B metabolites (10). They first identified the presence of the folic acid (vitamin B9) metabolite, 6-formyl pterin (6-FP), bound to MR1. However, 6-FP did not stimulate MAIT cells. They further identified several metabolites derived from the riboflavin (vitamin B2) biosynthesis pathway when MR1 was refolded in the presence of culture supernatant from Salmonella typhimurium, a bacterial strain known to stimulate MAIT cells. These riboflavin metabolites were able to activate human MAIT cells, as well as Jurkat cells engineered to express three different human MAIT TCRs. Strikingly, all of the microorganisms that had been previously shown to activate MAIT cells have a functional riboflavin biosynthesis pathway, and therefore produce these metabolites, while bacteria lacking this biochemical pathway are unable to stimulate MAIT cells. Among the 3 stimulatory compounds characterized to date, the two less active ones are generated by a known biochemical pathway from the immediate riboflavin precursor. However, the biochemical origin of the most stimulatory compound (1000 times more active) is still unclear (19). Altogether, these results demonstrate that a new class of antigen(s), in the form of vitamin metabolites, can be presented to, and be recognized by, the immune system. Because mammals do not synthesize riboflavin, this might represent a new mechanism of self/non-self discrimination that alerts the host to microbial infection. However, the ubiquitous nature of these small molecules that are found in many bacteria, including commensals, as well as their capacity to freely diffuse across epithelial layers, raises many questions regarding how the activation of MAIT cells might be regulated in vivo.
The identification of MAIT antigens, combined to the production of soluble MR1 molecules, led to the development of MR1-Ag tetramers (7). This new tool, which stains all MAIT cells in TCR transgenic mice as well as in humans, revealed some unexpected heterogeneity in TCR usage by MAIT cells. In addition to the typical TRAV1-2-TRAJ33 invariant α chain, 5–15% of human MAIT cells were found to use 2 other TRAJ segments ((TRAJ12 and TRAJ20) (7). The functional relevance of these “alternate” MAIT cells remains unknown.
There is little doubt that these new reagents, once available to the general research community, will be invaluable in the further characterization and tracking of MAIT cells in health and disease states.
MAIT TCR recognition
The molecular basis of MR1 antigen presentation and its recognition by the MAIT TCR was also recently solved (20–22). MR1 adopts the overall binary structure of a class I MHC fold associated with the β2m subunit. Its ligand-binding cavity is smaller than that of classical MHC class I molecules and is mostly closed due to the presence of aromatic and basic residues, with the exception of a small opening on the top of the groove (see figure). Plasticity in ligand conformation within the groove of MR1 molecules has been found for very similar ring-based ligands (see (20, 21) and figure), suggesting that the binding cavity might be able to accommodate a range of structures. In turn, this suggests that other, currently unknown, small molecules presented by MR1 might operate as antigens for MAIT cells as well.
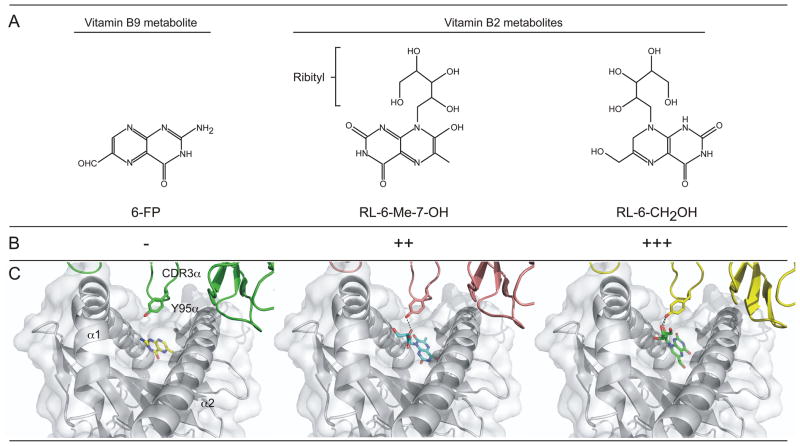
Figure 1.
Recognition by the MAIT T cell antigen receptor of vitamin metabolites presented by MR1 molecules. (A) Chemical structures of 3 vitamin metabolites, 6-formyl pterin (6-FP), 7-hydroxy-6-methyl-8-D-ribityllumazine (RL-6-Me-7-OH) and reduced 6-hydroxymethyl-8-D-ribityllumazine (RL-6-CH2OH) that have been shown to be presented by MR1 molecules. (B) Antigenic potency of these metabolites for MAIT cells. (C) The interaction between the MAIT TCR and MR1 molecules presenting the vitamin metabolites. They key role of CDR3α-encoded Tyr95 in interacting with the ribityl moiety found on stimulatory antigen is depicted.
Interestingly, a common feature of all riboflavin metabolites identified to date that stimulate MAIT cells is the presence of a ribityl group, which is formed by the removal of one of the terminal hydroxyl groups of ribitol. This moiety emerges from the opening in the MR1 groove and is the only part of the antigen recognized by the MAIT TCR (see figure). The MAIT TCR shows a highly conserved docking orientation even when different antigens are presented by MR1 (20–22). This suggests that the MAIT TCR might act as an innate-like recognition receptor similar to the TCR expressed by iNKT cells (2). However, in contrast with iNKT cells, the orientation of the MAIT TCR on MR1 is more akin to other classical αβ TCR-MHC-peptide complexes, with a perpendicular orientation that straddles the α1 and α2 helices of MR1, rather than the parallel binding of the iNKT TCR on CD1d. The nearly invariant TRAV1-2-TRAJ33 TCR chain largely dominates the recognition of the MR1 molecule and of the antigen(s), thereby providing an explanation for its selective usage by MAIT cells (17, 20–23). Furthermore, the TRAJ33-encoded tyrosine residue at position 95 of the CDR3α loop, which is conserved in all MAIT TCRs, appears to behave as a sensor of antigen presentation by MR1 molecules. This residue is ideally positioned directly over the opening of the MR1 binding cavity and makes extensive contacts with MR1 residues. Additionally, contacts between this residue and the antigen ribityl group directly correlates with increased TCR engagement and subsequent MAIT cell activation (20, 21).
Although the nearly invariant TCRα chain is found paired with a limited number of TCRβ chains (TRBV20 (Vβ2), TRBV6 (Vβ13) and TRBV2 (Vβ22) in humans and TRBV19 (Vβ6), TRBV13 (Vβ8) in mice), their CDR3β loops are highly variable and the role of the TCRβ chain in the recognition of the complex remains unclear. In some cases the TCRβ chain appears to play a passive role in the activation of MAIT cells (23), while in others it modulates the overall affinity of the MAIT TCR and/or plays an active role in directly recognizing the antigen (21). These results suggest that the CDR3β variability may directly impact the extent of antigen reactivity by MAIT cells (21).
MAIT cell development
MAIT cell development is incompletely understood. In both mouse and humans, it is generally accepted that MAIT cells develop in the thymus (3, 6, 24). However, it remains unclear whether the developmental process is identical between the two species.
In the mouse, the vast majority of MAIT cells are positively selected by MR1-expressing bone marrow-derived cells and not by the thymic epithelial cells (4). Based on data using TCR transgenic mice, it was shown that MAIT cells can be selected by MR1 expressed on double positive thymocytes (25). The idea that DP thymocytes represent the main MAIT-selecting cell type in the thymus is supported by the finding that mouse DP thymocytes express significant levels of MR1 molecules intracellularly (26). Therefore, in mice, MAIT cell development is clearly distinct from “conventional” T lymphocytes.
Positive selection of T lymphocytes on DP thymocytes was shown to impart the acquisition of “innate-like” characteristics (27) through a process that involves homodimeric interactions between SLAM family members expressed on cortical thymocytes, which signals through the SAP/Fyn/NFκB pathway (28). While both mouse MAIT and mouse iNKT cells likely encounter similar signals during their development, these two populations clearly diverge into unique developmental pathways. For example, the strong engagement of the iNKT TCR during positive selection (29–31), potentiated by the engagement of the SLAM family members (32), induces the transcription factor PLZF, which is in turn responsible for the effector phenotype of iNKT cells (28). Yet, MAIT cells in one of the tg models do not express PLZF (6). It is currently unclear what developmental clues are involved in setting the MAIT transcriptional program in mice. The memory/effector phenotype of mouse MAIT cells is acquired after birth in the periphery in a B cell and microbiota-dependent manner (4).
Human MR1 has also been detected primarily on CD3+ CD45+ hematopoietic T cells (33), suggesting that human MAIT cells might also be selected by DP thymocytes, similar to what has been described for mouse MAIT cells (25). This would be expected to drive the “innate-like” characteristics of MAIT cells and perhaps explain why MAIT cells in the human thymus express CD161, and both CD161 and IL-18Rα in human cord blood (6, 9, 33, 34). However, apart from these markers, human MAIT cells in the fetal thymus, spleen, mesenteric lymph nodes and cord-blood are otherwise immature functionally and phenotypically (24). Their maturation, which is associated with the acquisition of the PLZF transcription factor, occurs gradually in the mucosal tissues during fetal life and before exposure to environmental microbes and the commensal microflora (24). This process is likely independent of the SLAM/SAP pathway, as the number of MAIT cells was reported to be normal in five SAP-deficient patients (6) and SAP-deficient MAIT cells normally expressed PLZF (6).
MAIT cells and diseases
The role of MAIT cells is still enigmatic. The findings that MAIT cells recognize bacteria-infected cells in a MR1-dependent manner (15, 16) and their dependence on the microbiota for expansion in the periphery (4) pointed toward a potential role for MAIT cells in the immune response against bacteria. This was confirmed in several mouse models of infections using Escherichia, Mycobacteria, Klebsiella and Franciscella, in which a lack of MAIT cells correlated with increased bacterial loads by these species (12, 15, 35, 36). Furthermore, the number of MAIT cells in the blood of bacterially-infected patients was shown to be severely decreased (16) and correlated with the increased susceptibility of hospitalized patients to secondary nosocomial infections (37).
MAIT cells are apparently also involved in non-infectious inflammatory disorders. For example, MAIT cells were found enriched within the livers of patients with chronic hepatitis C (HCV) infection, autoimmune hepatitis, primary biliary cirrhosis, alcoholic liver disease, and nonalcoholic steatohepatitis (38, 39). Patients with inflammatory bowel diseases have a reduced frequency of MAIT cells in peripheral blood that is associated with an enrichment of MAIT cells in the inflamed tissue (40). MAIT cells have also been detected in the brain of patients with multiple sclerosis (41, 42) and in kidney and brain tumors (43, 44). A protective role for MAIT cells has been reported in the mouse model of autoimmune encephalomyelitis (14), while MAIT cells were proposed as one of the effector cells contributing to inflammation in a mouse model of arthritis (45). Finally, MAIT cell number was found to decline upon HIV infection (46–48).
In all of these situations, it is not obvious whether bacteria-derived vitamin B metabolites might be implicated in the activation of MAIT cells through their TCR. Microbiota imbalance (49–51) and/or increased permeability of the musosa (52) might indirectly influence MAIT cells in these instances. The underlying autoreactivity of some MAIT cells and/or their high expression levels of IL-12, IL-18 and IL-23 receptors might also play a role. Indeed, MR1-independent cytokine-driven activation of mice MAIT cells has been reported for IL-12 (36) and IL-23 (45). The combination of IL-12 and IL-18 was also shown to stimulate human MAIT cells (53). Altogether, these results suggest that MAIT cells could potentially be involved in a much broader range of diseases.
Conclusions
During the past few years a tremendous amount of knowledge, including some unexpected findings, has been acquired regarding the new population of T lymphocytes known as MAIT cells. First, MAIT cells represent a very sizable proportion of T lymphocytes in humans, where they sometimes are the majority of T cells found within a given organ. Second, the semi-invariant TCR expressed by MAIT cells recognizes the evolutionarily conserved monomorphic MHC-like molecule MR1 with a very conserved docking strategy. This strategy of conserved recognition is more reminiscent of pattern recognition receptor binding than the strategy of antigen discrimination usually carried out by conventional T cells. Third, MAIT cells have a memory-effector phenotype and can rapidly secrete the cytokines IFNγ and IL-17 as well as the chemokine CCL20, upon antigen recognition. Fourth, they recognize vitamin B metabolites generated by certain bacteria and yeasts when presented by MR1 molecules.
Thus, the immune system has evolved a previously unrecognized means of host alert against microbial infections through the recognition of highly conserved ligands by a large population of innate T cells.
Although the role of MAIT cells in health and diseases is starting to be revealed, many questions regarding their functions and how they might be manipulated still remain. For example, we do not yet know how MAIT cell responses affect the rest of the immune system and/or the microbiota. We also do not know whether the MAIT cell response can be manipulated and/or whether memory MAIT cells exist. However, based on their cytokine secretion profile, MAIT cells might be good candidate targets for new vaccines under development. Furthermore, because the proportion of MAIT cells in PBMCs appears inversely correlated with the infectious status of an individual, MAIT cells might represent a potentially attractive biomarker. Finally, the loss of MAIT cells following HIV infection, associated to their broad anti-bacterial activity, including against Mycobacterium Tuberculosis, might lead to strategies aimed at restoring MAIT cell numbers. Because PBMCs-derived MAIT cells are notoriously poor at proliferating in vitro, reprogramming of MAIT cells into induced pluripotent stem cells (iPSCs) followed by re-differentiation into MAIT cells was recently performed (54). This might provide a new avenue for therapies using MAIT cells.
At any rate, it is clear that immunologists and clinicians should check the MAIT.
Acknowledgments
I would like to thank current and past members of my laboratory for fruitful discussion and Jennifer Matsuda, Manfred Brigl and Olivier Lantz for critical reading of the manuscript. I apologize to my colleagues whose work was not cited due to space constraints.
This work was supported by a grant from the National Institutes of Health AI092108.
References
- 1.Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- αβ T cells demonstrates preferential use of several Vβ genes and an invariant TCR α chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol. 2012;12:845–857. doi: 10.1038/nri3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, Bendelac A, Bonneville M, Lantz O. An invariant T cell receptor α chain defines a novel TAP- independent major histocompatibility complex class Ib-restricted αβ T cell subpopulation in mammals. J Exp Med. 1999;189:1907–1921. doi: 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 5.Tsukamoto K, Deakin JE, Graves JA, Hashimoto K. Exceptionally high conservation of the MHC class I-related gene, MR1, among mammals. Immunogenetics. 2013;65:115–124. doi: 10.1007/s00251-012-0666-5. [DOI] [PubMed] [Google Scholar]
- 6.Martin E, Treiner E, Duban L, Guerri L, Laude Hln, Toly Cc, Premel V, Devys A, Moura IC, Tilloy F, Cherif Sp, Vera G, Latour S, Soudais C, Lantz O. Stepwise Development of MAIT Cells in Mouse and Human. PLoS Biol. 2009;7:e1000054. doi: 10.1371/journal.pbio.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reantragoon R, Corbett AJ, Sakala IG, Gherardin NA, Furness JB, Chen Z, Eckle SB, Uldrich AP, Birkinshaw RW, Patel O, Kostenko L, Meehan B, Kedzierska K, Liu L, Fairlie DP, Hansen TH, Godfrey DI, Rossjohn J, McCluskey J, Kjer-Nielsen L. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med. 2013;210:2305–2320. doi: 10.1084/jem.20130958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang XZ, Jo J, Tan AT, Sandalova E, Chia A, Tan KC, Lee KH, Gehring AJ, De Libero G, Bertoletti A. IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. J Immunol. 2013;190:3142–3152. doi: 10.4049/jimmunol.1203218. [DOI] [PubMed] [Google Scholar]
- 9.Dusseaux M, Martin E, Serriari N, Peguillet I, Premel V, Louis D, Milder M, Le Bourhis L, Soudais C, Treiner E, Lantz O. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117:1250–1259. doi: 10.1182/blood-2010-08-303339. [DOI] [PubMed] [Google Scholar]
- 10.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O’Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 11.Le Bourhis L, Dusseaux M, Bohineust A, Bessoles S, Martin E, Premel V, Core M, Sleurs D, Serriari NE, Treiner E, Hivroz C, Sansonetti P, Gougeon ML, Soudais C, Lantz O. MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS Pathog. 2013;9:e1003681. doi: 10.1371/journal.ppat.1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meierovics A, Yankelevich WJ, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci U S A. 2013;110:E3119–3128. doi: 10.1073/pnas.1302799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawachi I, Maldonado J, Strader C, Gilfillan S. MR1-restricted Vα19i mucosal-associated invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. J Immunol. 2006;176:1618–1627. doi: 10.4049/jimmunol.176.3.1618. [DOI] [PubMed] [Google Scholar]
- 14.Croxford JL, Miyake S, Huang YY, Shimamura M, Yamamura T. Invariant Vα 19i T cells regulate autoimmune inflammation. Nat Immunol. 2006;7:987–994. doi: 10.1038/ni1370. [DOI] [PubMed] [Google Scholar]
- 15.Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, Core M, Levy E, Dusseaux M, Meyssonnier V, Premel V, Ngo C, Riteau B, Duban L, Robert D, Huang S, Rottman M, Soudais C, Lantz O. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11:701–708. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 16.Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, Winata E, Swarbrick GM, Chua WJ, Yu YYL, Lantz O, Cook MS, Null MD, Jacoby DB, Harriff MJ, Lewinsohn DA, Hansen TH, Lewinsohn DM. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 2010;8:1–14. doi: 10.1371/journal.pbio.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young MH, U’Ren L, Huang S, Mallevaey T, Scott-Browne J, Crawford F, Lantz O, Hansen TH, Kappler J, Marrack P, Gapin L. MAIT cell recognition of MR1 on bacterially infected and uninfected cells. PLos One. 2013;8:e53789. doi: 10.1371/journal.pone.0053789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang S, Martin E, Kim S, Yu L, Soudais C, Fremont DH, Lantz O, Hansen TH. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc Natl Acad Sci U S A. 2009;106:8290–8295. doi: 10.1073/pnas.0903196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birkinshaw RW, Kjer-Nielsen L, Eckle SB, McCluskey J, Rossjohn J. MAITs. MR1 and vitamin B metabolites. Current opinion in Immunology. 2014;26:7–13. doi: 10.1016/j.coi.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Patel O, Kjer-Nielsen L, Le Nours J, Eckle SB, Birkinshaw R, Beddoe T, Corbett AJ, Liu L, Miles JJ, Meehan B, Reantragoon R, Sandoval-Romero ML, Sullivan LC, Brooks AG, Chen Z, Fairlie DP, McCluskey J, Rossjohn J. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat Commun. 2013;4:2142. doi: 10.1038/ncomms3142. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Sagaseta J, Dulberger CL, McFedries A, Cushman M, Saghatelian A, Adams EJ. MAIT recognition of a stimulatory bacterial antigen bound to MR1. J Immunol. 2013;191:5268–5277. doi: 10.4049/jimmunol.1301958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Sagaseta J, Dulberger CL, Crooks JE, Parks CD, Luoma AM, McFedries A, Van Rhijn I, Saghatelian A, Adams EJ. The molecular basis for Mucosal-Associated Invariant T cell recognition of MR1 proteins. Proc Natl Acad Sci U S A. 2013;110:E1771–1778. doi: 10.1073/pnas.1222678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reantragoon R, Kjer-Nielsen L, Patel O, Chen Z, Illing PT, Bhati M, Kostenko L, Bharadwaj M, Meehan B, Hansen TH, Godfrey DI, Rossjohn J, McCluskey J. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J Exp Med. 2012;209:761–774. doi: 10.1084/jem.20112095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leeansyah E, Loh L, Nixon DF, Sandberg JK. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nat Commun. 2014;5:3143. doi: 10.1038/ncomms4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seach N, Guerri L, Le Bourhis L, Mburu Y, Cui Y, Bessoles S, Soudais C, Lantz O. Double positive thymocytes select mucosal-associated invariant T cells. J Immunol. 2013;191:6002–6009. doi: 10.4049/jimmunol.1301212. [DOI] [PubMed] [Google Scholar]
- 26.Chua WJ, Kim S, Myers N, Huang S, Yu L, Fremont DH, Diamond MS, Hansen TH. Endogenous MHC-Related Protein 1 Is Transiently Expressed on the Plasma Membrane in a Conformation That Activates Mucosal-Associated Invariant T Cells. J Immunol. 2011;186:4744–4750. doi: 10.4049/jimmunol.1003254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Sofi MH, Yeh N, Sehra S, McCarthy BP, Patel DR, Brutkiewicz RR, Kaplan MH, Chang CH. Thymic selection pathway regulates the effector function of CD4 T cells. J Exp Med. 2007;204:2145–2157. doi: 10.1084/jem.20070321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol. 2013;25:161–167. doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seiler MP, Mathew R, Liszewski MK, Spooner CJ, Barr K, Meng F, Singh H, Bendelac A. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol. 2012;13:264–271. doi: 10.1038/ni.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bedel R, Berry R, Mallevaey T, Matsuda JL, Zhang J, Godfrey DI, Rossjohn J, Kappler JW, Marrack P, Gapin L. Effective functional maturation of invariant natural killer T cells is constrained by negative selection and T-cell antigen receptor affinity. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1320777110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dutta M, Kraus ZJ, Gomez-Rodriguez J, Hwang SH, Cannons JL, Cheng J, Lee SY, Wiest DL, Wakeland EK, Schwartzberg PL. A role for Ly108 in the induction of promyelocytic zinc finger transcription factor in developing thymocytes. J Immunol. 2013;190:2121–2128. doi: 10.4049/jimmunol.1202145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gold MC, Eid T, Smyk-Pearson S, Eberling Y, Swarbrick GM, Langley SM, Streeter PR, Lewinsohn DA, Lewinsohn DM. Human thymic MR1-restricted MAIT cells are innate pathogen-reactive effectors that adapt following thymic egress. Mucosal Immunol. 2013;6:35–44. doi: 10.1038/mi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker LJ, Kang YH, Smith MO, Tharmalingham H, Ramamurthy N, Fleming VM, Sahgal N, Leslie A, Oo Y, Geremia A, Scriba TJ, Hanekom WA, Lauer GM, Lantz O, Adams DH, Powrie F, Barnes E, Klenerman P. Human MAIT and CD8alphaalpha cells develop from a pool of type-17 precommitted CD8+ T cells. Blood. 2012;119:422–433. doi: 10.1182/blood-2011-05-353789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgel P, Radosavljevic M, Macquin C, Bahram S. The non-conventional MHC class I MR1 molecule controls infection by Klebsiella pneumoniae in mice. Molecular Immunology. 2011;48:769–775. doi: 10.1016/j.molimm.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Chua WJ, Truscott SM, Eickhoff CS, Blazevic A, Hoft DF, Hansen TH. Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect Immun. 2012;80:3256–3267. doi: 10.1128/IAI.00279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimaldi D, Le Bourhis L, Sauneuf B, Dechartres A, Rousseau C, Ouaaz F, Milder M, Louis D, Chiche JD, Mira JP, Lantz O, Pene F. Specific MAIT cell behaviour among innate-like T lymphocytes in critically ill patients with severe infections. Intensive Care Med. 2013 doi: 10.1007/s00134-013-3163-x. [DOI] [PubMed] [Google Scholar]
- 38.Billerbeck E, Kang YH, Walker L, Lockstone H, Grafmueller S, Fleming V, Flint J, Willberg CB, Bengsch B, Seigel B, Ramamurthy N, Zitzmann N, Barnes EJ, Thevanayagam J, Bhagwanani A, Leslie A, Oo YH, Kollnberger S, Bowness P, Drognitz O, Adams DH, Blum HE, Thimme R, Klenerman P. Analysis of CD161 expression on human CD8+ T cells defines a distinct functional subset with tissue-homing properties. Proc Natl Acad Sci U S A. 2010;107:3006–3011. doi: 10.1073/pnas.0914839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oo YH, Banz V, Kavanagh D, Liaskou E, Withers DR, Humphreys E, Reynolds GM, Lee-Turner L, Kalia N, Hubscher SG, Klenerman P, Eksteen B, Adams DH. CXCR3-dependent recruitment and CCR6-mediated positioning of Th-17 cells in the inflamed liver. J Hepatol. 2012;57:1044–1051. doi: 10.1016/j.jhep.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serriari NE, Eoche M, Lamotte L, Fumery M, Marcelo P, Chatelain D, Barre A, Nguyen-Khac E, Lantz O, Dupas JL, Treiner E. Innate Mucosal-Associated Invariant T (MAIT) cells are activated in Inflammatory Bowel Diseases. Clin Exp Immunol. 2014 doi: 10.1111/cei.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyazaki Y, Miyake S, Chiba A, Lantz O, Yamamura T. Mucosal-associated invariant T cells regulate Th1 response in multiple sclerosis. Int Immunol. 2011;23:529–535. doi: 10.1093/intimm/dxr047. [DOI] [PubMed] [Google Scholar]
- 42.Annibali V, Ristori G, Angelini DF, Serafini B, Mechelli R, Cannoni S, Romano S, Paolillo A, Abderrahim H, Diamantini A, Borsellino G, Aloisi F, Battistini L, Salvetti M. CD161(high)CD8+T cells bear pathogenetic potential in multiple sclerosis. Brain. 2011;134:542–554. doi: 10.1093/brain/awq354. [DOI] [PubMed] [Google Scholar]
- 43.Illes Z, Shimamura M, Newcombe J, Oka N, Yamamura T. Accumulation of Valpha7.2-Jalpha33 invariant T cells in human autoimmune inflammatory lesions in the nervous system. Int Immunol. 2004;16:223–230. doi: 10.1093/intimm/dxh018. [DOI] [PubMed] [Google Scholar]
- 44.Peterfalvi A, Gomori E, Magyarlaki T, Pal J, Banati M, Javorhazy A, Szekeres-Bartho J, Szereday L, Illes Z. Invariant Valpha7.2-Jalpha33 TCR is expressed in human kidney and brain tumors indicating infiltration by mucosal-associated invariant T (MAIT) cells. Int Immunol. 2008;20:1517–1525. doi: 10.1093/intimm/dxn111. [DOI] [PubMed] [Google Scholar]
- 45.Chiba A, Tajima R, Tomi C, Miyazaki Y, Yamamura T, Miyake S. Mucosal-associated invariant T cells promote inflammation and exacerbate disease in murine models of arthritis. Arthritis Rheum. 2012;64:153–161. doi: 10.1002/art.33314. [DOI] [PubMed] [Google Scholar]
- 46.Cosgrove C, Ussher JE, Rauch A, Gartner K, Kurioka A, Huhn MH, Adelmann K, Kang YH, Fergusson JR, Simmonds P, Goulder P, Hansen TH, Fox J, Gunthard HF, Khanna N, Powrie F, Steel A, Gazzard B, Phillips RE, Frater J, Uhlig H, Klenerman P. Early and nonreversible decrease of CD161++/MAIT cells in HIV infection. Blood. 2013;121:951–961. doi: 10.1182/blood-2012-06-436436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong EB, Akilimali NA, Govender P, Sullivan ZA, Cosgrove C, Pillay M, Lewinsohn DM, Bishai WR, Walker BD, Ndung’u T, Klenerman P, Kasprowicz VO. Low Levels of Peripheral CD161++CD8+ Mucosal Associated Invariant T (MAIT) Cells Are Found in HIV and HIV/TB Co-Infection. PLos One. 2013;8:e83474. doi: 10.1371/journal.pone.0083474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leeansyah E, Ganesh A, Quigley MF, Sonnerborg A, Andersson J, Hunt PW, Somsouk M, Deeks SG, Martin JN, Moll M, Shacklett BL, Sandberg JK. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. 2013;121:1124–1135. doi: 10.1182/blood-2012-07-445429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henao-Mejia J, Elinav E, Thaiss CA, Licona-Limon P, Flavell RA. Role of the intestinal microbiome in liver disease. J Autoimmun. 2013;46:66–73. doi: 10.1016/j.jaut.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 51.Kranich J, Maslowski KM, Mackay CR. Commensal flora and the regulation of inflammatory and autoimmune responses. Semin Immunol. 2011;23:139–145. doi: 10.1016/j.smim.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 53.Ussher JE, Bilton M, Attwod E, Shadwell J, Richardson R, de Lara C, Mettke E, Kurioka A, Hansen TH, Klenerman P, Willberg CB. CD161 CD8 T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur J Immunol. 2013 doi: 10.1002/eji.201343509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wakao H, Fujita H. Toward the realization of cell therapy: the advent of MAIT cells from iPSCs. Cell Cycle. 2013;12:2341–2342. doi: 10.4161/cc.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]